Abstract
Globally, heat stress seriously threatens productivity of cattle. The objective of this study was to identify novel miRNAs that regulated heat stress in feedlot cattle. Experiment was conducted under heat stress and normal conditions. With profiling miRNAs of each feedlot cattle, our results showed the level of miR-1246 was significantly increased in these heat-stressed cattle (P < 0.05). Furthermore, by using bioinformatics analysis and luciferase reporter assays combined with qPCR and western blot, we found miR-1246 negatively regulated poly (C) binding protein 2 (PCBP2) and cAMP response element binding protein-like 2 (CREBL2) mRNA and protein levels through binding to the 3′-UTR region (P < 0.05); further, it inhibited heat-induced apoptosis in lung cells. Finally, our results suggested that miR-1246 plays an important role in heat stress and it has the potential to be a novel modulation factor for heat stress in feedlot cattle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat stress most likely occurs in livestock which cannot adequately dissipate body heat to maintain thermal balance. In cattle, heat stress negatively affects food intake, growth, immunity, and reproduction efficiency. Moreover, some studies reported that numerous cattle were dead because of heat stress. For example, over 3000 feedlot cattle died in Queensland (Nienaber and Hahn 2007) and over 4000 cattle died in the Midwest of the USA (Mitlohner et al. 2001) during the period of heat wave. California heat wave had caused more than 25,000 deaths among dairy cows, and Iowa heat wave killed at least 4000 cattle in 2006 (Renaudeau et al. 2012). Collectively, all these responses lead to rising management costs and a decline in income. Some cooling methods were used to relieve heat stress, such as providing shades, fans, and sprinklers, but these did not solve problems from genetics. Therefore, improving the ability of heat tolerance of cattle could have cumulative and permanent effects. Up to now, many studies have focused on the molecular mechanism of heat stress, which indicated that heat stresses could evoke complex responses. Also, heat resisted was considered as an essential for safeguarding the survival of organisms and cellular integrity (Belhadj Slimen et al. 2016). Severe heat stress might play an adverse role in the pathogenesis of many lung diseases, such as respiratory distress syndrome, emphysema, asthma, bronchopulmonary dysplasia, and interstitial pulmonary fibrosis (Choi and Alam 1996; Kahle et al. 2015).Commonly, pulmonary integrity is considered a key factor for cattle high adaptiveness in harsh environment, while maintaining a balance of proliferation and apoptosis of irritated lung cells is very important. However, the mechanism of miRNAs regulate cattle lung cells during heat stress has not yet been understood.
MicroRNAs (miRNAs) are a class of small, highly conserved and noncoding RNAs that negatively regulate target genes by epigenetically regulating protein translation or degrading the 3′-untranslated region (UTR) of the complementary mRNAs (Ambros 2004; Bartel 2007). Growing evidence has indicated that unique miRNAs in serum and plasma are stable (Chen et al. 2008; Hunter et al. 2008), which could be served as novel disease markers (Kosaka et al. 2010; Wang et al. 2010; Nair et al. 2013). Notably, miRNAs play an important role in gene expression during environmental stress (Leung and Sharp 2010). Several miRNAs such as miR-398, miR-34, and miR-34b were highly expressed in heat stress condition (Yu et al. 2011; Guan et al. 2013; Isik et al. 2016; Chen et al. 2017). Studies on human suggested that serum miR-1246 could be a novel diagnostic and prognostic biomarker for colorectal cancer and esophageal squamous cell cancer (Takeshita et al. 2013; Ogata-Kawata et al. 2014). Besides, miR-1246 targeted RTKN2 gene to regulate UVB-induced apoptosis in HaCaT cells, indicating that miR-1246 could be a potential therapy target for the treatment of UVB-induced damage in skin cells (Li et al. 2014).
In this study, we performed miRNA profiling in the plasma of Angus and Droughtmasters beef cattle under heat stress condition and normal condition and found that miR-1246 was highly expressed in heat-stressed feedlot cattle. We further investigated whether miR-1246 could be used as a novel manipulation of these heat-stressed feedlot cattle.
Materials and methods
Ethics statement
All animal procedures were approved by the Southwest University Institutional Animal Care and Use Committee.
Experimental animals and samples
Five Red Angus and five Droughtmasters (average body weight of 450 kg with similar body condition and consistent feeding and management) were selected from Chongqing Improved Feedlot Cattle Farm, P. R. China and used in this study. Barn temperature and humidity index (THI) was detected at 2:00 p.m. (Sengar et al. 2018), as an indicator of heat-stressed condition. Average THI was calculated by the equation, THI = 0.72 (Tc + Tw) + 40.6 (Tc, dry bulb temperature; Tw, humidity bulb temperature) (Buzan et al. 2015). THI < 72 could be regarded as normal condition, while THI > 72 is regarded as heat stress condition. (72 to 79 as mild heat stress, 80 to 90 as moderate heat stress, and > 90 as severe heat stress.) (Armstrong 1994; Poisky and von Keyserlingk 2017). Rectal temperature and respiratory rate were detected respectively using thermometer and counter under heat stress and normal condition. Peripheral venous blood (10 mL) from each feedlot cattle was collected in EDTA tube under normal and heat stress. Blood was firstly kept at room temperature for about 1 h then centrifuged at 3000g for 15 min at the same temperature. Plasma was immediately separated, frozen at − 20 °C, then stored at − 80 °C before the isolation of RNA.
Small RNA library construction and sequencing
Total RNA was extracted from 200 μL plasma using Trizol reagent (TAKARA, Japan), following the manufacturer’s procedure. The total RNA quantity and purity were analysis of Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN score > 7.0; then, five samples of each group were pooled in one library. Approximately 1 μg of total RNA was used to prepare small RNA libraries and the single-end sequencing was performed on an Illumina Hiseq2500 at the LC-BIO (Hangzhou, China) following the vendor’s recommended protocol. The raw reads were firstly processed to remove adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA), and repeats. Subsequently, unique sequences with length in 18–26 nucleotides were mapped to specific species precursors in miRBase 20.0 by BLAST search to identify known miRNAs and novel 3p- and 5p- derived miRNAs. Differentially expressed miRNAs would be determined by comparing the miRNA expression levels between two samples. The expression of the miRNAs was normalized to obtain the expression in transcripts per million. Normalization method: (1) Find a common set of sequences among all samples. (2) Construct a reference data set. Each data in the reference set is the copy number median value of a corresponding common sequence of all samples. (3) Perform two-based logarithm transformation on copy numbers log2 (copy#) of all samples and reference data set. Δlog2(copy#). (4) Calculate the log2(copy#) differenceΔlog2(copy#) between individual sample and the reference data set. (5) Form a subset of sequences by selecting -Δlog2(copy#)-< 2, which means less than (22=) 4-fold change from the reference set. (6) Perform linear regressions between individual samples and the reference set on the subset sequences to derive linear equations iy = aix + bi, where ia and ib are the slope and interception, respectively, of the derived line, log2(copy#) copy of the reference set, and y is the expected log2(copy#) of sample I on a corresponding sequence. (7) Calculate the mid value xmid = (max(x) − min(x))/2 of the reference set. Calculate the corresponding expected log2(copy#) of sample i, yi, mid = aixmid+bi. Let yr, mid = xmid, let Δyi, =yr, mid-yi, mid, which is the logarithmic correction factor of sample i. We then derive the arithmetic correction factor fi = 2Δyi sample I. (8) Correct copy numbers of individual samples by multiplying corresponding arithmetic correction factor fi to original copy numbers. The significance threshold was set to be 0.01 and 0.05 in each test.
Target gene prediction
Prediction genes targeted by differentially expressed miRNAs were identified through TargetScan7.1, CoMeTa, and miRDB software. Then, the data predicted by all the algorithms were combined and the overlaps were calculated. The GO terms and KEGG pathway of these differentially expressed miRNA targets were also annotated.
Cell culture
HEK293 (human embryonic kidney) cells were obtained from Cell Bank on Type Culture Collection of Chinese Academy of Sciences, and BFR-L1 (gayal cattle lung epithelial) cells were obtained from Shanghai JiNing BioCo. HEK293 cells were cultured in DMEM/F12 (Gibco, Cat. no. C11330500BT) with 15% FBS (Gibco, Cat. no. 10099-141) and BFR-L1 cells were cultured in MEM (Gibco, Cat. no. 11095-080) with 15% FBS. Medium with cells were added to flasks (25 cm2, containing 6 mL medium, which include about 5 × 106 cells). 6-well plates (2 mL medium per well, which include about 2.5 × 106 cells). Twenty-four-well plates (500 μL medium per well, which include about 5 × 105 cells) and all cells were added 100 IU/mL penicillin/streptomycin and were incubated at 37 °C and 5% CO2.
Vector constructs
MiR-1246 (mimics) and negative control (NC) were synthesized by RiboBioCo (Guangzhou, China). The 3′UTR of CREBL2 and PCBP2 were sub-cloned into the psiCHECK-2 vector (Promega, USA) downstream of the Renilla luciferase reporter gene. Flanking sequence on each side was synthetized with a short extension containing cleavage sites for XhoI (CTCGAG) and NotI (GCGGCCGC), and the primer sequences are shown in Table 1.
Dual luciferase reporter assay
HEK 293 cells were cultured in 24-well plates and transfected with pluc-3′UTR+miR-1246 mimics (50 nM) or NC (50 nM) using Lipofectamine 2000 (Invitrogen, USA). After 48 h, the luciferase activity was determined using dual luciferase reporter reagent (Promega, USA, Cat. no. E2029), and the luciferase activity is presented as the ratio of Firefly luciferase against Renilla luciferase.
qRT-PCR
HEK293 cells were cultured in 6-well plate and transfected with 100 nm/L miR-1246 mimics or NC. After 48 h, total RNA was isolated using TRIZOL (TAKARA, Cat. no. 9767) according to the manufacturer’s instructions. Reverse transcription was performed using PrimeScript™RT reagent Kit (TAKARA, Cat. no. RR037A). mRNA levels were analyzed using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (TAKARA, Cat. no. DRR820A), and the primer sequences are shown in Table 1. Gene expression level was normalized to GAPDH and 2−ΔΔct method was used to quantify the relative expression level of genes.
Western blot analysis
HEK293 cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer and proteins in supernatants were quantified using the bicinchoninic acid (BCA) protein assay (Solarbio, China). Proteins were separated on 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, electroblotted onto a polyvinylidene difluoride (PVDF) membrane, and incubated with anti-CREBL2 antibody (1/1000; ProteinTechGroup, Cat. no. 18007-1-AP), anti-PCBP2 antibody (1/2000; ProteinTechGroup, Cat. no. 15070-1-AP), anti-GAPDH antibody (1/2000; ProteinTechGroup, Cat. no. 60004-1-Ig), followed by incubation with horseradish peroxidase-conjugated secondary antibody (Beyotime, China, Cat. no. A0216 or A0208). PVDF membranes were detected using chemiluminescent ECL reagent (Millipore, Cat. no. WBKLS0050). Images were acquired using Whole Body Imaging System (VilBer Fusion FX5, France).
Annexin V-FITC/PI fluorescein isothiocyanate staining
BFR-L1 cells were seeded in 6-well plates, transfected with miR-1246 mimics or NC, incubated at 42 °C for 2 h, then recovered at 37 °C for 1 h (Kapila et al. 2018). Treated cells were collected, washed, and stained using Annexin V-FITC/PI double staining apoptosis detection kit (Nanjing JianchengBioIn, Cat. no. G003). Apoptotic rates of BFR-L1 cells were determined by ELITE ESP (Beckman-Coulter, USA). Early apoptotic cells were positively stained for Annexin V-FITC only and nonviable apoptotic cells were positively stained for both Annexin V-FITC and PI represented. In addition, cells were incubated with DAPI (Beyotime, Cat. no. C1005) for 15 min for nuclei staining.
Statistical analysis
All data was analyzed by SPSS18.0 software. The differences between two groups were analyzed using Student’s t test. The differences for multiple comparisons were evaluated with one-way analysis of variance (ANOVA). All results were expressed as the mean ± standard deviation from at least three independent experiments. *P < 0.05 or **P < 0.01 was considered to indicate a significant difference.
Results
Changes of physiological traits during heat stress
To investigate the effect of heat stress on physiological traits of feedlot cattle, rectal temperature and respiratory rate of experiment cattle were detected. The results showed that THI of control group and experimental group were 62.51 ± 0.42 and 84.83 ± 0.29, respectively, which suggested that the experimental cattle were under heat stress and control cattle were in normal (Fig. 1a). Compared with normal feedlot cattle, the rectal temperature increased from 38.3 ± 0.11 to 39.3 ± 0.34 °C (0.03-fold) in Red Angus under heat stress (P < 0.01), but an elevation was not seen in Droughtmaster. During heat stress, mean respiratory rate increased from 32.00 ± 2.52 to 49.60 ± 2.61 (breaths per min) in Red Angus and increased from 26.2 ± 3.6 to 49.40 ± 2.19 in Droughtmaster (P < 0.01) (Fig. 1b, c).
Profiling of small RNAs in feedlot cattle
To identify the type of small RNA (sRNA) in feedlot cattle, two independent sRNA libraries were constructed from the peripheral blood by Illumina technology for heat stress and normal condition, respectively. Low-quality sequences (length filter, and junk reads) were removed and the remanent reads were in alignment with Repbase and small RNA databases (e.g., rRNA, tRNA, snoRNA, snRNA, et al.). For unique sequences, there were 94.13 and 93.29% clean reads in the A-normal and A-heat stress libraries, and 91.46 and 92.48% clean reads in D-normal and D-heat stress libraries (Fig. 2a, b). In addition, the length distribution of reads showed that clean reads was 20–24 nt in size (Fig. 2c), and the top abundance size was 22 nt, indicating that most of the sRNA belong to miRNAs.
Distribution and classification of ncRNAs. a, b Relative proportions of the different ncRNAs. The percent of rRNA, tRNA, snoRNA, snRNA, other RNAs and clean reads were showed in pie diagram. c Length distribution of unique small RNAs in four libraries. A-normal, normal Red Angus; A-heat stress, heat-stressed Red Angus; D-normal, normal Droughtmaster; D-heat stress, heat-stressed Droughtmaster
MiR-1246 is significant upregulated in Red Angus and Droughtmaster
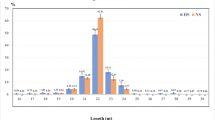
To investigate whether heat tolerance is related to the breed of feedlot cattle, we performed high-throughput miRNA profiling. In Red Angus, 365 miRNAs were detected and 63 miRNAs changed significantly; in Droughtmaster, 282 miRNAs were detected and 39 miRNAs changed significantly (including upregulated and downregulated miRNAs, fold change ≥ 2, P < 0.01, Supplementary tables 1 and 2). Co-miRNA expression patterns were analyzed using an unsupervised hierarchical clustering to screen how the differentially regulated miRNAs corresponded with different conditions in Red Angus and Droughtmaster. The results showed that although some of miRNAs remain unchanged in heat-stressed reaction among two kinds of cattle, all co-miRNAs had similar trend in expression model (upregulated or downregulated). In heat stress libraries, most of the differential miRNAs had lower expression levels in Droughtmaster than in Red Angus (Fig. 3a). Among the top 10 upregulated miRNAs identified from each group of cattle (read counts ≥ 100, fold change ≥ 2, P < 0.001, Fig. 3b, c), the expression of miR-1246 was significantly upregulated in both Red Angus and Droughtmaster.
MiR-1246 is significantly upregulated in Red Angus and Droughtmaster. a Clustering analysis of differentially expressed miRNAs in two feedlot cattle under heat stress and normal by heatmap. Red represents positively correlated and blue represents negatively correlated. b, c Read counts of top 10 miRNAs in Droughtmaster and Red Angus by high-throughput profiling (normalized reads)
Target gene prediction and pathway analysis of miR-1246
Next, TargetScan 7.1, CoMeTa, and miRDB software were used to predict target genes of miR-1246, and 47 genes were identified (Fig. 4a). Combined with the three software analytical results, including the seed matches in 3′UTR region, conserved seed matches in different species and minimum free energy analysis, only PCBP2 and CREBL2 were qualified, thus considered as target genes of miR-1246. KEGG enrichment analysis showed that PCBP2 and CREBL2 were mainly involved in cell apoptosis signal pathways, such as Hippo signaling pathway and cAMP signaling pathway (Fig. 4b, Supplementary table 3). In addition, our results showed that the two genes involved in heat stress-related reactions (Fig. 5a).
Prediction and pathway analysis of miR-1246 target genes. a Venn diagram indicated common and unique differential target genes. b Scatter plot of genes in the top 20 KEGG enrichments. In this diagram, correlated pathways of heat stress were analyzed using the molecule annotation system. X-axis represents the rich target genes of miR-1246, Y-axis represents associated pathway, P represents Benjamin corrected P value, and the black arrow indicates relation to genes within the functional pathway
miR-1246 targets PCBP2 and CREBL2. a PCBP2 and CREBL2 expression changes under normal and heat stress. Normal: BFR-L1 cells (lung cells) incubated at 37 °C. Heat stress: BFR-L1 cells (lung cells) incubated to temperature at 42 °C for 2 h and recovered at 37 °C for 1 h. b 3′-UTR of PCBP2 and CREBL2 were paired to the miR-1246 binding region. c Renilla/Firefly luciferase activity of candidate target genes co-transfected with mimics (miR-1246) or NC (miR-1246 negative control). d mRNA expression levels of PCBP2 and CREBL2 after cells transfected with mimics or NC. e, f Protein expression levels of PCBP2 and CREBL2 after cells transfected with mimics or NC. GAPDH was used as an internal control for mRNA and protein. Error bars represent SD (n = 3)
PCBP2 and CREBL2 are target genes of miR-1246
To identify whether PCBP2 and CREBL2 are the target genes of miR-1246, TargetScan7.1 software was used to analyze the sequences of binding site between miR-1246 and target genes 3′-UTR mRNA. The results showed miR-1246 binding sites located in the PCBP2 3′-UTR contained six nucleotides complement bases, and in CREBL2 3′-UTR had seven nucleotide complement bases (Fig. 5b). So we cloned the candidate binding sites from pig into a luciferase expression vector downstream of the Renilla luciferase stop codon. Dual luciferase assay showed that miR-1246 mimics significantly suppressed luciferase activity of the 3′-UTR of PCBP2 and CREBL2 (Fig. 5c). To further confirm that PCBP2 and CREBL2 are the target genes of miR-1246, levels of PCBP2 and CREBL2 mRNA and protein were detected. qPCR and western blot analysis showed that PCBP2 and CREBL2 mRNA and protein levels significantly decreased after transfection with miR-1246 mimics in HEK293 cells (Fig. 5d–f). Collectively, these results indicate that miR-1246 directly target PCBP2 and CREBL2.
MiR-1246 inhibits heat-induced cell apoptosis
Considering that the imbalance of cell apoptosis is caused by dysfunction of mRNA expression, which is the essential mechanism of heat-stressed injury. Therefore, we thought miRNA might participate in an important regulatory mechanism. To understand the role of miR-1246 during heat resistance in BFR-L1 cell lines, cells were incubated with heat stress temperature and then detected cell apoptosis by flow cytometry. The results showed that apoptotic cells significantly decreased in the miR-1246 mimics-transfected group compared to NC-transfected group (Fig. 6a). DAPI and FITC staining revealed that cells transfected with miR-1246 mimics had less condensation and fragmentation of the nuclei compared to the other groups (Fig. 6b).
MiR-1246 inhibits BFR-L1 cell apoptosis. a Flow cytometry analysis of BFR-L1 cell apoptosis. Lower left quadrant represents intact cells (FITC−/PI−), lower right quadrant represents early apoptotic cells (FITC+/PI−), upper right quadrant represents late apoptotic cells (FITC−/PI+), upper left quadrant represents necrotic cells (FITC+/PI+). b Cell apoptosis after mimics or NC transfection in BFR-L1 cells by DAPI and FITC/PI staining. Apoptosis cells showed fragmentation of the nuclei. Empty represents cells exposed in 42 °C, without transfection mimics or NC
Discussion
Stress is broadly defined as the state when cells deviate from the status quo due to sudden environmental changes or frequent fluctuations in environmental factors (Leung and Sharp 2010). Heat stress affects both productivity and physiology of cattle (Kishore et al. 2014), and the ability to recover from duration and intensity effect of it is an important factor of cattle to adapt. Additionally, rectal temperature and respiration rates are commonly used to assess cattle heat tolerance. In the present study, the physiological background of Red Angus and Droughtmaster respiratory trait was affected by heat stress, but molecule mechanism and cell phenotypic remain unknown. Further, we found that miR-1246 is significantly upregulated in the two kinds of heat-stressed cattle by high-throughput profiling. Research showed that dairy breeds are typically more sensitive to heat stress due to higher metabolism rate and higher-producing, the miR-1246 in heat-stressed dairy breeds was siginificantly up-regulated (Zheng et al. 2014). Based on this, we suppose that miR-1246 may be a novel anti-heat stress factor in cattle.
To elucidate the role of miR-1246 in heat stress, we focused on target genes of miR-1246 and found it could inhibit the expression of PCBP2 and CREBL2, further regulated lung cell apoptosis. Heat stress induces different forms of cell death, during which Hippo pathway plays an important roles in apoptotic cell death (Cheung et al. 2003; Harvey et al. 2003). In our study, the functional relationship between miR-1246 and PCBP2 was established. PCBP2 associates with the Hippo signaling pathway in mouse lung tissues could control cell growth and development (Li et al. 2016). Furthermore, PCBP2 contributes to miRNA processing in cells by interacting with both Dicer and precursor miRNA to enhance processing into mature miRNA (Li et al. 2012). Likewise, cAMP signaling pathway has been suggested to regulate transcription, cell survival, and cell cycle progression, which are usually connected with MAPK pathway during stress response (Kronstad et al. 1998). In the meanwhile, attenuated activation of MAPK in lungs could attenuate polymorphonuclear neutrophil proinflammatory activity and decrease the severity of acute lung injury (Kuo et al. 2011). On the one hand, increased temperature has been associated with increased mortality in multiple epidemiology studies, particularly in respiratory mortality (Basu and Malig 2011). Feedlot cattle exposed to elevated temperatures will induce an increase in the dissipation of body excess heat. In order to reduce the excessive heat load, they would like to evaporate water from the respiratory tract via panting. On the other hand, increased body temperature might be expected to exacerbate VILI by augmenting inflammation, inactivating surfactant, or increasing cardiac output (Broccard et al. 1998). Increased cardiac output directly induces carbon dioxide production and oxygen consumption. In order to keep the balance of blood–pH buffering system, the body needs to enhance respiratory rate to maintain HCO3−(bicarbonate) to control CO2 ratio. Additionally, respiratory trait is highly related to lung function. For example, the resistance of the lung falls as frequency increases over the range associated with normal breathing (Irvin and Bates 2003). Our study presented the first comprehensive view that miR-1246 regulate lung cells by inhibiting PCBP2 and CREBL2, of which involved in Hippo and cAMP signaling pathways during heat stress. We believe these results could provide an important theoretical basis to better understand the biology mechanism of heat-stressed feedlot cattle.
Conclusion
Taken together, our results indicate that miR-1246 is upregulated in high temperature and miR-1246 could suppress the expression of target genes PCBP2 and CERBL2, further inhibit lung cell apoptosis.
References
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Armstrong DV (1994) Symposium—nutrition and heat-stress heat-stress interaction with shade and cooling. J Dairy Sci 77:2044–2050
Bartel DP (2007) MicroRNAs: genomics, biogenesis, mechanism, and function (Reprinted from Cell, vol 116, pg 281–297, 2004). Cell 131:11–29
Basu R, Malig B (2011) High ambient temperature and mortality in California: Exploring the roles of age, disease, and mortality displacement. Environ Res 111:1286–1292
Belhadj Slimen I, Najar T, Ghram A, Abdrrabba M (2016) Heat stress effects on livestock: molecular, cellular and metabolic aspects, a review. J Anim Physiol Anim Nutr 100:401–412
Broccard AF, Hotchkiss JR, Kuwayama N, Olson DA, Jamal S, Wangensteen DO, Marini JJ (1998) Consequences of vascular flow on lung injury induced by mechanical ventilation. Am J Resp Crit Care 157:1935–1942
Buzan JR, Oleson K, Huber M (2015) Implementation and comparison of a suite of heat stress metrics within the Community Land Model version 4.5. Geosci Model Dev 8:151–170
Chen X, Ba Y, Ma LJ, Cai X, Yin Y, Wang KH, Guo JG, Zhang YJ, Chen JN, Guo X, Li QB, Li XY, Wang WJ, Zhang Y, Wang J, Jiang XY, Xiang Y, Xu C, Zheng PP, Zhang JB, Li RQ, Zhang HJ, Shang XB, Gong T, Ning G, Wang J, Zen K, Zhang JF, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997–1006
Chen KL, Fu YY, Shi MY, Li HX (2017) Down-regulation of miR-181a can reduce heat stress damage in PBMCs of Holstein cows (vol 52, pg 864, 2016). In Vitro Cell Dev-Anim 53:381–381
Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD (2003) Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113:507–517
Choi AMK, Alam J (1996) Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am J Resp Cell Mol 15:9–19
Guan QM, Lu XY, Zeng HT, Zhang YY, Zhu JH (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74:840–851
Harvey KF, Pfleger CM, Hariharan IK (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114:457–467
Hunter MP, Ismail N, Zhang XL, Aguda BD, Lee EJ, Yu LB, Xiao T, Schafer J, Lee MLT, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB (2008) Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3:1–11
Irvin CG, Bates JH (2003) Measuring the lung function in the mouse: the challenge of size. Respir Res 4:1–9
Isik M, Blackwell TK, Berezikov E (2016) MicroRNA mir-34 provides robustness to environmental stress response via the DAF-16 network in C. elegans. Sci Rep-Uk 6:1–11
Kahle JJ, Neas LM, Devlin RB, Case MW, Schmitt MT, Madden MC, Diaz-Sanchez D (2015) Interaction effects of temperature and ozone on lung function and markers of systemic inflammation, coagulation, and fibrinolysis: a crossover study of healthy young volunteers. Environ Health Perspect 123:310–316
Kapila N, Sharma A, Kishore A, Sodhi M, Tripathi PK, Mohanty AK, Mukesh M (2018) Impact of heat stress on cellular and transcriptional adaptation of mammary epithelial cells in Riverine Buffalo (Bubalus bubalis) (vol 11, e0157237, 2017). PLoS One 13:1–28
Kishore A, Sodhi M, Kumari P, Mohanty AK, Sadana DK, Kapila N, Khate K, Shandilya U, Kataria RS, Mukesh M (2014) Peripheral blood mononuclear cells: a potential cellular system to understand differential heat shock response across native cattle (Bos indicus), exotic cattle (Bos taurus), and riverine buffaloes (Bubalus bubalis) of India. Cell Stress Chaperon 19:613–621
Kosaka N, Iguchi H, Ochiya T (2010) Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 101:2087–2092
Kronstad J, De Maria A, Funnell D, Laidlaw RD, Lee N, Ramesh MMDM (1998) Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol 170:395–404
Kuo MY, Liao MF, Chen FL, Li YC, Yang ML, Lin RH, Kuan YH (2011) Luteolin attenuates the pulmonary inflammatory response involves abilities of antioxidation and inhibition of MAPK and NF kappa B pathways in mice with endotoxin-induced acute lung injury. Food Chem Toxicol 49:2660–2666
Leung AKL, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40:205–215
Li YJ, Lin L, Li ZG, Ye XC, Xiong K, Aryal B, Xu ZH, Paroo Z, Liu QH, He CA, Jin P (2012) Iron homeostasis regulates the activity of the microRNA pathway through poly(C)-binding protein 2. Cell Metab 15:895–904
Li W, Wu YF, Xu RH, Lu H, Hu C, Qian H (2014) miR-1246 releases RTKN2-dependent resistance to UVB-induced apoptosis in HaCaT cells. Mol Cell Biochem 394:299–306
Li FM, Bullough KZ, Vashisht AA, Wohlschlegel JA, Philpott CC (2016) Poly(rC)-binding Protein 2 regulates Hippo signaling to control growth in breast epithelial cells. Mol Cell Biol 36:2121–2131
Mitlohner FM, Morrow JL, Dailey JW, Wilson SC, Galyean ML, Miller MF, McGlone JJ (2001) Shade and water misting effects on behavior, physiology, performance, and carcass traits of heat-stressed feedlot cattle. J Anim Sci 79:2327–2335
Nair N, Kumar S, Gongora E, Gupta S (2013) Circulating miRNA as novel markers for diastolic dysfunction. Mol Cell Biochem 376:33–40
Nienaber JA, Hahn GL (2007) Livestock production system management responses to thermal challenges. Int J Biometeorol 52:149–157
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9:1–9
Poisky L, von Keyserlingk MAG (2017) Invited review: effects of heat stress on dairy cattle welfare. J Dairy Sci 100:8645–8657. https://doi.org/10.3168/jds.2017-12651
Renaudeau D, Collin A, Yahav S, de Basilio V, Gourdine JL, Collier RJ (2012) Adaptation to hot climate and strategies to alleviate heat stress in livestock production. Animal 6:707–728
Sengar GS, Deb R, Singh U, Raja TV, Kant R, Sajjanar B, Alex R, Alyethodi RR, Kumar A, Kumar S, Singh R, Jakhesara SJ, Joshi CG (2018) Differential expression of microRNAs associated with thermal stress in Frieswal (Bos taurus x Bos indicus) crossbred dairy cattle. Cell Stress Chaperon 23:155–170
Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H (2013) Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Brit J Cancer 108:644–652
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q (2010) Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 31:659–666
Yu J, Liu FH, Yin P, Zhu XY, Cheng GL, Wang N, Lu A, Luan WL, Zhang NW, Li JF, Guo KJ, Yin YL, Wang HC, Xu JQ (2011) Integrating miRNA and mRNA expression profiles in response to heat stress-induced injury in rat small intestine. Funct Integr Genomic 11:203–213
Zhao X, Zmijewski JW, Lorne E, Liu G, Park YJ, Tsuruta Y, Abraham E (2008) Activation of AMPK attenuates neutrophil proinflammatory activity and decreases the severity of acute lung injury. Am J Physiol Lung Cell 295:L497–L504
Zheng Y, Chen KL, Zheng XM, Li HX, Wang GL (2014) Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperon 19:973–981
Funding
This work was financially supported by the Chongqing Municipal Science and Technology Commission project (no. cstc2017jcyjBX0013) The Research and Innovation Project of Chongqing Graduate Students ( no.CYB18074).
Author information
Authors and Affiliations
Contributions
Y.H. and M.C.C. designed and performed the main experiments, Y.H. interpreted the results and wrote the manuscript. M.C.C. performed animal experiment; they equally contributed to this work. L.W. performed the qRT-PCR. T.H.Z. performed the vector construction. Z.G.L. performed the GO biological process enrichment and KEGG pathway network analyses. G.W.Z. amended the manuscript. F.Y.Z. designed the experiments and supervised the research. All authors read the final manuscript.
Corresponding author
Ethics declarations
All animal procedures were approved by the Southwest University Institutional Animal Care and Use Committee.
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hu, Y., Cai, MC., Wang, L. et al. MiR-1246 is upregulated and regulates lung cell apoptosis during heat stress in feedlot cattle. Cell Stress and Chaperones 23, 1219–1228 (2018). https://doi.org/10.1007/s12192-018-0927-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-018-0927-9