Abstract
Introduction
Neutrophils act as first responders during an infection, following signals from the pathogen as well as other host cells to migrate from blood vessels to the site of infection. This tightly regulated process is critical for pathogen clearance and, in many cases, eliminates the pathogen without the need for an additional immune response. It is, therefore, critical to understand what signals drive neutrophil migration to infection in a physiologically relevant environment.
Methods
In this study, we used an infection-on-a-chip model to recapitulate many important aspects of the infectious microenvironment including an endothelial blood vessel, an extracellular matrix, and the environmental fungal pathogen Aspergillus fumigatus. We then used this model to visualize the innate immune response to fungal infection.
Results
We found that A. fumigatus germination dynamics are influenced by the presence of an endothelial lumen. Furthermore, we demonstrated that neutrophils are recruited to and swarm around A. fumigatus hyphae and that the presence of monocytes significantly increases the neutrophil response to A. fumigatus. Using secreted protein analysis and blocking antibodies, we found that this increased migration is likely due to signaling by MIP-1 family proteins. Finally, we demonstrated that signal relay between neutrophils, mediated by LTB4 signaling, is also important for sustained neutrophil migration and swarming in response to A. fumigatus infection in our system.
Conclusions
Taken together, these results suggest that paracrine signaling from both monocytes and neutrophils plays an important role in driving the neutrophil response to A. fumigatus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance is a serious global health problem and it is estimated that deaths from resistant infections will surpass cancer deaths by 2050.37 Therefore, the development of new therapeutic interventions for these infections is critical. In healthy individuals, the innate immune response can clear many opportunistic infections on its own. Cells of the innate immune system are, therefore, an ideal therapeutic target for controlling infections. However, a better understanding of how the infectious microenvironment influences neutrophil dynamics during infection is necessary before immune cell-based treatments can be developed. One pathogen that is regularly cleared by the innate immune system in healthy individuals27 but causes serious disease in immunocompromised patients is the fungus Aspergillus.11
Invasive aspergillosis is a serious fungal infection that affects immunocompromised patients and is associated with high mortality.11 This infection is most often caused by the environmental fungus, Aspergillus fumigatus. Although humans inhale hundreds of Aspergillus fumigatus spores daily, these spores are quickly cleared by the innate immune system and do not cause disease.27 However, in immunocompromised individuals, especially those with neutropenia, these spores are not cleared and germinate into hyphae which makes containment of the infection difficult.35 While infection begins in the lungs, it can invade the vasculature and spread to other organs including the brain, heart, kidneys, and skin. The invasive hyphal growth can then lead to tissue and organ damage, ultimately ending in patient death. Options to treat these infections are limited31 and with increasing anti-fungal resistance to this pathogen23,24 a better understanding of the immune cell dynamics in response to Aspergillus is necessary.
Many different experimental platforms have been used to investigate interactions between neutrophils and Aspergillus fumigatus. A considerable amount of work has been done in animal models to visualize the innate immune response to infection with A. fumigatus.4,19,34 Using zebrafish models, which allow for direct visualization of neutrophil and macrophage dynamics, researchers have identified unique roles for neutrophils and macrophages in clearing A. fumigatus infections.34 In addition to in vivo models, several in vitro devices have been developed to look at the role of neutrophil-fungal interactions in fungal growth and neutrophil function.2,9,16,17,18 While these studies have reported important findings on the role of neutrophils in Aspergillus infection, they cannot provide insight into the role of the 3D microenvironment in directing the neutrophil response. Current in vitro platforms lack the complexity to investigate the role of the endothelial blood vessel or myeloid cells on the neutrophil response and in vivo models are too complex to tease apart these individual interactions. Therefore, a new in vitro model that incorporates important aspects of the infectious microenvironment is needed to directly visualize the neutrophil response to Aspergillus infection and determine the role of multicellular interactions in driving this response.
We have developed an in vitro fungal infection model to investigate neutrophil migration to the fungal pathogen Aspergillus fumigatus. We focused on the neutrophil response to the fungus following germination, as this is the most detrimental phase of an infection with A. fumigatus, occurring throughout the body and causing tissue damage. Importantly, this model recapitulates many common features of the infectious microenvironment including incorporation of a model endothelial blood vessel, extracellular matrix, live fungus, and primary human innate immune cells. Using this model, we showed that the presence of an endothelial lumen drives fungal germination which in turn stimulates neutrophil migration. Furthermore, we showed that this neutrophil migration is driven by signaling from both neutrophils themselves as well as other cells in the infectious microenvironment. Finally, this model illustrates the power of physiologically relevant in vitro systems for studying the neutrophil response to fungal infection.
Methods
LumeNEXT Fabrication
The LumeNEXT devices were fabricated as previously described.15 Briefly, the devices were prepared using standard photo- and soft-lithography techniques. Silicon masters were fabricated using photolithography. The devices were designed with Adobe Illustrator and patterned onto high-resolution photomasks in which the features were transparent, and the remaining area was black. The device layers were formed by spin-coating the negative photoresist, SU-8, onto silicon wafers. Following a soft bake of the wafer, the pattern was transferred by shining UV light through the photomask onto the photoresist causing it to cure. A postexposure hard bake was done and the remaining SU-8 was removed by washing with developer, propylene glycol methyl ether acetate (Sigma). Finally, the wafer was washed with acetone and isopropyl alcohol.
The devices were prepared using soft lithography. PDMS (polydimethylsiloxane) (Dow) was poured over the silicon master on a hot plate and baked for 4 h at 60 °C. PDMS rods were fabricated using 23 gauge needles resulting in lumens with an inner diameter of approximately 340 µm.
The top and bottom layers were bonded to the glass coverslide using oxygen plasma (Diener Electronic Femto Plasma Surface System) and the PDMS rods were inserted into the unit, completing the device.
iEC Culture
iCell-endothelial cells (iECs) were purchased from Cellular Dynamics International. iECs were maintained in Vasculife Basal maintenance media from LifeLine Cell Technologies supplemented with iCell-Endothelial Cell Medium Supplement (CDI). Cell culture treated flasks were functionalized with 30 µg/mL bovine fibronectin (Sigma) for 1 h at room temperature prior to cell seeding. iECs were passaged at 80% confluency and used from passage three to passage six.
Device and Collagen Preparation
LumeNEXT devices were prepared as previously described.3 Briefly, devices were treated with 1% polyethylenimine followed by 0.1% glutaraldehyde (Sigma) and washed with DI water. Collagen I (Corning) was prepared at 4.5 mg/mL and neutralized to pH 7.2. The collagen was pipetted into the device and polymerized for a minimum of 2 h at 37 °C. After polymerization, the PDMS rods were pulled from the chambers leaving behind a hollow lumen. Lumens were functionalized with 30 µg/mL bovine fibronectin and seeded with iECs at 2 × 104 cells/µL. Devices were rotated to allow adherence of iECs to all sides of the lumen. After 2 h, unadhered cells were aspirated and replaced with fresh media. Lumens were cultured for 2 days with media exchanges twice daily. All devices were subjected to the same functionalization and media changes, including those without endothelial cells.
Neutrophil/Monocyte Isolation
All blood samples were drawn from healthy individuals according to our institutional review board approved protocol per the Declaration of Helsinki. Donors were provided a description of the procedure and risks prior to the procedure. They were given time to ask questions and then signed an informed consent prior to the blood draw. Blood was drawn into Potassium/EDTA vacutainer tubes to prevent coagulation.
Peripheral neutrophils were isolated using the MACSxpress Neutrophil Isolation Kit according to the manufacturer’s directions (Miltneyi Biotec). Residual erythrocytes were removed by magnetic depletion using the MACSxpress Erythrocyte Depletion Kit (Miltenyi Biotec). Neutrophils were pelleted at 200×g for 5 min, resuspended in 1 mL PBS, and counted. Neutrophils were stained with calcein AM at 10 nM (Thermofisher) and resuspended in PBS at 15 × 106 cells/mL. Experiments with neutrophils were run immediately after isolation.
Monocytes were isolated using the MACSxpress Monocyte Isolation Kit II. Blood was diluted 1:1 with PBS and layered on top of 15 mL Lymphoprep (Axis-Shield) in a SepMate tube (Stem Cell Technologies). The blood/PBS mixture was spun for 10 min at 1200×g. The PBMC layer was transferred to a 50 mL conical by quickly pouring off the supernatant. The cells were washed 2x in 50 mL PBS and spun down at 300×g for 5 min. Monocytes were then isolated using the MACSxpress Monocyte Isolation Kit II according to the manufacturer’s directions. Finally, monocytes were pelleted at 300×g for 5 min, resuspended in 1 mL PBS, and counted. Monocytes were resuspended to a final concentration of 3 × 106 cells/mL in iEC media and used immediately following isolation.
Aspergillus fumigatus Culture
The Aspergillus fumigatus strain TBK5.134 was used in all experiments. TBK5.1 was grown on solid glucose minimal medium (GMM) at 37 °C in darkness. Conidial suspensions were prepared using a modified version of a previously published protocol.19 Briefly, Aspergillus was plated at a concentration of 1 × 106 conidia/10 cm plate on solid GMM and grown for 3–4 days. Fresh conidia were then harvested in 0.01% Tween water by scraping with an L-spreader. The spore suspension was passed through sterile Miracloth into a 50 mL conical tube and the volume was adjusted to 50 mL with 0.01% Tween water. The spore suspension was centrifuged at 900×g for 10 min at room temperature. The spore pellet was resuspended in 50 mL PBS. The spore suspension was then vacuum filtrated using a Buchner filter funnel with a glass disc containing 10–15 µm diameter pores. The filtered suspension was centrifuged at 900×g for 10 min and resuspended in 1 mL PBS. Conidia were counted using a hemacytometer and the concentration was adjusted to 1.5 × 108 spores/mL. Conidial stocks were stored at 4 °C and used up to one month after harvesting.
Pseudomonas Aeruginosa Culture
P. aeruginosa PAK strain was used. One colony from an LB plate was grown overnight in 5 mL LB. The next day the culture was diluted 1:5 in fresh media and grown for 1.5 h. 1 mL of bacterial culture was pelleted by centrifugation and resuspended in 100µL iEC media. The optical density was measured and diluted in iEC media to OD = 5.
Neutrophil Migration
Aspergillus fumigatus spores were resuspended at 1 × 105 spores/mL in iEC media and 1µL was added to the top port of the device. Devices were incubated for 6 h to allow germination to begin before neutrophils were added.
For neutrophil-only experiments, neutrophils were diluted 1:1 with iEC media and plated in lumens at a final concentration of 7.5 × 106 cells/mL. For neutrophil/monocyte experiments, neutrophils and monocytes were combined 1:1 and plated in lumens at a final concentration of 7.5 × 106 neutrophils/mL and 1.5 × 106 monocytes/mL. Devices were imaged immediately following cell seeding.
For blocking antibody experiments, neutrophils and monocytes were resuspended in anti-mouse IgG1 (Control) (R&D), 10 µg/mL anti-IL6Ra (R&D), 0.5 µg/mL anti-IL1β (R&D), 250 µg/mL anti-IL1Ra (R&D), 10 µg/mL anti-MIP1α, or 10 µg/mL anti-MIP1β. Concentrations were chosen to exceed the reported ND50. For inhibitor experiments, neutrophils were resuspended in PBS with DMSO (Control) or 1 µM MK-886 (Millipore). For all experiments, cells were incubated with inhibitor or control for 20 min at room temperature prior to plating.
For experiments with P. aeruginosa, neutrophils were seeded in the lumens with or without monocytes. P. aeruginosa was then added to the top port of the device and lumens were immediately imaged.
Image Acquisition
Germination and migration experiments were imaged on an inverted microscope (Nikon Eclipse TE300) using a Nikon 10x/0.45(NA) objective, a motorized stage (Ludl Electronica Products), and the acquisition software Metamorph (Molecular Devices). Images were acquired every 20 min for 24 h. All imaging was done at 37 °C. Neutrophil-fungal interactions were also imaged on a Zeiss Z.1 Observer-based inverted spinning disk confocal microscope (CSU; Yokogawa) run with Zen software. Images were collected every 20 min for 24 h using a Photometrics Evolve EMCCD camera.
Image Processing and Data Analysis
Images were transferred to FIJI (ImageJ) for data analysis. Migrated neutrophils were counted using the Analyze Particles function in FIJI. Briefly, the fluorescent images were leveled individually using the Adjust Window/Level function. The cells were outlined using the Find Edges function. Binary images were created using the Threshold function and compared to original fluorescent images to check for consistency. A box was drawn from the lumen edge towards the source of A. fumigatus to select an area with a height equal to 400 µm. The Analyze Particles function was used to count the number of cells in the region that fell between 10 and 150 pixels2 to eliminate artifacts. The average number of migrated neutrophils was determined and then pooled using the least squared adjusted means function in R version 3.
Germination distance was measured by thresholding the fluorescent images individually in FIJI. The distance between the furthest germinated hyphae and the lumen edge was measured at every timepoint.
Migration distance was determined by measuring the distance of all neutrophils from the lumen edge at each timepoint. The average distances per replicate were determined and then pooled using the least squared adjusted means function in R version 3.
Lumens were defined as having a swarm if they contained at least 4 neutrophils in close proximity to each other and a hypha, or if they contained a neutrophil “cloud” as seen in Figure 7Aii. The percentage of lumens with a swarm was calculated by dividing all lumens analyzed by the number with swarms.
Magpix Secretion Analysis
Multiplexed protein secretion analysis was performed on lumen-conditioned media from five experimental conditions using the MagPix Luminex Xmap System (Luminex) with the Human Discovery Immunotherapy Fixed Panel (R&D Systems) per the manufacturer’s protocol. Lumens were seeded with iECs, A. fumigatus, neutrophils, and/or monocytes according to the conditions and incubated for 20 h. For each replicate, media from six lumens per condition was collected, pooled, and centrifuged to remove cellular debris. Data are from five replicates. Samples were frozen at – 20 °C from the time of collection until the assay was run. Secreted protein levels from each replicated were normalized to endothelial lumens alone. The heat map was generated using Multi Experiment Viewer software and displays log2 fold change over the condition with endothelial lumens alone.
Statistical Analysis
For all statistical analyses, at least three independent experiments were performed. Experimental Ns are noted in figure legends. For quantification of migration experiments, pooled data from three or more replicates was compared between experimental conditions using analysis of variance. Results are summarized in terms of least-squared adjusted means (lsmeans) and standard error (SEM). Statistical analyses and graphical representations were done in R version 3.4 and Microsoft Excel.
Results and Discussion
Characterization of an Aspergillus fumigatus Infection Model
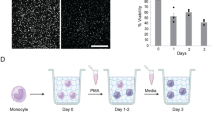
In this study, we developed an in vitro fungal infection model comprising many important features of the infectious microenvironment including a model endothelial blood vessel, an extracellular matrix, primary human immune cells, and the fungal pathogen Aspergillus fumigatus (Fig. 1a). To model a blood vessel, we used the recently developed LumeNEXT device15 to create an endothelial-lined lumen, in a collagen matrix. To model the fungal infection, spores were seeded in the top port of the device and germinated toward the lumen. Neutrophils and/or monocytes were then seeded in the interior of the lumen and migrated across the endothelium and through the collagen matrix, towards the germinating fungus (Fig. 1a).
Infection-on-a-Chip Model of A. fumigatus Infection. (a) Schematic of the infection-on-a-chip model shows neutrophils (green) leaving the endothelial lumen and migrating to germinated A. fumigatus hyphae (purple). (c) A. fumigatus germination dynamics measured as distance from the lumen over time. Purple lines indicate replicates with an endothelium present, blue lines indicate replicates without an endothelium present. Bold lines are the average over all replicates. Data shown from 15 replicates across 5 independent experiments. (b) Representative images showing A. fumigatus with and without an endothelium. Scale bar = 100 µm.
We were first interested in using this model to determine how multicellular interactions control the infectious microenvironment during A. fumigatus infection. We have previously shown that the presence of an endothelial lumen significantly increased the recruitment of neutrophils to a bacterial infection.13 Therefore, we were interested in investigating the role of the endothelium in our fungal infection model. Surprisingly, we found that A. fumigatus germination dynamics were significantly altered in the presence of an endothelial lumen (Figs. 1b and 1c). When an endothelial lumen was present, fungal hyphae reached the imaging window, approximately 300 microns from the lumen edge, 17 h after the spores were seeded. The hyphae continued to grow, breaching the lumen barrier within another 4 h, on average (Fig. 1c). In contrast, A. fumigatus spores seeded in devices without an endothelial lumen failed to germinate within the 30-h imaging timespan. These spores did eventually germinate, with significant hyphal growth seen 72 h after spore seeding (observed in 16/16 lumens) but the dynamics were significantly delayed in devices without an endothelium. These results indicate that the endothelium itself is supplying a signal to the fungus that increases the rate of germination. Several studies have looked at endothelial cell- A. fumigatus hyphal interactions at the stage where hyphae breach the endothelial layer and have found upregulation of pro-inflammatory factors such as IL-8 and TNFα6,18,25 but the effect of the endothelium on A. fumigatus germination remains unclear.
A. fumigatus Induces Neutrophil Extravasation and Migration
To investigate the neutrophil response to A. fumigatus, fungal spores were first added to the device and incubated for several hours, allowing the germination process to begin. The endothelial lumens were then seeded with primary human neutrophils and imaged. We found that neutrophils efficiently migrated across the endothelial lumen towards the fungal hyphae (Fig. 2a and Supplemental Video 1). We have previously shown that neutrophils do not migrate out of the endothelial lumen to media alone13; therefore, this response is due to the presence of the fungus. We quantified neutrophil migration in two ways: by counting the number of neutrophils outside the endothelial lumen and by measuring the distance of each neutrophil from the lumen edge. The number of migrated neutrophils steadily increased over the imaging window with a sharp increase in the rate of neutrophil migration at 4 h post germination. The increased neutrophil migration is illustrated by the significantly higher number of migrated neutrophils at 6, 7, and 8 h post germination compared to all other timepoints (Fig. 2b). This is further illustrated by comparing the cell flux, or time rate of change for neutrophil migration, before and after 4 h post germination. The cell flux from − 5 to 4 h post germination was calculated to be 1.46 cells/h compared to 5.29 cells/h from 4 to 8 h post germination. The average distance from the lumen also increased over time with neutrophils reaching significantly further distances at 7 and 8 h post germination (Fig. 2c). We found that the neutrophil response to Aspergillus was not limited to A. fumigatus as neutrophils also migrated to Aspergillus nidulans hyphae (Supplemental Video 2).
A. fumigatus Germination Drives Neutrophil Migration. (a) Representative images of neutrophils migrating from the endothelial lumen (white line) to germinating A. fumigatus. Source of A. fumigatus shown in purple. (b) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time germination began in the upper port of the device. (c) The distance from the lumen edge was measured for all neutrophils outside the lumen every hour. The data was then adjusted for the time that germination began in the upper port of the device. (b, c) Each point represents the least-squared mean (+/-) standard error of the mean (SEM). Data quantified for 11 replicates across 5 independent experiments. Asterisks represent significance compared to earlier time points. *p < 0.05, **p < 0.005.
Our results show that neutrophils respond robustly to A. fumigatus hyphae, with extensive neutrophil migration out of the lumen towards the fungus several hours post germination, when the hyphal branches near the endothelial lumen. This is consistent with in vitro and in vivo data showing that while macrophages respond to and phagocytose Aspergillus conidia, neutrophils primarily respond to growing hyphae.14,34 This demonstrates the relevance of our model in recapitulating an accurate neutrophil response to A. fumigatus infection.
Monocytes Increase the Neutrophil Response to A. fumigatus
We were next interested in improving the relevance of our model by including other cells known to be present in the infectious microenvironment and investigating their role in the neutrophil response to infection. We were specifically interested in investigating the role of monocytes, another type of innate immune cell known to produce neutrophil activating signals. Monocytes have been shown to both augment neutrophil activity towards A. fumigatus and phagocytose A. fumigatus conidia while undergoing differentiation.10 To investigate the role of monocyte signaling in the neutrophil response to infection, monocytes were seeded in the lumen along with neutrophils and the neutrophil response was quantified. We found that significantly more neutrophils migrated out of the lumen when monocytes were present compared to neutrophils alone (Figs. 3a and 3b). This trend is evident following germination and was significantly different at 6 h post germination, with almost twice as many neutrophils migrated in the presence of monocytes (Fig. 3b). This result is consistent with in vivo data that show the presence of monocytes increases the rate and number of neutrophils accumulating in the lung of mice in response to the inflammatory molecule LPS.28 Additionally, it indicates that paracrine signaling is important for the neutrophil response to A. fumigatus.
Monocytes Increase the Neutrophil Response to A. fumigatus. (a) Representative images of neutrophils migrating from the endothelial lumen (white line) to germinating A. fumigatus with and without monocytes. Source of A. fumigatus shown in purple. (b) The number of neutrophils outside the lumen migrating to A. fumigatus was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time that germination began in the upper port of the device. (c) The number of neutrophils outside the lumen migrating to P. aeruginosa was quantified every hour using particle analysis in FIJI. (b, c) Each point represents the least-squared mean (+/-) standard error of the mean (SEM). Data quantified for 9 replicates across 4 independent experiments. Asterisks represent significance between conditions at each point. **p < 0.005.
We were surprised by this dramatic response and were, therefore, interested in determining if monocytes would increase the neutrophil response to other infections. We therefore investigated the role of monocytes in the neutrophil response to infection with the bacterium Pseudomonas aeruginosa. We have previously shown that neutrophils will efficiently migrate to this bacterium in our in vitro infection model.13 Interestingly, we found that the presence of monocytes did not increase the neutrophil response to P. aeruginosa (Fig. 3c). Instead we saw a trend of fewer neutrophils migrating out of the lumen in the presence of monocytes, though this difference was insignificant. Collectively, these results indicate that paracrine signaling between neutrophils and monocytes is important for the neutrophil response to A. fumigatus and that this effect is not seen in response to P. aeruginosa infection. We were, therefore, interested in determining which signals were contributing to this increased migration.
Secretion of Several Pro-inflammatory Proteins are Altered in the Presence of Neutrophils and/or Monocytes
Given our data showing increased neutrophil migration in the presence of monocytes and our previous data showing neutrophil activation by an endothelium,13,29 we sought to identify factors, produced by the endothelial and innate immune cells in the infection model, that could be contributing to neutrophil migration. We used a multiplexed ELISA (MagPix) panel to compare the levels of pro- and anti-inflammatory proteins in lumen-conditioned media (Fig. 4a). The total protein levels measured represent the current protein level at 16 h post neutrophil seeding, before the hyphae breached the endothelial lumen. Thus, the protein levels show a snapshot of both secretion and consumption by all cells in the system at that timepoint. While the contributions of each cell type cannot be separated, we can gain insight by comparing between conditions. All secretion levels were normalized to the levels secreted by endothelium without an infection.
Secretion of Several Pro-inflammatory Proteins are Altered in the Presence of Neutrophils and/or Monocytes. A multiplexed enzyme-linked immunosorbent assay screen (Luminex) was run on lumen-conditioned media from 5 conditions with the indicated combinations of endothelium, neutrophils, and/or monocytes. (a) The log2 fold change of secreted protein levels over the endothelial lumen-only condition are plotted as a heat map. Blue indicates expression below the endothelial lumen-only condition and red indicates increased expression. Condition combinations are indicated at the top of the heat map. Factors measured are on the left of the heat map. (b) The levels of MIP-1α, MIP-1β, IL-1Ra, and IL-6 expressed as a fold change over the endothelial lumen-only condition were plotted. Bars represent least-squared mean (+/-) SEM. Condition combinations are indicated below all the graphs. All data represent 5 independent experiments with media from 3 lumens pooled per replicate. Asterisks show significance relative to all other conditions. *p < 0.05, **p < 0.005, *** < 0.0005.
Our analysis revealed several interesting secretion patterns (Fig. 4a). First, we saw very little change in endothelial cell protein secretion in the presence of A. fumigatus compared to uninfected devices. This is in contrast to our previous findings that showed significant changes in endothelial cell secretion in the presence of a bacterial infection.13 Second, we saw significantly increased secretion of MIP-1 family proteins in the conditions containing monocytes. The macrophage inflammatory proteins MIP-1α and MIP-1β were both significantly upregulated in the presence of innate immune cells. MIP-1α showed almost a 1.5-fold increase in the presence of neutrophils and over a 2-fold increase in the presence of both neutrophils and monocytes (Fig. 4b). MIP-1β had more than a 2-fold increase in both conditions with monocytes (Fig. 4b). Genomic analysis has shown that in the presence of A. fumigatus, human monocytes upregulated genes encoding IL-1β, MIP-1α (CCL3), and MIP-1β (CCL4) among others.8 Furthermore, MIP-1α and MIP-1β were first identified as macrophage-secreted proteins that stimulated neutrophil chemokinesis and reactive oxygen species generation.36,39 It is, therefore, likely that this increased in MIP-1α and MIP-1β secretion in conditions with monocytes is in part responsible for the increased neutrophil migration to A. fumigatus.
IL-1 family signaling was also altered in the presence of innate immune cells. The most extreme change was seen in the secretion of the IL-1 receptor antagonist (IL-1Ra) which was upregulated almost 20-fold in the presence of monocytes and over 70-fold in the presence of neutrophils and in the presence of both neutrophils and monocytes (Fig. 4b). The exceptionally high levels of IL-1Ra are interesting as IL-1Ra is known to inhibit the pro-inflammatory roles of IL-1 family proteins.32 In fact, a modified version of the IL-1Ra, Anakinra, is used to treat a number of inflammatory diseases including rheumatoid arthritis.5 Secreted IL-1β was slightly higher in the presence of monocytes. In contrast, the level of secreted IL-1α was decreased to 0.5-fold in all immune cell conditions (Fig. 4a). This decrease in secreted IL-1α could be due to internalization by the neutrophils and monocytes; however, IL-1Ra is known to act as antagonist to IL-1α binding so the high levels of IL-1Ra should significantly reduce the amount of binding and internalization. Therefore, an alternative explanation is that IL-1α is downregulated by endothelial cells in the presence of neutrophils and monocytes. Overall, the secretion results are in agreement with previous data showing slight increases in MIP-1α and IL-1β secretion by an endothelial lumen in the presence of A. fumigatus hyphae in an in vivo bronchiole model.2
Finally, the protein levels of IL-6 were decreased to approximately 0.6-fold in conditions containing neutrophils (Fig. 4b). We previously found that IL-6 signaling was critical for neutrophil migration to a bacterial infection in our model.13 In this system, the decrease in IL-6 signaling in the presence of innate immune cells, primarily neutrophils, likely indicates consumption by these cells.
Monocyte Secretion of MIP-1α Drives Neutrophil Migration to A. fumigatus
Given the secretion data showing increased protein levels of MIP-1 and IL-1 family proteins in the presence of monocytes or neutrophils, respectively, we hypothesized that these signals could be contributing to the neutrophil response to A. fumigatus. To test this hypothesis, we pre-incubated neutrophils and monocytes with either a control IgG1 or a blocking antibody against MIP-1α, MIP-1β, IL-1β, or IL-1R antagonist and ran the experiment in the continued presence of the antibody. In all cases, the blocking antibody was used in excess of the reported inhibitory concentration. Inhibition of IL-1 family proteins (IL-1β or IL-1R antagonist) had no significant effect on neutrophil migration to A. fumigatus (Supplemental Figure 1A and B); however, inhibition with MIP-1 family proteins did have a significant effect. Incubation of neutrophils and monocytes with MIP-1α blocking antibody significantly reduced neutrophil migration to A. fumigatus compared to an IgG1 control antibody (Figs. 5a and 5b). This trend was also observed when neutrophils and monocytes were incubated with MIP-1β blocking antibody (Fig. 5c). These results indicate that the increased migration of neutrophils in the presence of monocytes is likely due in part to increased secretion of MIP-1α and MIP-1β, in agreement with previous data showing neutrophil activation by MIP-1α and MIP-1β.36,39
Monocyte Secretion of MIP-1α Drives Neutrophil Migration to A. fumigatus. (a) Representative images of neutrophils migrating from the endothelial lumen (white line) to germinating A. fumigatus. Source of A. fumigatus shown in purple. (b) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time that germination began in the upper port of the device. (c) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time that germination began in the upper port of the device. (b, c) Each point represents the least-squared mean (+/-) standard error of the mean (SEM). Data quantified for 9 replicates across 3 independent experiments (b) or 6 replicates across 2 independent experiments (c). Asterisks represent significance between conditions at each point. ***p < 0.0005.
Neutrophil Migration to A. fumigatus Depends on IL-6 Signaling
The Magpix data also showed a decrease in protein levels of IL-6 in all conditions containing neutrophils, indicating consumption. Furthermore, we have previously shown that IL-6 signaling is critical for neutrophil migration to P. aeruginosa.13 Therefore, we hypothesized that IL-6 signaling could also be important for neutrophil migration to A. fumigatus. To test this hypothesis, we pre-incubated neutrophils with either the control IgG1 or anti-IL-6R blocking antibody before plating the cells in the lumen. We then measured the neutrophil response to A. fumigatus. We found that fewer neutrophils migrated out of the lumen, toward A. fumigatus in the presence of the blocking antibody compared to the control (Fig. 6a). We found that the presence of the blocking antibody decreased the number of migrated neutrophils, particularly at timepoints after 4 h post germination, where the cell flux increases in the control condition (Fig. 6b). This result, in combination with our previous findings, indicates a universal role for IL-6 signaling in driving the neutrophil response to infection. This is particularly interesting as IL-6 inhibitors are used clinically for the treatment of several diseases7,12,20,38 and use of these inhibitors is associated with a higher risk of infection, including fungal infections.33
Neutrophil Migration to A. fumigatus Depends on IL-6 Signaling. (a) Representative images of neutrophils migrating from the endothelial lumen (white line) to germinating A. fumigatus. Source of A. fumigatus shown in purple. (b) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time germination began in the upper port of the device. Each point represents the least-squared mean (+/-) standard error of the mean (SEM). Data quantified for 8 replicates across 3 independent experiments.
LTB4 Signaling Drives Neutrophil Migration and Swarming in Response to A. fumigatus
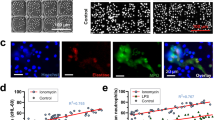
In addition to receiving signals from other cell types in the infectious environment, it is also known that neutrophils can relay signals to each other during an infection.1 Furthermore, we found that in our fungal infection model, neutrophils regularly coated the hyphal branches and created clusters of neutrophils, known as swarms (Figs. 7ai and 7ii, white arrows indicate swarms). Interestingly, these swarms resembled those seen in vivo during the neutrophil response to A. fumigatus infection in zebrafish.34 It is known that neutrophil swarming is driven in part by LTB4 signaling.21,22 Therefore, we hypothesized that neutrophil migration to A. fumigatus depended on LTB4 signaling. To test this hypothesis, we inhibited neutrophils with MK886, a potent inhibitor of leukotriene biosynthesis.26,30 We found that neutrophils migrated significantly less in the presence of this inhibitor (Fig. 7b). The trend of fewer neutrophils migrating in the presence of MK886 began at 2 h post germination and reached significance at 6 h post germination (Fig. 7c). In addition to the role of LTB4 in neutrophil migration, we were also interested in its role in neutrophil swarming in the device; therefore, we quantified the number of devices in which at least one neutrophil swarm could be identified. We found that neutrophil swarming was present in about 91 percent of control devices but only 56 percent of devices with MK886 (Fig. 7d). Altogether, this data indicates that LTB4 signaling between neutrophils is critical for neutrophil migration and swarming to A. fumigatus.
LTB4 Signaling Drives Neutrophil Migration and Swarming in Response to A. fumigatus. (a) Representative images showing neutrophils coating A. fumigatus hyphae. i. A. fumigatus in purple, neutrophils in green. ii. Neutrophil swarms shown in white, indicated by arrows. (b) Representative images of neutrophils migrating from the endothelial lumen (white line) to germinating A. fumigatus. Source of A. fumigatus shown in purple. (c) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time that germination began in the upper port of the device. Each point represents the least-squared mean (+/-) standard error of the mean (SEM). (d) Swarms were identified in each condition and the percentage of lumens with at least one swarm was determined. (c, d) Data quantified for 9 replicates across 4 independent experiments. *p < 0.05, **p < 0.005.
Neutrophils have been shown to coat A. fumigatus hyphae in several in vitro and in vivo models.9,17,19,34 This function has been shown to reduce Aspergillus hyphal growth34 and cause evasive action from the hyphae.9 Conversely, it has been shown that blocking neutrophil access to germinating hyphae causes increased hyphal growth as well as worse infection and outcomes.34 Collectively, these results indicate that neutrophil swarming around hyphae it is a critical function for neutrophil defense against A. fumigatus infection. It is known that a secondary neutrophil-generated gradient of LTB4 is critical for the formation of these swarms21,22 and our results confirm that reduction in LTB4 signaling reduces the ability of neutrophils to form swarms around A. fumigatus hyphae. It is also known that LTB4 signal relay increases neutrophil chemotaxis1,26 and our results confirm reduced neutrophil recruitment without LTB4 signaling. Taken together, our results support the hypothesis that effective neutrophil recruitment and swarming depend on LTB4 signaling during an A. fumigatus infection.
Conclusions
In this study we developed an infection-on-a-chip model that recapitulates many important factors of the in vivo infectious microenvironment during an A. fumigatus infection. Using this model, we were first able to show that the presence of an endothelium had a significant impact on the germination dynamics of A. fumigatus. This result indicates a multi-kingdom signaling interaction and invites further study.
Our model also allowed us to investigate the dynamics of the neutrophil response to A. fumigatus hyphae and determine what signal events were critical for this recruitment. Importantly, neutrophil recruitment to A. fumigatus in the model resembled the in vivo neutrophil response in many ways. First, neutrophils were recruited to A. fumigatus hyphae as described by other studies.4,34 Additionally, neutrophils were seen to swarm around the hyphae as seen in both in vivo and in vitro models of A. fumigatus infection.9,17,34
Finally, unlike in vivo models where complex cellular interactions make investigating the effect of individual signals more complicated, our model allowed us to investigate the role of multiple cell types in driving neutrophil migration and determine which signals were upregulated in the presence of neutrophils, monocytes, or the endothelium. Harnessing this ability allowed us to show that MIP-1 family proteins are upregulated by innate immune cells, particularly monocytes, and are important for an efficient neutrophil response. Furthermore, we were able to show that in similar bacterial infections, IL-6 is critical for the neutrophil response in fungal infections. However, our findings also show that distinct mechanisms mediate neutrophil responses to A. fumigatus and P. aeruginosa infections, as monocytes play a key role for the fungal but not bacterial response.
Collectively, these results show the importance of studying neutrophil migration in a physiologically relevant model system. In the future, this system could be used to investigate the interesting finding that the presence of an endothelium influences A. fumigatus germination. It could also be used to test the efficacy of new pharmaceuticals in driving neutrophil response to A. fumigatus or inhibiting A. fumigatus germination and growth. Multicellular models, such as the one in this study, hold immense promise for elucidating the complex signaling mechanisms that contribute to the innate immune response to infection and will allow for closer investigation and discovery of these signaling pathways.
References
Afonso, P. V., M. Janka-Junttila, Y. J. Lee, C. P. McCann, C. M. Oliver, K. A. Aamer, W. Losert, M. T. Cicerone, and C. A. Parent. LTB4 is a signal-relay molecule during neutrophil chemotaxis. Dev. Cell 22:1079–1091, 2012.
Barkal, L. J., C. L. Procknow, Y. R. Alvarez-Garcia, M. Niu, J. A. Jimenez-Torres, R. A. Brockman-Schneider, J. E. Gern, L. C. Denlinger, A. B. Theberge, N. P. Keller, E. Berthier, and D. J. Beebe. Microbial volatile communication in human organotypic lung models. Nat. Commun. 8:1770, 2017.
Ingram PN, Hind LE, Jiminez-Torres JA, Huttenlocher A, Beebe DJ. An accessible organotypic microvessel model using iPSC-derived endothelium. Adv Healthc Mater 7: 2018.
Bhatia, S., M. Fei, M. Yarlagadda, Z. Qi, S. Akira, S. Saijo, Y. Iwakura, N. van Rooijen, G. A. Gibson, C. M. St Croix, A. Ray, and P. Ray. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PLoS ONE 6:e15943, 2011.
Bullock, J., S. A. A. Rizvi, A. M. Saleh, S. S. Ahmed, D. P. Do, R. A. Ansari, and J. Ahmed. Rheumatoid Arthritis: a brief overview of the treatment. Med Princ Pract 27:501–507, 2018.
Chiang, L. Y., D. C. Sheppard, F. N. Gravelat, T. F. Patterson, and S. G. Filler. Aspergillus fumigatus stimulates leukocyte adhesion molecules and cytokine production by endothelial cells in vitro and during invasive pulmonary disease. Infect. Immun. 76:3429–3438, 2008.
Choy, E. H., D. A. Isenberg, T. Garrood, S. Farrow, Y. Ioannou, H. Bird, N. Cheung, B. Williams, B. Hazleman, R. Price, K. Yoshizaki, N. Nishimoto, T. Kishimoto, and G. S. Panayi. Therapeutic benefit of blocking interleukin-6 activity with an anti-interleukin-6 receptor monoclonal antibody in rheumatoid arthritis: a randomized, double-blind, placebo-controlled, dose-escalation trial. Arthritis Rheum. 46:3143–3150, 2002.
Cortez, K. J., C. A. Lyman, S. Kottilil, H. S. Kim, E. Roilides, J. Yang, B. Fullmer, R. Lempicki, and T. J. Walsh. Functional genomics of innate host defense molecules in normal human monocytes in response to Aspergillus fumigatus. Infect. Immun. 74:2353–2365, 2006.
Ellett, F., J. Jorgensen, G. H. Frydman, C. N. Jones, and D. Irimia. Neutrophil interactions stimulate evasive hyphal branching by Aspergillus fumigatus. PLoS Pathog. 13:e1006154, 2017.
Espinosa, V., A. Jhingran, O. Dutta, S. Kasahara, R. Donnelly, P. Du, J. Rosenfeld, I. Leiner, C. C. Chen, Y. Ron, T. M. Hohl, and A. Rivera. Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS Pathog. 10:e1003940, 2014.
Gregg, K. S., and C. A. Kauffman. Invasive Aspergillosis: epidemiology, clinical aspects, and treatment. Semin Respir Crit Care Med 36:662–672, 2015.
Hennigan, S., and A. Kavanaugh. Interleukin-6 inhibitors in the treatment of rheumatoid arthritis. Ther. Clin. Risk Manag. 4:767–775, 2008.
Hind, L. E., P. N. Ingram, D. J. Beebe, and A. Huttenlocher. Interaction with an endothelial lumen increases neutrophil lifetime and motility in response to P aeruginosa. Blood 132:1818–1828, 2018.
Hohl, T. M., H. L. Van Epps, A. Rivera, L. A. Morgan, P. L. Chen, M. Feldmesser, and E. G. Pamer. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 1:e30, 2005.
Jimenez-Torres, J. A., S. L. Peery, K. E. Sung, and D. J. Beebe. LumeNEXT: a practical method to pattern luminal structures in ECM gels. Adv Healthc Mater 5:198–204, 2016.
Jones, C. N., L. Dimisko, K. Forrest, K. Judice, M. C. Poznansky, J. F. Markmann, J. M. Vyas, and D. Irimia. Human neutrophils are primed by chemoattractant gradients for blocking the growth of Aspergillus fumigatus. J. Infect. Dis. 213:465–475, 2016.
Jones, C. N., F. Ellett, A. L. Robertson, K. M. Forrest, K. Judice, J. M. Balkovec, M. Springer, J. F. Markmann, J. M. Vyas, H. S. Warren, and D. Irimia. Bifunctional small molecules enhance neutrophil activities against Aspergillus fumigatus in vivo and in vitro. Front. Immunol. 10:644, 2019.
Kamai, Y., A. S. Lossinsky, H. Liu, D. C. Sheppard, and S. G. Filler. Polarized response of endothelial cells to invasion by Aspergillus fumigatus. Cell. Microbiol. 11:170–182, 2009.
Knox, B. P., Q. Deng, M. Rood, J. C. Eickhoff, N. P. Keller, and A. Huttenlocher. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot. Cell 13:1266–1277, 2014.
Kumari, N., B. S. Dwarakanath, A. Das, and A. N. Bhatt. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 37:11553–11572, 2016.
Lammermann, T. In the eye of the neutrophil swarm-navigation signals that bring neutrophils together in inflamed and infected tissues. J. Leukoc. Biol. 100:55–63, 2016.
Lammermann, T., P. V. Afonso, B. R. Angermann, J. M. Wang, W. Kastenmuller, C. A. Parent, and R. N. Germain. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498:371–375, 2013.
Lestrade, P. P., R. G. Bentvelsen, A. Schauwvlieghe, S. Schalekamp, W. van der Velden, E. J. Kuiper, J. van Paassen, B. van der Hoven, H. A. van der Lee, W. J. G. Melchers, A. F. de Haan, H. L. van der Hoeven, B. J. A. Rijnders, M. T. van der Beek, and P. E. Verweij. Voriconazole resistance and mortality in invasive Aspergillosis: a multicenter retrospective cohort study. Clin. Infect. Dis. 68:1463–1471, 2019.
Lestrade, P. P. A., J. F. Meis, W. J. G. Melchers, and P. E. Verweij. Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin. Microbiol. Infect. 25:799–806, 2019.
Lopes Bezerra, L. M., and S. G. Filler. Interactions of Aspergillus fumigatus with endothelial cells: internalization, injury, and stimulation of tissue factor activity. Blood 103:2143–2149, 2004.
Majumdar, R., A. Tavakoli Tameh, and C. A. Parent. Exosomes mediate LTB4 release during neutrophil chemotaxis. PLoS Biol. 14:e1002336, 2016.
Margalit, A., and K. Kavanagh. The innate immune response to Aspergillus fumigatus at the alveolar surface. FEMS Microbiol. Rev. 39:670–687, 2015.
Maus, U. A., K. Waelsch, W. A. Kuziel, T. Delbeck, M. Mack, T. S. Blackwell, J. W. Christman, D. Schlondorff, W. Seeger, and J. Lohmeyer. Monocytes are potent facilitators of alveolar neutrophil emigration during lung inflammation: role of the CCL2-CCR2 axis. J. Immunol. 170:3273–3278, 2003.
McMinn, P. H., L. E. Hind, A. Huttenlocher, and D. J. Beebe. Neutrophil trafficking on-a-chip: an in vitro, organotypic model for investigating neutrophil priming, extravasation, and migration with spatiotemporal control. Lab Chip 19:3697–3705, 2019.
Needleman, P., J. Turk, B. A. Jakschik, A. R. Morrison, and J. B. Lefkowith. Arachidonic acid metabolism. Annu. Rev. Biochem. 55:69–102, 1986.
Perlin, D. S., E. Shor, and Y. Zhao. Update on Antifungal Drug Resistance. Curr. Clin. Microbiol. Rep. 2:84–95, 2015.
Perrier, S., F. Darakhshan, and E. Hajduch. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS Lett. 580:6289–6294, 2006.
Rose-John, S., K. Winthrop, and L. Calabrese. The role of IL-6 in host defence against infections: immunobiology and clinical implications. Nat. Rev. Rheumatol. 13:399–409, 2017.
Rosowski, E. E., N. Raffa, B. P. Knox, N. Golenberg, N. P. Keller, and A. Huttenlocher. Macrophages inhibit Aspergillus fumigatus germination and neutrophil-mediated fungal killing. PLoS Pathog. 14:e1007229, 2018.
Segal BH, Bow EJ and Menichetti F. Fungal infections in nontransplant patients with hematologic malignancies. Infect Dis Clin North Am 16: 935-964, vii, 2002.
Sherry, B., P. Tekamp-Olson, C. Gallegos, D. Bauer, G. Davatelis, S. D. Wolpe, F. Masiarz, D. Coit, and A. Cerami. Resolution of the two components of macrophage inflammatory protein 1, and cloning and characterization of one of those components, macrophage inflammatory protein 1 beta. J. Exp. Med. 168:2251–2259, 1988.
Tagliabue, A., and R. Rappuoli. Changing priorities in vaccinology: antibiotic resistance moving to the top. Front. Immunol. 9:1068, 2018.
Trikha, M., R. Corringham, B. Klein, and J. F. Rossi. Targeted anti-interleukin-6 monoclonal antibody therapy for cancer: a review of the rationale and clinical evidence. Clin. Cancer Res. 9:4653–4665, 2003.
Wolpe, S. D., G. Davatelis, B. Sherry, B. Beutler, D. G. Hesse, H. T. Nguyen, L. L. Moldawer, C. F. Nathan, S. F. Lowry, and A. Cerami. Macrophages secrete a novel heparin-binding protein with inflammatory and neutrophil chemokinetic properties. J. Exp. Med. 167:570–581, 1988.
Acknowledgments
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences R35 GM1 18027 01, as well as the National Institutes of Health, National Institutes of Allergy and Infectious Diseases through R01 AI134749, T.S. was supported by the National Institute on Aging of the National Institutes of Health under Award Number T32AG000213.
Conflict of interest
David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso Inc., Salus Discovery LLC, LynxBiosciences Inc, Turba LLC, Stacks to the Future, LLC, and Onexio Biosystems, LLC. The authors declare no competing financial interests. LEH, MG, TJS, NPK, and AH do not have any conflicts to disclose.
Ethical Approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
Research Involving Human/Animal Participants
No animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Michael R. King oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1. IL-1 family protein signaling does not significantly alter neutrophil response to
A. fumigatus. (A and B) The number of neutrophils outside the lumen was quantified every hour using particle analysis in FIJI. The data was then adjusted for the time that germination began in the upper port of the device. Each point represents the least-squared mean plus standard error of the mean (SEM). (A) Data quantified for 6 replicates across 2 independent experiments. (B) Data quantified for 9 replicates across 3 independent experiments. *p < 0.05, **p < 0.005. (TIFF 1529 kb)
Supplemental Video 1. Neutrophil migration to A. fumigatus hyphae. Representative movie showing neutrophils coating A. fumigatus hyphae, A. fumigatus in purple, neutrophils in green. (AVI 1533 kb)
Supplemental Video 2. Neutrophil migration to A. nidulans. Representative movie showing neutrophils migrating to a source of A. nidulans. Phase video on left shows hyphal growth, fluorescent video on right shows neutrophils in white. (AVI 3998 kb)
Rights and permissions
About this article
Cite this article
Hind, L.E., Giese, M.A., Schoen, T.J. et al. Immune Cell Paracrine Signaling Drives the Neutrophil Response to A. fumigatus in an Infection-on-a-Chip Model. Cel. Mol. Bioeng. 14, 133–145 (2021). https://doi.org/10.1007/s12195-020-00655-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-020-00655-8