Abstract
Medicinal plants are rich source of bioactive compounds which are important for human healthcare. The production of bioactive compounds such as artemisinin is limited in plants. Novel antimalarial drug artemisinin was obtained from leaves of Artemisia annua L. plants. The content of artemisinin in the natural plant is 0.01–1.1 % on dry weight basis. Due to its low content, the drug could not be commercialized in the global market to explore the artemisinin-based combined therapies in the world. The economic limitations of chemical synthesis is unfeasible due to its complexity and cost effectiveness. In this article, we have studied the effect of rate limiting enzymes/genes expression, HMG-Co A reductase and amorpha-4,11-diene synthase on physiochemical changes in terms of total biomass, chlorophyll, total protein, artemisinin content and yield in transgenic lines of A. annua L. plants. The shoot length (42.00 %), number of leaves (86.89 %), the number of root (39.82 %) root length (83.61 %), total biomass accumulation (100 %), total chlorophyll level (138.14 %), total protein level (73.36 %) in TR4 at in vitro level while artemisinin (93.54 %) and its yield (237.26 %) were maximum in TR5 at polyhouse conditions, followed by TR1, TR2, TR3, TR7 and TR9 as compared to non-transgenic Artemisia annua L. plants. A similar trend in leaf biomass, stem biomass and leaf:stem ratio was also recorded in polyhouse condition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
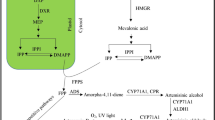
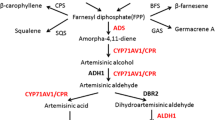
Artemisinin is biosynthesized mainly in the leaves of A. annua L., a novel antimalarial drug used in artemisinin-based combination therapies (ACTs) recommended by WHO. The low yield of artemisinin (0.1–1.1 %) is a serious impediment in the commercialization of this drug in the global market. The chemical synthesis of artemisinin is also very complicated and economically non-feasible due to poor yields (Abdin et al. 2003; Ravindranathan et al. 1990). The biosynthetic pathway of artemisinin belongs to the isoprenoid metabolite pathway, the key rate limiting enzymes involved in the biosynthesis of artemisinin include: 3-hydroxy-3-methylglutaryl-CoA reductase (first rate limiting enzymes from mevalonate pathway), and amorpha-4,11-diene synthase catalyzes the important steps of the artemisinin pathway leading to the highly specific olefinic sesquiterpene skeletons and have been postulated as the regulatory step in the biosynthesis of artemisinin. Various studies have also been conducted in different laboratories to reveal the biosynthetic pathway of artemisinin and its regulation by using the molecular stresses (biotic, abiotic stresses) and genetic engineering approaches involved in the improvement of artemisinin concentration in the Artemisia annua L. plants (Abdin and Alam 2015; Alam et al. 2014; Alam and Abdin 2011; Nafis et al. 2010; Jha et al. 2010; Zhang et al. 2009; Sa et al. 2001; Avery et al. 1992; Akhila et al. 1990; Xu et al. 1986).
Significant efforts such as chemical synthesis, plant cell suspension cultures, hairy root system and fermentation of the engineered microorganism have been made to enhance the production of artemisinin. However, none of these methods are commercially feasible to fill the gap between supply and demand (Sharaf-Eldin and Elkholy 2009; Alam and Abdin 2011; Liu et al. 2010). In recent past, researchers tried to improve artemisinin concentration by targeting the rate limiting enzymes of biosynthetic pathway and enhance the level of artemisinin in A. annua L. plants. In this prospect, we have study the relation of rate limiting enzymes, biomass, protein and physiological changes in transgenic Artemisia annua L. plants for enhanced production of artemisinin.
2 Materials and methods
2.1 Plant material and culture conditions
Previously developed transgenic lines harboring HMG-CoA reductase gene (hmgr) isolated from C. roseus (L) G. Don (Acc. No. AY623812) and amorpha-4,11-diene synthase (ads) gene (Acc. No. 432667), isolated from Artemisia annua L. plants were over-expressed in A. annua L. plants in our laboratory (Alam and Abdin 2011) and were maintained in rooting medium (MS + 0.5 mg/L NAA) supplemented with 0.8 % agar. The pH of the medium was adjusted to 5.8 before the addition of agar and the medium was autoclaved at 1 bar for 20 min. The cultures were maintained at 25 ± 2 °C under 16-h photoperiods. The high artemisinin yielding lines (TR1, TR2, TR3, TR4, TR5, TR7 and TR9) and non-transgenic A. annua L. were used in the present study in vitro and polyhouse condition for their growth and development.
2.2 Acclimatization and hardening of plantlets
The survival and establishment of plantlets were evaluated after transplanting them in pots containing vermiculite, perlite and soilrite (1:1:1) and kept in a transgenic containment facility at 25 ± 2 °C under 5000 Lux light intensity (using fluorescent lamps and 16/8 h light/dark period of further growth).
2.3 Data observation of in vitro developed transgenic and non-transgenic lines of A. annua L. plants
2.3.1 Shoot and root length
At every sampling, five replicates were taken from each transgenic and non-transgenic (control plantlets) lines. The length of roots and shoots were measured and expressed in cm.
2.3.2 Biomass accumulation
To record the dry weight, the plantlets were cut at the root–shoot junction and the separated parts of transgenic and non-transgenic lines were dried separately in hot air oven at 50 °C for 48 h. Thereafter, the dry weight (g) was determined to digital balance by unitary method.
2.3.3 Estimation of chlorophyll content
The total chlorophyll content of the leaves was estimated according to the method of Hiscox and Israelstam (1979). The chlorophyll was extracted from 100 mg of leaf sample using 7 ml dimethyl sulfoxide (DMSO) in the test tube and kept in the oven at 65 °C for 2.30 h, so as to complete the leaching of the pigments. The reaction mixture was transferred to a graduated tube and final volume was made up to 10.0 ml by adding DMSO. The chlorophyll content was then measured immediately. The absorbance of the reaction mixture was recorded at 645 and 660 nm by UV–Vis spectrophotometer (Spectroscan 80 DV, USA). Total chlorophyll content was calculated according to the method of Arnon (1949).
2.3.4 Estimation of total soluble protein
The total soluble protein content of the leaves of transgenic and non-transgenic lines was determined using the Bradford’s (1976) method.
2.4 Data observation of polyhouse-grown transgenic and non-transgenic lines of A. annua L. plants
The polyhouse-grown transgenic and non-transgenic lines of Artemisia annua L. were harvested at pre-flowering stage to observe the plant height, leaves biomass, stem biomass and leaf:stem ratio. Dried samples were kept in plastic bags at room temperature for analysis of artemisinin.
2.4.1 Biochemical analysis of A. annua L. plants grown in polyhouse
Artemisia annua L. leaves (5 g) used for enzyme essay from transgenic and non-transgenic lines were quickly frozen in liquid nitrogen and homogenized in a pre-cooled mortar containing 50 mM Mopso (pH 7.0), 20 % (v/v) glycerol, 50 mM sodium ascorbate, 50 mM NaHSO3, 10 mM MgCl2, 5 mM DTT, 1 g polyvinylpolypyrrolidone (PVPP) with a spatula tip of purified sea sand and 5 g of polystyrene resin Amberlite XAD-4. Similarly, HMGR assay, leaves, (2 g) from transgenic and non-transgenic lines of A. annua L. were washed and chopped into small pieces (1–2 mm). Each sample was homogenized in 5 volumes of ice-cold buffer (100 mM K-Phosphate, pH 7.2, 30 mM EDTA, 0.35 mM sucrose, 20 mM β-mercaptoethanol and 0.3 % BSA) and homogenate was squeezed through two layers of muslin cloth and filtered. The enzyme activities of amorpha-4,11-diene synthase (ADS) and HMGR were determined by using the method of Alam and Abdin (2011).
2.4.2 Estimation of artemisinin
1 g dry leaf material of A. annua L. plants was used for the estimation of artemisinin content using the method as described by Zhao and Zeng (1986). The derivatized artemisinin was analyzed and quantified using reverse phase column (C18, 5 µm, 4.6 × 250 mm) with mobile phase methanol: 100 mM K-Phosphate buffer (pH 6.5) in the ratio of 60:40. The artemisinin was quantified with the help of HPLC.
3 Results
3.1 Growth and physicochemical analyses of in vitro and polyhouse-grown A. annua L. plants
To observe the impact of hmgr and ads transgenes containing transgenic lines on the shoot growth of transgenic A. annua, the shoot lengths of both transgenic and non-transgenic plants were measured with a scale in centimeter and the number of leaves per plant after 2nd, 4th and 6th week of culture. The maximum shoot length 9.33 ± 0.088 cm (42.00 % higher) and maximum number of leaves per plant 52.33 ± 1.76 (86.89 % higher) were recorded in one of the transgenic lines, TR4, as compared to the non-transgenic plants (Fig. 1a, b) in 6th week of culture on rooting medium followed by other transgenic lines (TR1, TR2, TR3, TR5, TR7, TR8 and TR9). Similarly, the root length and the number of roots per plant were recorded in both transgenic and non-transgenic plants after 2nd, 4th and 6th week of culture on rooting medium. The maximum root length 9.83 ± 0.080 cm (39.82 % higher) and maximum number of root per plant 28.0 ± 0.943 (83.61 % higher) were recorded in plants of one of the transgenic lines, TR4 as compared to the non-transgenic plants in 6th week of culture on rooting medium (Fig. 2a, b) followed by other transgenic lines (TR1, TR2, TR3, TR5, TR7, TR8 and TR9). Significant differences in the biomass accumulation per plant were also observed in transgenic and non-transgenic A. annua L. plants after 2nd, 4th and 6th week of culture. The maximum biomass 0.46 ± 0.006 dry wt. (g DW plant−1; 100 % higher) was recorded in TR4 transgenic line followed by other transgenic lines (TR1, TR2, TR3, TR5, TR7, TR8 and TR9) compared with non-transgenic lines in 6th week of culture (Fig. 3). Some transgenic A. annua L. lines maintained higher chlorophyll and soluble protein contents as compared to the non-transgenic plants. The maximum increase in chlorophyll (8.43 ± 0.26 mg/g fresh weight basis) and soluble protein (23.56 ± 0.32 mg/g fw) were, however, found in TR4 (138.13 % and 73.36 % higher, respectively) followed by other transgenic lines (TR1, TR2, TR3, TR5, TR7, TR8 and TR9) as compared to non-transgenic plants on 6th week of culture on rooting medium (Figs. 4, 5). In polyhouse condition the plant height (150.0 ± 1.53–180 ± 1.16 cm), leaf biomass (30.0 ± 0.71–75.5 ± 0.48 g plant−1 dry weight basis), stem biomass (64.3 ± 0.57–111.7 ± 1.29 g plant−1 dry weight basis) and leaf:stem ratio (0.37 ± 0.21–0.82 ± 0.66) were varied in these transgenic lines compare with non-transgenic plant height (140.2 ± 1.07 cm), leaf biomass (25.0 ± 0.21 g plant−1 dry weight basis), stem biomass (62.4 ± 0.42 g plant−1 dry weight basis) and leaf:stem ratio (0.40 ± 0.22) (Table 1). The maximum biomass of leaf and stem was observed in TR5 lines of A. annua L. plants in polyhouse conditions.
3.2 Biochemical analysis of A. annua L. plants grown in polyhouse
The in vitro raised A. annua L. plants were transferred to polyhouse for hardening and further growth as well as development. The survival and establishment of plantlets were evaluated after transplanting them in pots containing soil, sand and peat moss (3:3:1). The plantlets were watered and covered with transparent plastic bags to maintain high relative humidity. The percentage survival of the hardened plants was recorded after 1 month of transfer. The leaves of plants were showing yellowing during gradual reduction of humidity and plants had 85 % survival rate. This step was very critical; some plants died during this period due to desiccation. The data for biochemical parameters were recorded on pre-flowering stage. The mevalonate content in the leaves of transgenic line (TR1, TR2, TR3, TR4, TR5, TR7, TR8 and TR9) and non-transgenic plants of A. annua L. was analyzed and expressed as μg g−1 dw (Fig. 6). It was found 12.40 ± 0.100 μg g−1 dw (58.97 % higher) in TR4 followed by TR1, TR2, TR3, TR4, TR7, TR8 and TR9 when compared with the non-transgenic (Fig. 7). Further, the enzymatic analyses of HMGR 13.41 ± 0.220 nmol MVA formed h−1 mg−1 (55.21 % higher) and ADS µM h−1 mg−1 3.22 ± 0.130 (65.21 % higher) in transgenic (TR5) lines followed by TR1, TR2, TR3, TR4, TR7, TR8 and TR9 as compared to the non-transgenic plants (Fig. 7).
3.3 Artemisinin content and yield
The artemisinin content in the leaves of transgenic (TR) as well as non-transgenic (W) plants of A. annua L. and artemisinin yield (mg plant−1) was determined in the plants grown in the polyhouse at pre-flowering stage. These were found to be 93.54 % (1.258 %, artemisinin content dry wt. basis) and 237.26 % (12.31 ± 0.208 mg/g, yield) higher in TR5 followed by other transgenic lines (TR1, TR2, TR3, TR4, TR7, TR8 and TR9) as compared to the non-transgenic (3.65 ± 0.173 mg/g) plants (Fig. 7).
4 Discussion
4.1 Influence of transgenes hmgr and ads on growth of A. annua L. plants
The impact of hmgr and ads genes on physiological as well as biochemical parameters were assessed in transgenic and non-transgenic A. annua L. plants after 2nd, 4th and 6th weeks of culture. In our study, we have observed that the shoot length, number of roots, chlorophyll and soluble protein contents of all transgenic plants of A. annua L. were higher (Figs. 1, 3). The chlorophyll and protein content were recorded maximum in TR4 followed by TR1, TR2, TR3, TR5, TR7 TR8 and TR9 in the 6th week as compared to non-transgenic plants. The transgenic lines with higher expression of hmgr and exhibiting increased activity of HMGR enzyme have also found to have higher chlorophyll and soluble protein contents and accumulated more biomass when compared with other transgenic and non-transgenic A. annua L. plants (Figs. 1a, b, 2). This could be due to the expression of hmgr only because of the more HMGR activity led to the higher substrate pool for the downstream enzymes from MVA pathway such as isopentenyl transferase (IPT) which involved in cytokinins biosynthesis (Sa et al. 2001).
4.2 HMG-CoA reductase, amorpha-4,11-diene synthase activities, mevalonate and artemisinin contents
In the recent past, there is a now strong evidence that mevalonate biosynthesized from mevalonate pathway have been utilized in the biosynthesis of not only MVA pathway but also other isoprenoids such as artemisinin, sterol, etc. (Zhang et al. 2009; Arsenault et al. 2010; Mauji Ram et al. 2010), which becomes a limiting factor for the artemisinin biosynthesis. Keeping in view our study, we have attempted to over-express the hmgr encoding HMGR, the rate limiting enzyme of the mevalonate pathway led to enhance the HMG-CoA reductase activity (55.21 %) and mevalonate content (58.97 %) in one of the transgenic lines (TR5) followed by another (TR1, TR2, TR3, TR4, TR7 TR8 and TR9) of Artemisia annua L. plants. Similarly, ads, the gene encoding ADS, the enzyme catalyzing the first rate limiting step in artemisinin biosynthesis, linking it to the mevalonate pathway (Arsenault et al. 2010; Koobkokkruad et al. 2008) had also been over-expressed simultaneously with hmgr led to higher amorpha-4,11-diene synthase activity (65.13 %) in TR5 followed by another (TR1, TR2, TR3, TR4, TR7 TR8 and TR9) (Figs. 6, 7) as well as artemisinin content (93.54 %) and yield (237.26 %) when compared with the non-transgenic A. annua L. plants and in polyhouse compared to wild-type plants. A large variation in artemisinin content has been observed in the leaves of plants from different origins by different investigators. The high artemisinin yielding varieties are the product of recurrent and mass selection of breeding lines as well as transgenic strains of A. annua (Alam et al. 2014; Paul et al. 2010). Artemisinin production by A. annua is usually in the range of 0.01–0.4 %, but some clones produce over 1 % (Delabays et al. 1993). Thus we have proved that the yield obtained by us in transgenic plants of A. annua L. was recorded more than others (Koobkokkruad et al. 2008; Aquil et al. 2009).
The developmental stage of A. annua plant at which maximum artemisinin concentration could be found is also an important aspect. Majority of investigators have reported the highest artemisinin content at the pre-flowering stage (Liersch et al. 1986; Jha et al. 2010). In the present study also, the highest concentration of artemisinin in the plant was observed at the pre-flowering stage (Fig. 7). It is due to higher expression of hmgr and ads transcript levels which led to the increased HMGR and ADS in transgenic lines of A. annua L. (Alam and Abdin 2011; Nafis et al. 2010; Aquil et al. 2009). It is quite interesting that the differences in physiological and biochemical parameters of TR4 and TR5 were more significance with each other. But when the transgenic plantlets were shifted to a transgenic containment facility, the activity of the ADS, HMGR and yield of artemisinin was enhanced in TR5 line. It might be due to environmental effects regulating any abiotic responsive genes that promote the increased expression level of hmgr and ads at this level which was ultimately responsive for higher artemisinin accumulation in TR5 lines. However, these differences could probably be due to the genotypic differences in A. annua plants as well as the differences in environmental conditions.
The higher artemisinin contents in A. annua L. plants and other organisms like tobacco, yeast, etc., may also be achieved by simultaneous control of multi-steps in MVA/MEP and artemisinin biosynthetic pathways using transcriptional regulators, knock down the gene(s) expression, gene/tissue specific promoters in Artemisia annua L. plants. This could lead to more artemisinin biosynthesis in genetically engineered strain of Artemisia annua L. plants affect the cost of ACTs for malaria caused by multi-drug resistant P. falciparum sp. in the endemic area of the world. The higher biomass accumulation of transgenic Artemisia annua L. in this study is a good sign for Artemisia annua L. breeders to cultivate and produce more crops for their economic value. Finally, it is proved that if the other enzymes from the artemisinin biosynthetic pathway over-expressed or down regulates, the level of artemisinin might be improved with transgenic plants and the cost of ACTs will be reduced for poor people.
References
Abdin MZ, Alam P (2015) Genetic engineering of artemisinin biosynthesis: prospects to improve its concentration. Act Physiol Plant. doi:10.1007/s11738-015-1771-5
Abdin MZ, Israr M, Rehman RU, Jain SK (2003) Artemisinin, a novel antimalarial drug: biochemical and molecular approaches for enhanced production. Planta Med 69:289–293
Akhila A, Kumkum R, Thakur RS (1990) Biosynthesis of artemisinic acid in Artemisia annua. Phytochemistry 29:2129–2132
Alam P, Abdin MZ (2011) Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin content. Plant Cell Rep 10:1919–1928
Alam P, Kamaluddin Khan MA, Mohammad A, Khan R, Abdin MZ (2014) Enhanced artemisinin accumulation and metabolic profiling of transgenic Artemisia annua L. plants over-expressing by rate limiting enzymes from isoprenoid pathway. J Plant Interact. doi:10.1080/17429145.2014.893030
Aquil S, Husaini AM, Abdin MZ, Rather GM (2009) Overexpression of the HMG-CoA reductase gene leads to enhanced artemisinin biosynthesis in transgenic artemisia annua plants. Planta Med 75:1–6
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Po1ypheno1oxidase in Beta vulgaris. Plant Physiol 24:1–15
Arsenault PR, Vail DR, Wobbe KK, Weathers PJ (2010) Effect of sugars on artemisinin production in Artemisia annua L.; transcription and metabolite measurements. Molecules 15:2302–2318. doi:10.3390/molecules15042302
Avery MA, Chong WKM, Jennings-White C (1992) Stereo selective total synthesis of (+)-artemisinin, the antimalarial constituent of Artemisia annua L. J Am Chem Soc 114:974–979
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Delabays N, Benakis A, Collet G (1993) Selection and breeding for high artemisinin (qinghaosu) yielding strains of Artemisia annua. Acta Hortic 330:203–206
Hiscox JD, Israelstam GF (1979) A method for extraction of chlorophyll from leaf tissue without maceration. Can J Bot 59:463–469
Jha P, Maujiram Khan MA, Kiran U, Mahamooduzzafar Abdin MZ (2010) Impact of organic manure and chemical fertilizers on artemisinin content and yield in Artemisia annua L. Ind Crop Prod 33:296–301
Koobkokkruad T, Chochai A, Kirdmanee C, De-Eknamkul W (2008) Effects of low-dose gamma irradiation on artemisinin content and amorpha-4,11-diene synthase activity in Artemisia annua L. J Radiat Biol 84(11):878–884
Liersch R, Soicke H, Stehr C, Tullner HU (1986) Formation of artemisinin in Artemisia annua during vegetation period. Planta Med 52:387–390
Liu B, Wang H, Du Z, Li G, Ye H (2010) Metabolic engineering of artemisinin biosynthesis in Artemisia annua L. Plant Cell Rep 30:689–694
Mauji Ram MA, Jha P, Khan S (2010) HMG-CoA reductase limits artemisinin biosynthesis and accumulation in Artemisia annua L. plants. Acta Physiol Plant 32:859–866
Nafis T, Akmal M, Mauji Ram Alam P, Ahlawat S, Mohammad A, Abdin MZ (2010) Enhancement of artemisinin content by constitutive expression of HMG CoA Reductase gene in high yielding strain of Artemisia annua L. plant. Biotechnol Rep 5:53–60
Paul S, Khanuja SPS, Shasany AK, Gupta MM, Darokar MP, Saikia D, Gupta AK (2010) Enhancement of artemisinin content through four cycles of recurrent selection with relation to heritability, correlation and molecular marker in Artemisia annua L. Planta Med 76:1468–1472
Ravindranathan T, Anil Kumar M, Menon RB, Hiremath SV (1990) Stereo selective synthesis of artemisinin. Tetrahedron Lett 31:755–758
Sa G, Ma M, Ye HC, Liu BY, Li GF, Chong K (2001) Effects of ipt gene expression on the physiological and chemical characteristics of Artemisia annua L. Plant Sci 160:691–698
Sharaf-Eldin M, Elkholy S (2009) Artemisinin production from different shoot culture systems of Artemisia annua L. Aust J Basi Appl Sci 3:2212–2216
Xu XX, Zhu J, Huang DZ, Zhou WS (1986) Total synthesis of arteannuin and deoxyarteannuin. Tetrahedron 42:819–828
Zhang L, Jing F, Li F, Li M, Wang Y, Wang G, Sun X, Tang K (2009) Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA mediated gene silencing. Biotechnol Appl Biochem 52:199–207
Zhao SS, Zeng MY (1986) Determination of Qinghaosu in Artemisia annua L. by high performance liquid chromatography. Chin J Pharm Ana 6:3–5
Acknowledgments
P.A. is thankful to Jamia Hamdard and DST, New Delhi, India, for providing Junior Research Fellowship and UGC for Dr. D.S. Kothari Post-Doctoral fellowship.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Alam, P., Kamaluddin, Sharaf-Eldin, M.A. et al. The effect of over-expression of rate limiting enzymes on the yield of artemisinin in Artemisia annua . Rend. Fis. Acc. Lincei 27, 311–319 (2016). https://doi.org/10.1007/s12210-015-0481-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12210-015-0481-7