Abstract
In compressed sensing magnetic resonance imaging (CS-MRI), undersampling of k-space is performed to achieve faster imaging. For this process, it is important to acquire data randomly, and an optimal random undersampling pattern is required. However, random undersampling is difficult in two-dimensional (2D) Cartesian sampling. In this study, the effect of random undersampling patterns on image reconstruction was clarified using phantom and in vivo MRI, and a sampling pattern relevant for 2D Cartesian sampling in CS-MRI is suggested. The precision of image restoration was estimated with various acceleration factors and extents for the fully sampled central region of k-space. The root-mean-square error, structural similarity index, and modulation transfer function were measured, and visual assessments were also performed. The undersampling pattern was shown to influence the precision of image restoration, and an optimal undersampling pattern should be used to improve image quality; therefore, we suggest that the ideal undersampling pattern in CS-MRI for 2D Cartesian sampling is one with a high extent for the fully sampled central region of k-space.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Compressed sensing magnetic resonance imaging (CS-MRI) allows scan times to be shortened by reducing the quantity of sampled data [1,2,3,4,5,6,7,8,9,10,11]. In MRI, a decrease in the sampling of data, known as undersampling, generally causes a decrease in spatial resolution or aliasing artifacts, and the quality of reconstructed images is subject to deterioration. However, CS-MRI permits image reconstruction without deteriorations in image quality [12, 13], although it is important that data are acquired randomly.
In MRI, sampled data are stored in k-space, with the data in the central region of k-space contributing to the contrast of the reconstructed image, while data in the edge region contribute to spatial resolution. When the sampled data are randomly reduced in CS-MRI, the sampling pattern can affect the quality of the reconstructed image; an inappropriate sampling pattern will result in the degradation of image quality. Therefore, the choice of sampling pattern is an important factor in CS-MRI.
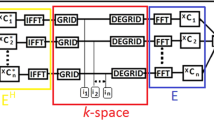
Two-dimensional (2D) and three-dimensional (3D) Cartesian sampling are used in MRI. In 2D Cartesian sampling, the data acquisition is performed in both phase-encode and read-out directions, with these directions being orthogonal. However, in 3D Cartesian sampling, an extra phase-encode direction is added. In CS-MRI, the reduced sampling of data is not performed in the read-out direction; thus, 3D Cartesian sampling is suitable for CS-MRI, because it is easier to perform random undersampling in 3D Cartesian sampling than in 2D Cartesian sampling. Many studies have reported on CS-MRI with 3D data acquisition [1, 2, 7,8,9,10, 14]. In these studies, satisfactory reconstruction images were obtained by applying various undersampling strategies, such as uniform distribution, variable density distribution, and Poisson-disk distribution, and a shorter scan time was achieved using a high acceleration factor. In contrast, in studies using CS-MRI with 2D data acquisition [3, 4, 6, 13], reducing the scan time, which is a principal benefit of CS-MRI, is difficult, because the undersampling strategy is rigid and it is difficult to utilize a high acceleration factor. However, 2D sampling of data is often performed in the clinical setting, so the use of CS-MRI with 2D data acquisition is useful. To apply CS-MRI for 2D data more effectively, it would be helpful to investigate the use of CS-MRI with 2D sampling data.
The aim of this study was to clarify the influence of the random undersampling pattern on CS-MRI with 2D Cartesian sampling. Images reconstructed with 2D CS-MRI using various undersampling patterns were evaluated. The random undersampling pattern strategy and the estimation method were performed as mentioned below.
2 Materials and methods
A 3-Tesla MRI system (Discovery 750w, GE Healthcare, Wisconsin, USA) was used for data acquisition in this study. The data used in this study were from the imaging of a phantom and in vivo imaging of the human brain. The imaging data were acquired using several patterns of random undersampling for k-space, and image reconstruction was then performed using the CS-MRI from these acquisitions. The reconstructed images were evaluated for quality and precision of restoration.
2.1 Random undersampling pattern
Figure 1 shows the details of the sampling patterns used in this study. The data of the central region of the k-space were acquired fully, while those of the edge region were randomly sampled according to a Gaussian distribution [4, 15]. The mean value and the standard deviation (SD) of the Gaussian distribution were 0 and 0.21, respectively. The SD corresponded to the full width at half maximum (FWHM), which equaled to 0.5. In the case of a large FWHM, the sampling data converged around the center of the k-space, with a few data on the edge. In contrast, a small FWHM vitiated the randomness of the data sampling. Therefore, a moderate FWHM (0.5) was chosen in this study. The random sampling was performed in the phase-encode direction only. Undersampled data were attainted by reducing the acquired data to 50%, 40%, and 30%, with acceleration factors corresponding to 2×, 2.5×, and 3.3×, respectively. For each acceleration factor, the extent of the central fully sampled region (hereinafter, referred to as CFSR extent) was varied from 20 to 80% in 10% intervals, and various sampling patterns were obtained. The influence of the random undersampling was observed by evaluating the quality of the images reconstructed using these sampling patterns.
Pattern mask for undersampling in k-space. The acquired data increase as the acceleration factor increases, with the central fully sampled data increasing as the CFSR extent increases. The vertical and horizontal directions in the mask correspond to the phase-encode and frequency-encode directions, respectively
2.2 CS-MRI
In CS-MRI, the reconstructed image is usually obtained by solving the unconstrained optimization problem. In this study, a nonlinear conjugate gradient descent algorithm [16] was used to solve the following equation:
where x is the reconstructed image, y is the acquired undersampled k-space data, Fu is the partial Fourier transform, TV is the total variation as a sparsity transform, and λ is a regularization weight for the total variation term. We chose λ = 0.01 to determine the trade-off between data consistency and sparsity. The conjugate gradient method requires the computation of \(\nabla f\left( x \right),\) which is defined as follows:
where \(F_{{\text{u}}}^{*}\) represents the complex conjugate of \({F_{\text{u}}}\), and \(\nabla f(x)\) and \(\nabla \|{\rm TV}x\|_1\) represent the finite differences of the object function and TV. \(\nabla \|{\rm TV}x\|_1\) was approximated as follows [17]:
where smoothing parameter \(\varepsilon =1 \times {10^{ - 4}}\) was used.
Algorithm for CS-MRI
Number of iterations: n = 50 was used for each CS reconstruction:
Iterations: for n = 0, 1, 2, …, do the following
Step 1: update the step size \({\alpha ^n}\)
we used back tracking line search [12],
where the line search parameters a = 0.05 and b = 0.6 were used. Then, \({\alpha ^n}\) is given by \({\alpha ^n}=t\).
Step 2: update the image with
Step 3: calculate the residual image
Step 4: calculate \({\beta ^n}\) used to find the searching direction
Step 5: calculate the new searching direction for the next iteration
The reconstruction was carried out with code developed in C++ (Visual Studio 2015, Microsoft Corporation, Redmond, Washington, USA), on a computer equipped with an Intel 2.5 GHz processor and with 8GB of RAM.
2.3 Phantom study
A quality control phantom (90-401 SYSTEM2, Nikko Fins Industries, Tokyo, Japan) comprising acrylic and polyvinyl alcohol (PVA) gel was scanned. The scanning parameters were as follows: pulse sequence, 2D fast spin echo (FSE); trajectory of k-space, Cartesian; TR/TE, 600/13 ms; echo train length, 16; field of view (FOV), 220 × 220 mm; matrix size in frequency direction, 256; slice thickness, 8 mm; bandwidth, 100 kHz. A parallel imaging technique was not used, and a single-channel birdcage coil was employed. The scan was repeated ten times to avoid the sources of measurement error such as signal inhomogeneity from the scanner, coil, and phantom.
To evaluate the precision of image restoration in the phantom study, the root-mean-square error (RMSE) and a structural similarity (SSIM) index [18] were measured on the images reconstructed with each sampling pattern, because the RMSE was generally applied for CS-MRI [1, 7, 19] and the SSIM [1, 10, 19] normalizes the image luminance and contrast, and is a good image quality index. The RMSE and the SSIM were calculated from the following equations:
where x is the image reconstructed from fully sampled data, y is the reconstructed image from undersampled data, and N is the total number of data points in the reconstructed image:
where x and y are the local areas in the image reconstructed from the fully sampled data and undersampled data, respectively, µx and µy are the averages of x and y, respectively, σx and σy are the variances of x and y, σxy is the covariance of x and y, and C1 and C2 were constant values used to avoid instability. In this study, the local SSIM values were calculated within a local 8 × 8 square window, with the window being moved pixel-by-pixel over the entire image.
In CS-MRI, a reduction in the data in the high-frequency region of the k-space due to undersampling causes a decrease in the spatial resolution. Therefore, to investigate spatial resolution quantitatively, a profile curve was drawn on the edge of the acrylic and PVA gel in the reconstructed image, and a modulation transfer function (MTF) was measured [20]. Because the undersampling was carried out in the phase-encode direction only, the profile curve was drawn horizontally to the phase-encode direction.
2.4 In vivo study
Thirty-nine patients (20 male and 19 female patients; age range 88–24 years; mean age 66.1 ± 15.3 years) who underwent brain MRI for evaluation of cerebrovascular disorders, brain tumor, vertigo, or other reasons during May 2017 were included; this study was approved by the institutional review broad of our facility. FSE T2-weighted imaging (T2WI) and FSE T1-weighted imaging (T1WI) were performed using a 12-channel phased-array head coil. The scan parameters for T1WI were TR/TE, 500/12.9 ms; echo train length 3, while for T2WI, they were TR/TE, 3000/90.8 ms; echo train length, 16. The following parameters were the same for both sequences: FOV, 220 × 220 mm; matrix size in frequency-encode direction, 256; slice thickness, 5 mm; bandwidth, 15.63 kHz. No parallel imaging technique was used.
The RMSE and the SSIM were measured in a similar manner to the phantom study. Furthermore, visual assessments were performed by a radiologist and two radiological technologists, each of whom had more than 15 years of experience in MRI. The reconstructed images were scored on a four-point scale with respect to aliasing artifacts and depiction of structure: 0 = nondiagnostic, conspicuous artifact, indistinct depiction; 1 = poor, moderate artifact, moderately indistinct depiction; 2 = adequate, mild artifact, slightly indistinct depiction; 3 = good, no artifact, distinct depiction. In the visual assessment, the images reconstructed from full sampling data were added as the reference images. A score was determined for each image by consensus between the three assessors.
2.5 Data analysis
A Kruskal–Wallis test was performed for multiple comparisons between the images reconstructed using the various sampling patterns by varying the CFSR. A P value < 0.05 was considered statistically significant. The analyses were carried out using JMP12.1 (SAS Institute, Cary, NC, USA).
3 Results
3.1 Phantom study
Figure 2a shows the RMSE for each acceleration factor. The RMSE increased with large acceleration factors. When the acceleration factor was 3.3×, the RMSE decreased as the CFSR extent increased, with the RMSE reaching a minimum when the CFSR extent was equal to 50%. The RMSE then increased slowly with further increases in CFSR extent. The same tendency was observed when the acceleration factors were 2.5× and 2×; however, the difference in the RMSE for each CFSR extent was small. Table 1 shows the P value for each acceleration factor. There was no significant difference between the different CFSRs when acceleration factors were 2.5× and 2×. In the case of an acceleration factor of 3.3×, there was no significant difference between the moderate and high CFSRs (40–80%).
The results of the SSIM index analyses (Fig. 2b) show that when the acceleration factor was high, the SSIM was small, with the SSIM increasing as the CFSR extent increased at each acceleration factor, achieving a maximum when the CFSR extent was equal to 80%. With an acceleration factor of 3.3×, there was conspicuous deterioration of the SSIM when a low CFSR extent was used. Table 2 shows the P values for the results of SSIM for each acceleration factor. For each acceleration factor, significant difference was almost recognized; however, for high CFSR (60–80%), there was no significant difference.
Figure 3 shows the MTF, which deteriorated when the acceleration factor was high. At each acceleration factor, the MTF decreased as the CFSR extent increased; however, the variation in the MTF was minimal when an acceleration factor of 2× was used.
MTF for the 2×, 2.5×, and 3.3× acceleration factors used in the phantom study. The black, gray, and light gray lines show the 20, 30, and 40% CFSR extents, respectively. The black, gray, and light gray dashed lines show the 50, 60, and 70% CFSR extents, respectively. The black dotted line shows the 80% CFSR extent
Figure 4 shows the reconstructed images and Fig. 5 shows enlarged images. Aliasing artifacts are recognizable in the image with an acceleration factor of 3.3× and a CFSR extent of 20%, although such artifacts were seldom observed in the other images. The depiction of a pin pattern was obscured as the acceleration factor and CFSR extent increased (Fig. 5), with the blurring of the pin pattern occurring in the phase-encode direction (vertical direction on the images).
Enlarged portions of the phantom images. The depiction of the pin pattern is focused and the area of the pin pattern is expanded. The arrangement of the images is the same as in Fig. 4
3.2 In vivo study
Figure 6a, c shows the RMSE of the T1WI and T2WI. In both cases, the RMSE increased with increasing acceleration factor, while, at each acceleration factor, the RMSE decreased as the CFSR extent increased. The RMSE was the lowest when the CFSR extent was near 50%. Tables 3, 4 show the P values in T1WI and T2WI. When the acceleration factor was 2×, there was no significant difference between low (20–40%) and high CFSRs (70–80%).
Figure 6b, d shows the SSIM indices of the T1WI and T2WI. Similar to the phantom study, the SSIM indices showed corruption in both the T1WI and T2WI when the acceleration factor was large, while at each acceleration factor, the SSIM improved with higher CFSR extents. Tables 5, 6 show the P values of results for the SSIM in T1WI and T2WI. When the acceleration factors were 2.5× and 2×, there was significant difference between low (20–40%) and high CFSRs (70–80%).
Table 7 lists the results of the visual assessments. In both the T1WI and T2WI, the scores for artifact and the depiction of structure were low when a high acceleration factor was used. Furthermore, the score was high when the CFSR extent was high at each acceleration factor. The images reconstructed from full sampling data had the highest score in both T1WI and T2WI. Tables 8, 9, 10, 11 show the P values in the visual assessment with respect to aliasing artifact and depiction of structure for T1WI and T2WI, respectively. There was a significant difference between the full sampling images and CS-MRI images. For each acceleration factor, a significant difference was recognized for almost CFSR; however, no significant difference was observed when the CFSRs were close to each other.
Figures 7 and 8 show reconstructed T1WI and T2WI of the brain. Similar to the phantom images, aliasing artifacts were conspicuous when the acceleration factor was high and the CFSR extent was low. This tendency was especially prominent with an acceleration factor of 3.3 × according to the data.
Reconstructed T2WI of the brain for each condition. The arrangement of the images is the same as in Fig. 7
4 Discussion
In this study, to clarify the influence of the random undersampling pattern in CS-MRI, the RMSE and SSIM were estimated to determine the precision of image restoration, while the MTF was measured to determine spatial resolution. Furthermore, visual assessments were performed for the qualitative evaluation of T1WI and T2WI of the brain. The results of the RMSE analysis show that the optimum CFSR extent was near 50% at each acceleration factor. However, according to the results of the SSIM indices, the optimal CFSR extent was 80%. These results indicate that the optimal CFSR extent can vary according to the evaluation method used. Furthermore, the MTF was improved when the CFSR extent was small.
A large CFSR extent results in a decline in the spatial resolution; therefore, the MTF was improved by the use of a low CFSR extent, because the data in the edge region of k-space increased. Conversely, a small CFSR extent resulted in aliasing artifacts in the reconstructed image. For the RMSE, the optimal CFSR extent was near 50%, because the occurrence of aliasing artifacts and the decline in spatial resolution were moderate. Regarding the SSIM, as the precision of image restoration depends on the degree of the aliasing artifact rather than the spatial resolution, the appropriate CFSR extent was 80%. On the visual assessments, the high CFSR extent provided a high score, similar to the SSIM. In a previous study, it was reported that there was a correlation between the visual assessment and the SSIM [19], and our results are in accordance with this. Therefore, we conclude that the optimal CFSR extent was 80% in this study.
The previous studies have recommended the use of a low CFSR [4], which is in contrast with the results of this study. This may be attributed to a number of reasons. First, the evaluation method used in this study differed from those used in the previous studies. In the previous studies, only the concordance correlation coefficient was applied and visual assessment was not performed for the evaluation of image quality. For CS-MRI, only quantitative evaluation is not sufficient for the evaluation of the image quality [19], and visual assessment is also important. Second, it appears that the reconstruction algorithm used in our study was different, although the algorithm used in the previous study was unclear. In the case of low CFSR, the sampling data of the edge in the k-space were relatively higher compared to that with high CFSR. As a result, the aliasing artifact was conspicuous. In CS-MRI, it is essential to suppress the aliasing artifact, and it is believed that the reconstruction algorithm used in the previous study was better. In any case, we believe that a high CFSR should be employed to obtain good image quality using 2D CS-MRI when using the reconstruction algorithm used in this study.
Parallel imaging, such as sensitivity encoding (SENSE) or generalized autocalibrating partially parallel acquisitions (GRAPPA), is a fast imaging technique generally used in the clinical environment. It provides good image quality using 2D data, even if the acceleration factor is greater than 2. In contrast, as observed in this study, the image quality of 2D CS-MRI deteriorated when the acceleration factor was greater than 2. Therefore, we cannot assume that, for 2D data, CS-MRI is superior to parallel imaging. However, CS-MRI can be applied in combination with parallel imaging [11]. Thus, we believe that a combination of CS-MRI and parallel imaging may be useful when using 2D data.
Our study has several limitations. First, the image restoration was performed using the conjugate gradient method. There are various methods for image restoration, such as the fast iterative shrinkage threshold algorithm [19] or the split Bregman algorithm [9]. Since the suitable technique for 2D CS-MRI is not yet established, the conjugate gradient method that was reported initially was used in this study. Therefore, it is debatable whether other methods would present the same results, and further investigations with other algorithms would prove meaningful in the future. Second, only brain images were used, and the influence of contrast differences in the precision of image restoration was only evaluated according to T1WI and T2WI. Investigations using the images of other regions, such as the spine or abdomen, are important, because the structure within an image can affect the precision of image restoration. Third, in this study, the sampling pattern of the edge of the k-space (high-frequency region) exhibited a Gaussian distribution, and the parameters, such as the SD, were fixed. To improve the spatial resolution, the number of phase encode in the high-frequency region should be increased. It was surmised that the optimal CFSR depended on the sampling pattern of the high-frequency region, even if the acceleration factor was the same. Therefore, the optimization of parameters in Gaussian distribution would be useful in future.
5 Conclusions
In this study, the influence of the random undersampling pattern on the quality of a reconstructed image with CS-MRI using 2D data was clarified. The results demonstrate that the undersampling pattern has a considerable effect on the reconstructed images. Based on the results of this study, when using undersampling for 2D CS-MRI, we recommend the use of a high CFSR to improve the precision of image restoration.
References
Mann LW, Higgins DM, Peters CN, Cassidy S, Hodson KK, Coombs A, Taylor R, Hollingsworth KG. Accelerating MR imaging liver steatosis measurement using combined compressed sensing and parallel imaging: a quantitative evaluation. Radiology. 2016;278(1):247–56.
Li B, Li H, Kong H, Dong L, Zhang J, Fang J. Compressed sensing based simultaneous black- and gray-blood carotid vessel wall MR imaging. Magn Reson Imaging. 2017;38:214–23.
Kido T, Kido T, Nakamura M, Watanabe K, Schmidt M, Forman C, Mochizuki T. Compressed sensing real-time cine cardiovascular magnetic resonance: accurate assessment of left ventricular function in a single-breath-hold. J Cardiovasc Magn Reson. 2016;18(1):50.
Han S, Cho H. Temporal resolution improvement of calibration-free dynamic contrast-enhanced MRI with compressed sensing optimized turbo spin echo: the effects of replacing turbo factor with compressed sensing accelerations. J Magn Reson Imaging. 2016;44(1):138–47.
Kim SG, Feng L, Grimm R, Freed M, Block KT, Sodickson DK, Moy L, Otazo R. Influence of temporal regularization and radial undersampling factor on compressed sensing reconstruction in dynamic contrast enhanced MRI of the breast. J Magn Reson Imaging. 2016;43(1):261–9.
Nam S, Hong SN, Akçakaya M, Kwak Y, Goddu B, Kissinger KV, Manning WJ, Tarokh V, Nezafat R. Compressed sensing reconstruction for undersampled breath-hold radial cine imaging with auxiliary free-breathing data. J Magn Reson Imaging. 2014;39(1):179–88.
Rapacchi S, Han F, Natsuaki Y, Kroeker R, Plotnik A, Lehrman E, Sayre J, Laub G, Finn JP, Hu P. High spatial and temporal resolution dynamic contrast-enhanced magnetic resonance angiography using compressed sensing with magnitude image subtraction. Magn Reson Med. 2014;71(5):1771–83.
Wang H, Miao Y, Zhou K, Yu Y, Bao S, He Q, Dai Y, Xuan SY, Tarabishy B, Ye Y, Hu J. Feasibility of high temporal resolution breast DCE-MRI using compressed sensing theory. Med Phys. 2010;37(9):4971–81.
Smith DS, Welch EB, Li X, Arlinghaus LR, Loveless ME, Koyama T, Gore JC, Yankeelov TE. Quantitative effects of using compressed sensing in dynamic contrast enhanced MRI. Phys Med Biol. 2011;56(15):4933–46.
Worters PW, Sung K, Stevens KJ, Koch KM, Hargreaves BA. Compressed-sensing multispectral imaging of the postoperative spine. J Magn Reson Imaging. 2013;37(1):243–8.
Zhang T, Chowdhury S, Lustig M, Barth RA, Alley MT, Grafendorfer T, Calderon PD, Robb FJ, Pauly JM, Vasanawala SS. Clinical performance of contrast enhanced abdominal pediatric MRI with fast combined parallel imaging compressed sensing reconstruction. J Magn Reson Imaging. 2014;40(1):13–25.
Lustig M, Donoho D, Pauly JM. Sparse MRI. The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–95.
Haldar JP, Hernando D, Liang ZP. Compressed-sensing MRI with random encoding. IEEE Trans Med Imaging. 2011;30(4):893–903.
Pandey A, Yoruk U, Keerthivasan M, Galons JP, Sharma P, Johnson K, Martin DR, Altbach MI, Bilgin A, Saranathan M. Multiresolution imaging using golden angle stack-of-stars and compressed sensing for dynamic MR urography. J Magn Reson Imaging. 2017;46(1):303–11.
Sharma SD, Fong CL, Tzung BS, Law M, Nayak KS. Clinical image quality assessment of accelerated magnetic resonance neuroimaging using compressed sensing. Invest Radiol. 2013;48(9):638–45.
Kawata S, Nalcopglu O. Constrained iterative reconstruction by the conjugate gradient method. IEEE Trans Med Imaging MI. 1985;-4(2):65–71.
Zeng GL. Medical image reconstruction. a conceptual tutorial. New York: Springer; 2010. pp. 131–134 (146–147).
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13(4):600–12.
Akasaka T, Fujimoto K, Yamamoto T, Okada T, Fushumi Y, Yamamoto A, Tanaka T, Togashi K. Optimization of regularization parameters in compressed sensing of magnetic resonance angiography: can statistical image metrics mimic radiologists’ perception? PLoS One. 2016;11(1).
Steckner MC, Drost DJ, Prato FS. Computing the modulation transfer function of a magnetic resonance imager. Med Phys. 1994;21(3):483–9.
Acknowledgements
We thank Karl Embleton, Ph.D., from Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures involving human participants were in accordance with the ethical standards of the Institutional Review Board (IRB) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The IRB waived the requirement for patients’ informed consent. This study does not involve any experiments involving animals.
Informed consent
The requirement for informed consent was waived by the institutional review board because of the retrospective nature of the analysis.
Conflict of interest
The authors declare that they have no conflict of interest in this article.
About this article
Cite this article
Kojima, S., Shinohara, H., Hashimoto, T. et al. Undersampling patterns in k-space for compressed sensing MRI using two-dimensional Cartesian sampling. Radiol Phys Technol 11, 303–319 (2018). https://doi.org/10.1007/s12194-018-0469-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12194-018-0469-y