Abstract
Human blood coagulation factor VIII (hFVIII) is used in hemostatic and prophylactic treatment of patients with hemophilia A. Biotechnological innovations have enabled purification of the culture medium of rodent or human cells harboring the hFVIII expression cassette. However, cell lines express hFVIII protein derived from an exogenous expression vector at a lower level than most other proteins. Here, we describe hFVIII production using piggyBac transposon and the human-derived expi293F cell line. Use of a drug selection protocol, rather than transient expression protocol, allowed cells harboring hFVIII expression cassettes to efficiently produce hFVIII. In heterogeneous drug-selected cells, the production level was maintained even after multiple passages. The specific activity of the produced hFVIII was comparable to that of the commercial product and hFVIII derived from baby hamster kidney cells. We also applied codon optimization to the hFVIII open reading frame sequences in the transgene, which increased production of full-length hFVIII, but decreased production of B-domain-deleted human FVIII (BDD-hFVIII). Low transcriptional abundance of the hF8 transgene was observed in cells harboring codon-optimized BDD-hFVIII expression cassettes, which might partially contribute to decreased hFVIII production. The mechanism underlying these distinct outcomes may offer clues to highly efficient hFVIII protein production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human blood coagulation factor VIII (hFVIII) is infused into patients with Hemophilia A to prevent spontaneous hemorrhage and excessive bleeding after trauma [1]. The major source of hFVIII agents is human blood in the old days, but biotechnological innovations have enabled their purification in large quantities from the culture medium of rodent or human cells harboring the hFVIII expression cassette [2]. However, the recombinant hFVIII protein derived from an exogenous expression vector is expressed in cell lines at a lower level than most other proteins, even when compared to proteins with similar molecular weights and patterns of post-translational modifications [3]. This poor expression is attributed to limitations in several steps, from transcription to secretion in cells [4, 5].
Recombinant hFVIII has been produced in both rodent (Baby Hamster Kidney (BHK) and Chinese hamster ovary (CHO) cells) and human (HEK293) cell lines [4, 6]. These two types of products share essentially the same pro-coagulant activity [7], but a rodent cell line has non-human post-translational modifications that are potentially immunogenic to humans [4, 8,9,10,11].
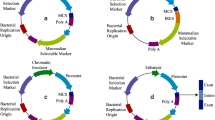
We sought for the leading edge of high expression system for FVIII production using a human cell line. In the current study, we present a novel expression system to efficiently produce recombinant B-domain-deleted hFVIII protein (BDD-hFVIII). To achieve high expression of BDD-hFVIII, which contains SQ linker amino acid sequences instead of the B domain [12], we applied the following two systems. The first is the piggyBac transposon system, which allows efficient integration of intact expression cassettes into the genome at multiple copy numbers [13, 14]. The system requires transposase to cut DNA sequences containing expression cassette(s) flanked by transposon sequences from the plasmid vector and paste them into the genome. The integrated DNA sequences comprised two exogenous expression cassettes: CAG promoter-driven FVIII-encoding and PGK promoter-driven puromycin-resistant protein-encoding cDNAs. The second is a system for human protein production, the expi293 expression system, which enables high-density suspension culture of an expi293F cell line with optimized materials and methods, thereby giving high expression yields [15]. We expected these systems to theoretically facilitate high production of BDD-hFVIII in the human cell line in a synergistic manner.
Materials and methods
Plasmid construction
The human FVIII expression vector was constructed as follows: pPB-CAG-EBNXN [16], which contains the constitutively active CAG promoter to express the gene of interest [17], was used as a backbone of the piggyBac transposon vector. To obtain pPB-CAG-EGFP-BSPX, the EGFP coding region was amplified by PCR from pCX-EGFP [18] using following primers and cloned into the BglII-XhoI site of the pPB-CAG-EBNXN: 5'-GGCAGATCTGCGATCGCCATGGTGAGCAAGGGCGAGGAGCTG-3' and 5'-CGGCTCGAGGTTTAAACTTACTTGTACAGCTCGTCCATGCC-3'. As a result, SgfI and PmeI sites were introduced at the upstream and downstream of the EGFP, respectively. To obtain pPB-CAG-EGFP-BSPX-PGK-puro, the PGK-puro-pA expression cassette derived from pPGKpuro (gift from Dr. K. Oishi) was cloned into the PstI-SalI site of the pPB-CAG-EGFP-BSPX. Plasmids containing human FVIII cDNA (pF1KE2421) were purchased from Kazusa DNA Research Institute (Chiba, Japan). The EGFP-coding region flanked by the SgfI and PmeI sites in the pPB-CAG-EGFP-BSPX-PGK-puro was replaced with human FVIII cDNAs. Codon-optimized sequences for full-length hFVIII were selected using GeneArt GeneOptimizer software (Thermo Fisher).
For codon-optimized sequences that encode amino acid sequences corresponding to full-length hFVIII and HSQ FVIII, a vector backbone was obtained by digestion of pPB-full-length hF8 with SgfI and PmeI, and recombined with the relevant DNA fragments (GeneArt Strings DNA Fragments, Thermo Fisher) using an in-fusion HD cloning kit (Takara). The DNA sequences derived from polymerase chain reaction (PCR)-amplified or synthetic fragments in the constructed plasmids were sequenced using the BigDye Terminator v1.1 cycle sequencing kit and Applied Biosystems 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA). Plasmids for transfection were purified using a PureLink HiPure Plasmid Midi prep kit (K210005; Thermo Fisher).
Cell culture
expi293F cells (Thermo Fisher) were cultured in Erlenmeyer flasks with 30 ml of expi293 Expression Medium (Thermo Fisher), which was horizontally shaken by an orbital shaker (Waken B Tech) at 130 rpm inside an incubator maintained at 8% carbon dioxide (CO2) and 37 °C. Transfection was performed according to the manufacturer’s protocol (expi293 Expression System; Thermo Fisher). For pPB-BDDhF8, pPB-BDDhF8 (optimized), pPB-BDDhF8 (optimized)-woodchuck hepatitis post-transcriptional regulatory element (WPRE), pPB-full-length hF8, and pPB-full-length hF8 (optimized), these individual protein expression plasmids and pCMV-hyPBase for transposase expression [14] were transfected with a weight ratio of 3:1. The cells were passaged every 3–4 days according to the experimental schedule.
FVIII protein purification
A Poly Prep chromatography column (731-1550, Bio-Rad) was filled with 1 ml volume of affinity resin (VIII select, GE Healthcare). The column was equilibrated with 10 ml of wash buffer 1 (20 mM histidine, 20 mM calcium chloride, 300 mM sodium chloride, 0.02% Tween 80, pH 6.5). Then, 0.45 µm-filtered cell culture supernatant was loaded onto the column. The column was washed sequentially with 5 ml of wash buffer 1 and 5 ml of wash buffer 2 (20 mM histidine, 20 mM calcium chloride, 1 M sodium chloride, 0.02% Tween 80, pH 6.5). The FVIII protein binding to the affinity resin was eluted with 7 ml of elution buffer (20 mM histidine, 20 mM calcium chloride, 1.5 M sodium chloride, 0.02% Tween 80, 50% ethylene glycol, pH 6.5). Using centrifugal filter column (Amicon ultra-4 3k, Millipore), the solvent of the eluate was replaced by 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) buffer 1 (20 mM HEPES, 0.1 M NaCl, 0.01% Tween 20, 0.01% NaN3) and simultaneously FVIII protein was concentrated.
For size-exclusion chromatography using AKTA purifier (Cytiva, Tokyo, Japan), the affinity-purified FVIII protein solution was loaded onto a size-exclusion column (Superdex 200 Increase 10/300 GL, GE Healthcare) and run with HEPES buffer 2 (20 mM HEPES, 1 M NaCl, 0.01% Tween 20, 0.01% NaN3) at a flow rate of 0.5 ml/min. The absorbance at 280 nm was used to detect the proteins. Each fraction separated every 0.5 ml was subjected to one-stage clotting assays, and the fractions containing concentrated hFVIII were determined and mixed. Using a centrifugal filter column (Amicon ultra-4 3k, Millipore), the solvent of the purified solution was replaced with HEPES buffer 1 and the hFVIII protein was simultaneously concentrated.
Measurement of FVIII activity
FVIII coagulation activity (FVIII:C) was measured in one-stage clotting assays using commercial FVIII-deficient plasma (Sysmex, Kobe, Japan) and a STart4 Hemostasis Analyzer (Diagnostica Stago, Asnieres, France). Normal human plasma (Coagtrol N, Sysmex) was used as the standard for units per ml (U/ml) calculation. The samples were diluted with Owren’s verbal buffer (Sysmex) at 1/10 or 1/100 concentrations and subjected to the assays.
Enzyme-linked immunosorbent assay (ELISA) for FIII protein quantity
FVIII protein levels were measured using a sandwich ELISA [19]. Each well of ELISA plate (F96 Polysorp Nunc-Immuno plate, 475094, Thermo Fisher) was coated with 1:5000-diluted monoclonal anti-heavy chain antibody (VIII-2236 [20]) in carbonate–bicarbonate buffer (0.017 M Na2CO3, 0.034 M NaHCO3, pH 9.6) overnight at 4 °C. The wells were washed three times with wash buffer (phosphate-buffered saline, 0.05% Tween 20). Each well was treated with blocking solution (10% nonfat dry milk dissolved in wash buffer) for 2 h at 37 °C. The wells were washed three times with wash buffer. (10% human serum albumin, 0.5% Tween 20, and 17 mM TBS buffer, pH 7.6), and coated in. After removal and further washing of the plate, 0.05 ml the samples or standards were added and incubated overnight at 4 °C. The samples were composed of serially diluted, purified FVIII protein solutions. The standards comprised purified full-length hFVIII (Kogenate FS, Bayer) of known concentrations varying from 0.16 to 2.5 nM. After washing, 1 g biotinylated R8B12 (20 g/ml) was added to each well. Each well was incubated with 1:400-diluted peroxidase-conjugated monoclonal anti-light chain antibody (VIII-3776 [21]) in the blocking solution for 1.5 h at 37 °C. The chromogenic reaction was initiated by the addition of a peroxidase substrate (KPL TMB Microwell Peroxidase Substrate System, 5120-0053, Seracare), incubated for 5 min at 20 °C, and stopped by the addition of 2 N H2SO4. Colorimetric measurements were recorded at 450 nm using a microplate reader (Maltiscan JX, Thermo Fisher).
FXa generation assay
All reactions were performed in HBS buffer (20 mM HEPES, pH 7.2, 0.1 M NaCl, 0.01% Tween 20, 0.01% NaN3) at 22 °C. Purified FVIII was incubated with PL in HBS buffer for 30 s before thrombin was added and incubated for 30 s. Thrombin activity was stopped by incubation with hirudin for 30 s. After the reaction mixture was incubated with FIXa for 30 s, FXa generation was initiated by adding FX and performed for 60 s. The final concentration of each substance in the reaction was 1 nM purified FVIII, 20 µM PL, 30 nM thrombin, 10 U/ml hirudin, 40 nM FIXa, and 300 nM FX. EDTA (final concentration, 0.1 M) was added to stop the reaction. One-sixth of the generated FXa reacted with the chromogenic substrate S-2222 (final concentration: 0.46 mM), and the absorbance at 405 nm was measured every 15 s. The FXa generated was calculated as the initial rate of absorbance and quantified according to the standard curve.
Quantitative reverse transcription PCR (qRT-PCR)
DNase-treated RNA was purified from cultured cells using NucleoSpin RNA (Takara, Japan), and 1 μg of the RNA was subjected to cDNA synthesis in a 20 μl total reaction volume using PrimeScript RT Master Mix (Perfect Real Time) (Takara) according to the manufacturer’s instructions. The cDNAs were subjected to qPCR using TB Green Premix Ex TaqII (Tli RNase H Plus) (Takara) on a Step One Plus (Thermo Fisher) according to the manufacturer’s instructions. The reaction solution (total reaction volume of 20 μl) comprised 10 μl of TB Green Premix ExTaq II (2×), 0.8 μl each of forward and reverse primers (400 nM each), 0.4 μl of ROX reference dye (50×), 6 μl of nuclease-free water, and 2 μl of cDNA solution diluted at 1:20. The thermal cycles were as follows: 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C, and 30 s at 60 °C. Product specificity was evaluated using melting curves. Relative quantity was calculated according to the standard curve for each primer pair. Each sample was analyzed in triplicate. Quantitative values obtained from three independent biological samples were analyzed. Each quantitative value corresponding to the 5′- and 3′-untranslated regions (UTRs) of the F8 transgene was normalized to that corresponding to the 18S rRNA or human Actb gene and evaluated. Statistical analyses were performed using an unpaired t test. The primer pairs used were as follows: 5′-TGGGCAACGTGCTGGTTATTG-3′ and 5′-TGCATGGCGATCGCAGATCTG-3′ for the 5′-UTR region of the F8 transgene; 5′-TTGCCAGCCATCTGTTGTTTG-3′ and 5′-AGAATGACACCTACTCAGACAATG-3′ for the 3′-UTR region of the F8 transgene; 5′-GTCTGTGATGCCCTTAGATG-3′ and 5′-AGCTTATGACCCGCACTTAC-3′ for 18S rRNA; and 5′-CTCTTCCAGCCTTCCTTCCT-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′ for human Actb.
Data analysis
Excel (Microsoft) or GraphPad Prism 9 software (GraphPad Software, San Diego, CA, USA) was used for all data analysis.
Results
Transient expression protocol yields moderate level of BDD-hFVIII
As the expi293 expression system is designed for the transient expression of exogenous proteins at high levels, we examined whether the standard protocol provided by the manufacturer yields BDD-hFVIII efficiently. The cells were transfected with pPB-BDDhF8 (Fig. 1a) and pCMV-hyPBase plasmids (day 0) and exposed to the enhancer solution provided by the manufacturer the following day (Fig. 1b). The count of viable cells continued to grow without the enhancer, whereas it stopped growing around day 3 and thereafter decreased in its presence (Fig. 1c). During this time course, exposure to the enhancer negatively affected cell viability (Fig. 1d). FVIII:C during the periods were in the range of 1.0–3.5 U/ml/24 h similarly in the both groups (Fig. 1e), whereas the exposure of the enhancer elevated the efficiency of FVIII production from day 4–5 to reach into the range of 0.7–1.0 U/106 cells/24 h, compared to the range of 0.1–0.2 U/106 cells/24 h without the enhancer (Fig. 1f). Overall, these results showed that the protocol for transient expression moderately improved BDD-hFVIII production from the human cell line compared to a previous report [22], but its cellular condition appeared to be inappropriate.
Transient expression protocol provided by the manufacturer yields moderate levels of B-domain-deleted-human blood coagulation factor VIII (BDD-hFVIII) protein in expi293 expression system. A schematic representation of the piggyBac transposon vector construct with the BDD-hFVIII expression cassette is shown (a). The timeline of the applied transient expression protocol is shown at each time point of treatment and sample collection (b). Each graph shows the growth curve of live cells (cells/ml) (c), cell viability curve (%) (d), FVIII:C (U/ml/24 h) profiles in the medium at each time point (e), and efficiency profiles of FVIII production (U/106/24 h) (f) in the culture of expi293F cells harboring the BDD-hFVIII expression cassette. Each value is the average of biologically triplicated samples, represented with error bars indicating the standard error. The statistical significance of the differences between the groups was calculated using the unpaired t test. *p < 0.05 and **p < 0.01
Drug-resistant heterologous cells yield highly and effectively BDD-hFVIII
As the transient expression protocol may result in ineffective production of BDD-hFVIII, we examined the effect of puromycin selection to eliminate cells without valid exogenous expression cassettes (Fig. 2a–d). Two days after transfection (day −2), puromycin was added to the culture medium (day 0), and the cells were passaged every three days (Fig. 2a). Although the viability was below 93% on day 3 in three groups of puromycin concentration, it remained above 93% thereafter (Fig. 2b). The number of live cells was in the range of 0.5–1.2 × 106 cells/ml on days 3 and 6, whereas it shifted to the range of 1.2–2.4 × 106 cells/ml at and after day 9, indicating that most of the cells without valid exogenous expression cassettes were eliminated (Fig. 2c). In terms of the efficiency of FVIII production during the time course, the experimental groups without puromycin showed stable production of at least ~ 2 U/106 cells/24 h until day 9–10, whereas those treated with puromycin showed 0.2–0.4 U/106 cells/24 h at day 3–4, which increased gradually thereafter and reached to the range of 4.5–9.0 U/106 cells/24 h on average (Fig. 2d). The change in the value in the group treated with 0.5 μg/ml puromycin was slower than that in the other groups with higher concentrations. Therefore, nine-day selection with 1.0–1.5 μg/ml puromycin was the shortest time to achieve maximum BDD-hFVIII production in the selected heterogeneous cells.
Enhanced production of BDD-hFVIII protein by drug selection and its persistence in the high passage cells. The timeline of the applied expression protocol followed by puromycin selection is shown at each time point of puromycin addition and sample collection (a). The cells were passaged every three days at a cell density of 2 × 105/ml. Each graph shows the growth curve of live cells (cells/ml) (b), cell viability curve (%) (c), and efficiency profiles of FVIII production (U/106/24 h) (d) by the culture of expi293F cells harboring the BDD-hFVIII expression cassette. Each value represents the average of the biologically duplicated samples from these value plots
High-density culture improves the production of BDD-hFVIII
As shown in Fig. 2, low density culture enables high efficiency of BDD-hFVIII production, but is not suitable to obtain high concentration of the protein (max 2 U/ml/24 h). Next, we examined whether high-density cultures enabled us to obtain higher concentrations of BDD-hFVIII (Fig. 3a–e and Fig. S1a). On day 9 of selection, cells at a density of 106 cells/ml (day 0) were cultured with puromycin, and all the increased cells were passaged daily with fresh culture medium (Fig. 3a). The cells grew with healthy viability even when exposed to high density (Fig. 3a, c). During growth, FVIII:C reached 17–27 U/ml/24 h on average until day 4 and dropped to 8 U/ml/24 h on average on day 5 (Fig. 3d). In contrast, the efficiency of FVIII production was gradually decreased from 17 to 0.6 U/106 cells/24 h during the time course (Fig. 3e). These results demonstrated that the high-density culture in our protocol yielded a relatively high concentration of BDD-hFVIII but diminished the efficiency of FVIII production as the cell density increased.
Codon optimization negatively affects BDD-hFVIII production from drug-resistant cells. The timeline of the expression protocol using puromycin-resistant cells is shown at each time point of treatment and sample collection (a). Each graph shows the growth curve of live cells (cells/ml) (b), cell viability curve (%) (c), FVIII:C (U/ml/24 h) profiles in the medium at each time point (d), and efficiency profiles of FVIII production (U/106/24 h) (e) in the culture of expi293F cells harboring each BDD-hFVIII expression cassette. Specific activity of each culture medium at day 4 relevant to expi293F cells harboring pPB-BDDhF8, pPB-BDDhF8 (optimized), or pPB-BDDhF8 (optimized)-woodchuck hepatitis post-transcriptional regulatory element (WPRE) was determined by one-stage coagulation assay and quantification by sandwich enzyme-linked immunosorbent assay (ELISA) (f). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of F8 transgene-derived mRNAs was performed on days 10 (g) and 0 (h) of puromycin selection. Each value is the average of biologically triplicated samples represented by error bars indicating standard error (b–e, g, h) and standard deviation (f). The statistical significance of the differences between groups was calculated using the unpaired t test. *p < 0.05 and **p < 0.01
Codon optimization negatively affects BDD-hFVIII production from drug-resistant cells
Codon optimization of human F8 cDNA effectively facilitates translation and protein folding [23]. We examined whether codon optimization of cDNA encoding BDD-hFVIII resulted in higher protein production than that of the naive cDNA sequences (Fig. 3a–e, and Fig. S1b, c). As the cells grew, the count of live cells and the viability in the groups harboring optimized cDNA showed a profile similar to that of the non-optimized group (Fig. 3b, c). However, FVIII:C (Fig. 3d), FVIII:Ag (Fig. S2a), and efficiency of FVIII production (Fig. 3e) showed lower values at individual time points in the optimized cDNA groups than in the non-optimized cDNA groups. We also examined the effect of WPRE [24, 25] on codon-optimized mRNA encoding BDD-hFVIII. The insertion of WPRE into the expression cassettes (Fig. S3) improved both FVIII:C (Fig. 3d) and FVIII production efficiency (Fig. 3e) at individual time points compared to the values in the non-WPRE groups, but did not come close to those in the non-optimized non-WPRE groups. Next, we examined whether the lower FVIII production in the codon-optimized groups was due to decreased specific activity of the secreted FVIII protein. The values from the codon-optimized groups showed similar to or slightly higher specific activity (U/mg) than that of the non-optimized groups (Fig. 3f), indicating that the FVIII protein activity was not compromised, but rather improved, by codon optimization, which is consistent with a previous report demonstrating that the biochemical properties of codon-optimized BDD-hFVIII were similar to or slightly higher than that of non-optimized protein [26]. To confirm the genomic copy number of FVIII expression cassette in each group of the cells, we performed genomic qPCR, and demonstrated that the copy number of FVIII expression cassette do not significantly differ between the three groups (Fig. S2b). To clarify whether the decreased production of FVIII results from low transcript abundance, we performed qRT-PCR for transcript quantities of F8 transgene-derived mRNAs and compared the codon-optimized groups with the non-optimized ones (Fig. 3g, h). The target amplicons were set in the 5′ and 3′ UTRs of the F8 transgene shared within different vectors, and the human Actb gene and 18S rRNA were used as control amplicons for normalization. On day 10 of puromycin selection, the relative abundance of codon-optimized F8 transcripts tended to be lower than that of non-optimized transcripts, except for that of the 5′-UTR normalized to Actb (Fig. 3g). To exclude the possibility that puromycin selection caused a skewed heterologous cell population, cells at day 0 of puromycin selection were examined for non-biased transcript abundance. As was the case on day 10 of puromycin selection, the relative abundance of codon-optimized F8 transcripts tended to be lower than that of non-optimized transcripts, except for that of the 5′UTR normalized to Actb (Fig. 3h). Therefore, the low production of BDD-hFVIII by codon optimization may be, in part, caused by the decreased mRNA level.
The expression system produces BDD-hFVIII with normal specific activity
After purification of BDD-hFVIII from the culture medium through steps that comprise affinity and size-exclusion chromatography, we examined the specific activity of BDD-hFVIII produced from this expression system using FXa generation assay, and compared it with both therapeutic and BHK cells-derived BDD-hFVIII proteins. This expression system-derived BDD-hFVIII had similar or slightly higher specific activity than that of either the therapeutic or BHK cells-derived BDD-hFVIII proteins (Fig. 4). Thus, the results demonstrated that this expression system could produce macroscopically intact BDD-hFVIII proteins, despite their high expression.
FXa generation assay for 1 nM of purified FVIII. Specific activity of purified BDD-hFVIII obtained from our protocol was similar to that of therapeutic full-length hFVIII and BDD-hFVIII derived from the Baby Hamster Kidney (BHK) cell line. Quantification of each purified hFVIII protein was performed using sandwich ELISA. Purified BDD-hFVIII produced from the expi293F cell line was biologically triplicated to perform the FXa generation assay (rep1–3). Each assay was performed in triplicates for each purified protein sample. Error bars represent the standard deviation. The statistical significance of the differences between the groups was calculated using the unpaired t test. *p < 0.05 and **p < 0.01
Codon optimization facilitates full-length hFVIII production from drug-resistant cells
Next, we examined whether codon optimization improved the production of full-length hFVIII in our expression system (Figs. 3e, 5a). As was the case with experiments for the BDDhF8 expression vector, the cells selected with puromycin for nine days were applied (Fig. 5a). Along the course of cell growth, the live cell count and viability showed similar profiles among the three groups for full-length hFVIII expression (Fig. 5b, c), even when compared to those for BDD-hFVIII expression (Fig. 3b, c). However, in the case of full-length hFVIII expression, codon optimization increased maximally two-fold on average in FVIII:C (Fig. 5d), FVIII:Ag (Fig. S2c), and efficiency of FVIII production (Fig. 5e) at individual time points, which was in contrast to the case of BDD-hFVIII expression (Fig. 3d, e). We also examined whether the increased production of FVIII resulted from improved transcript abundance, using qRT-PCR. The results revealed that transcriptional abundance in the codon-optimized group was equal to or slightly lower than that in the intact group (Fig. 5f). Therefore, the improved production of full-length hFVIII may be attributed to alterations at the translational or post-translational levels.
Codon optimization facilitates full-length hFVIII production from drug-resistant cells. The timeline of the expression protocol using puromycin-resistant cells is shown at each time point of treatment and sample collection (a). Each graph shows the growth curve of live cells (cells/ml) (b), cell viability curve (%) (c), FVIII:C (U/ml/24 h) profiles in the medium at each time point (d), and efficiency profiles of FVIII production (U/106/24 h) (e) in the culture of expi293F cells harboring each full-length hFVIII expression cassette. qRT-PCR analysis of F8 transgene-derived mRNA was performed on day 10 of puromycin selection (f). Each value is the average of biologically triplicated samples, represented with error bars indicating the standard error. The statistical significance of the differences between groups was calculated using the unpaired t test. *p < 0.05 and **p < 0.01
Discussion
In the present study, we achieved a high-level range of BDD-hFVIII production using our expression system, which is comparable to the level achieved by methotrexate-driven gene amplification [27] or lentiviral vector platform [28]. Our novel expression system has the following advantages. First, the piggyBac transposon system simply requires plasmid construction, followed by cationic lipid-mediated DNA delivery into target cells, whereas lentiviral vector systems are more complicated and require several steps with specific care until transfection. Furthermore, during transient expression or drug selection, the culture of expi293 cells integrated by piggyBac transposon resulted in higher production of FVIII than that by random integration (Figs. S4 and S5). Second, cell cloning is not necessarily required to obtain purified BDD-hFVIII at the laboratory level because of the persistently high production of hFVIII over a long period. Third, the expi293F cells we used are a human-derived cell line that can produce BDD-hFVIII with human-type post-translational modification [15]. Several rodent cell-derived recombinant hFVIII therapeutics contain a small amount of unsulfured hFVIIIs at Tyr1680 [29] which leads to an inability to bind to the von Willebrand factor (vWF), and their infusions to hemophilia A patients result in faster clearance from the blood [30] and are at risk of FVIII inhibitor development [31,32,33].
A previous report described that human cell-derived recombinant hFVIII has a similar specific FVIII activity to several rodent cell-derived recombinant hFVIII [7]. Consistent with this report, we also revealed a similar specific activity of the BDD-hFVIII proteins produced by our expression system to that of BHK-derived BDD-hFVIII (Fig. 4). For detailed coagulation characteristics, chromogenic assay experiments, inactivation by activated protein C, and binding potential with vWF are required.
Codon optimization is a commonly used method to boost protein production by efficient translation. In the present study, codon optimization of BDD-hFVIII did not lead to a higher yield of protein production compared with its wild-type sequence counterpart, but negatively affected production (Fig. 3d, e). This negative effect is probably not due to misdirected protein folding and/or aberrant post-translational modification, which can influence the specific activity of FVIII. This notion is supported by the experiments for specific activity of FVIII in the culture medium, showing that the specific activity of BDD-hFVIII produced from the optimized sequence was the same as or slightly higher than that of the non-optimized sequence (Fig. 3f). This observation is consistent with a previous report demonstrating that the biochemical properties, including specific activity, of BDD-hFVIII produced from codon-optimized sequences were similar to those of the protein from the wild-type sequence [26].
We focused on the abundance of F8 transgene-derived mRNA to clarify whether a distinct F8 sequence contributed to FVIII production by codon optimization. We used two different housekeeping genes, 18S rRNA and Actb, to normalize the relative values obtained by qRT-PCR. 18S rRNA has been reported to be a more stable housekeeping gene than Actb [34]. According to the data normalized to 18S rRNA, codon-optimized F8 transcripts were less abundant than their non-optimized counterparts (Fig. 3g). Essentially, the same result was obtained in non-selected heterogeneous cells on day 0 of puromycin selection (Fig. 3h). Nevertheless, in terms of the efficiency of FVIII production, the non-selected heterogeneous cells at day 2 after transfection did not show a significant difference between codon-optimized and non-optimized groups (Fig. S6e, f). This might imply that lower transcriptional abundance does not largely contribute to FVIII production as a final outcome through complex cellular mechanisms. Therefore, we could not rule out the possibility that the observed negative impact of codon optimization on FVIII production results not from low transcript abundance but from heterogeneous cells containing those with low expression potential, which predominated during puromycin exposure. Distinct synonymous replacement by codon optimization may have a negative effect. In general, codon optimization does not necessarily enhance protein production. Different optimization algorithms result in different outputs [35]. The observed negative impact on FVIII production might result from repressive nucleic sequences that affect transcription, stability of the transcript, and/or its transport to the cytoplasm.
In contrast to codon-optimized BDD-hFVIII expression, the culture of expi293F cells harboring codon-optimized full-length hF8 expression cassette yielded higher efficiency in full-length hFVIII production compared with the non-optimized counterpart (Fig. 5). The transcript abundance of expi293F cells harboring the codon-optimized full-length hF8 expression cassette was similar to or slightly lower than that of the non-optimized counterpart. This might imply that an improvement in translational efficiency contributed to a higher production of full-length hFVIII protein encoded by the codon-optimized sequence than that of the non-optimized counterpart.
A more detailed characterization of these distinct outcomes may provide insight into how to overcome the low production of the hFVIII protein.
References
Manco-Johnson MJ, Abshire TC, Shapiro AD, Riske B, Hacker MR, Kilcoyne R, et al. Prophylaxis versus episodic treatment to prevent joint disease in boys with severe hemophilia. N Engl J Med. 2007;357(6):535–44.
Kingdon HS, Lundblad RL. An adventure in biotechnology: the development of haemophilia A therapeutics—from whole-blood transfusion to recombinant DNA to gene therapy. Biotechnol Appl Biochem. 2002;35(2):141–8.
Cao W, Dong B, Horling F, Firrman JA, Lengler J, Klugmann M, et al. Minimal essential human factor VIII alterations enhance secretion and gene therapy efficiency. Mol Ther Methods Clin Dev. 2020;19:486–95.
Kumar SR. Industrial production of clotting factors: challenges of expression, and choice of host cells. Biotechnol J. 2015;10(7):995–1004.
Soukharev S, Hammond D, Ananyeva NM, Anderson JA, Hauser CA, Pipe S, et al. Expression of factor VIII in recombinant and transgenic systems. Blood Cells Mol Dis. 2002;28(2):234–48.
Swiech K, Picanco-Castro V, Covas DT. Production of recombinant coagulation factors: are humans the best host cells? Bioengineered. 2017;8(5):462–70.
Sandberg H, Kannicht C, Stenlund P, Dadaian M, Oswaldsson U, Cordula C, et al. Functional characteristics of the novel, human-derived recombinant FVIII protein product, human-cl rhFVIII. Thromb Res. 2012;130(5):808–17.
Butler M, Spearman M. The choice of mammalian cell host and possibilities for glycosylation engineering. Curr Opin Biotechnol. 2014;30:107–12.
Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol. 2010;28(8):863–7.
Durocher Y, Butler M. Expression systems for therapeutic glycoprotein production. Curr Opin Biotechnol. 2009;20(6):700–7.
Ghaderi D, Zhang M, Hurtado-Ziola N, Varki A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol Genet Eng Rev. 2012;28:147–75.
Sandberg H, Almstedt A, Brandt J, Gray E, Holmquist L, Oswaldsson U, et al. Structural and functional characteristics of the B-domain-deleted recombinant factor VIII protein, r-VIII SQ. Thromb Haemost. 2001;85(1):93–100.
Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–83.
Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A. 2011;108(4):1531–6.
Jain NK, Barkowski-Clark S, Altman R, Johnson K, Sun F, Zmuda J, et al. A high density CHO-S transient transfection system: comparison of ExpiCHO and Expi293. Protein Expr Purif. 2017;134:38–46.
Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6(5):363–9.
Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–9.
Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407(3):313–9.
Jenkins PV, Freas J, Schmidt KM, Zhou Q, Fay PJ. Mutations associated with hemophilia A in the 558–565 loop of the factor VIIIa A2 subunit alter the catalytic activity of the factor Xase complex. Blood. 2002;100(2):501–8.
Muto A, Yoshihashi K, Takeda M, Kitazawa T, Soeda T, Igawa T, et al. Anti-factor IXa/X bispecific antibody (ACE910): hemostatic potency against ongoing bleeds in a hemophilia A model and the possibility of routine supplementation. J Thromb Haemost. 2014;12(2):206–13.
Sugita C, Yamashita A, Moriguchi-Goto S, Furukoji E, Takahashi M, Harada A, et al. Factor VIII contributes to platelet-fibrin thrombus formation via thrombin generation under low shear conditions. Thromb Res. 2009;124(5):601–7.
Swiech K, Kamen A, Ansorge S, Durocher Y, Picanco-Castro V, Russo-Carbolante EM, et al. Transient transfection of serum-free suspension HEK 293 cell culture for efficient production of human rFVIII. BMC Biotechnol. 2011;11:114.
Ward NJ, Buckley SM, Waddington SN, Vandendriessche T, Chuah MK, Nathwani AC, et al. Codon optimization of human factor VIII cDNAs leads to high-level expression. Blood. 2011;117(3):798–807.
Donello JE, Loeb JE, Hope TJ. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol. 1998;72(6):5085–92.
Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73(4):2886–92.
Shestopal SA, Hao JJ, Karnaukhova E, Liang Y, Ovanesov MV, Lin M, et al. Expression and characterization of a codon-optimized blood coagulation factor VIII. J Thromb Haemost. 2017;15(4):709–20.
Orlova NA, Kovnir SV, Gabibov AG, Vorobiev II. Stable high-level expression of factor VIII in Chinese hamster ovary cells in improved elongation factor-1 alpha-based system. BMC Biotechnol. 2017;17(1):33.
Spencer HT, Denning G, Gautney RE, Dropulic B, Roy AJ, Baranyi L, et al. Lentiviral vector platform for production of bioengineered recombinant coagulation factor VIII. Mol Ther. 2011;19(2):302–9.
Grancha S, Navajas R, Maranon C, Paradela A, Albar JP, Jorquera JI. Incomplete tyrosine 1680 sulphation in recombinant FVIII concentrates. Haemophilia. 2011;17(4):709–10.
Chun H, Pettersson JR, Shestopal SA, Wu WW, Marakasova ES, Olivares P, et al. Characterization of protein unable to bind von Willebrand factor in recombinant factor VIII products. J Thromb Haemost. 2021;19(4):954–66.
Gangadharan B, Ing M, Delignat S, Peyron I, Teyssandier M, Kaveri SV, et al. The C1 and C2 domains of blood coagulation factor VIII mediate its endocytosis by dendritic cells. Haematologica. 2017;102(2):271–81.
Hartholt RB, van Velzen AS, Peyron I, Ten Brinke A, Fijnvandraat K, Voorberg J. To serve and protect: The modulatory role of von Willebrand factor on factor VIII immunogenicity. Blood Rev. 2017;31(5):339–47.
Muczynski V, Casari C, Moreau F, Ayme G, Kawecki C, Legendre P, et al. A factor VIII-nanobody fusion protein forming an ultrastable complex with VWF: effect on clearance and antibody formation. Blood. 2018;132(11):1193–7.
Kuchipudi SV, Tellabati M, Nelli RK, White GA, Perez BB, Sebastian S, et al. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol J. 2012;9:230.
Kimchi-Sarfaty C, Schiller T, Hamasaki-Katagiri N, Khan MA, Yanover C, Sauna ZE. Building better drugs: developing and regulating engineered therapeutic proteins. Trends Pharmacol Sci. 2013;34(10):534–48.
Acknowledgements
This work was partly supported by a Grant-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to Takuji Yoshimura (grant number 17K08632), Keiji Nogami (grant numbers 18K07885 and 21K07804), and Investigator-Research Support grant (Sanofi AS).
Funding
This study were funded by Japan Society for the Promotion of Science (Grant Nos. 17K08632, 18K07885, 21K07804), Sanofi AS (Investigator-Research Support grant).
Author information
Authors and Affiliations
Contributions
TY conceived and designed the research, performed experiments, analyzed and interpreted the data, prepared figures, and wrote and edited the manuscript; KaH performed experiments; NS helped in vector construction; KO supplied resource; KyH supplied resource; MS supervised the research; KN supervised the research and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
TY has received research funding from Bioverativ Inc./Sanofi S.A.; KaH, KO, and KyH have no conflict of interest; NS teaches a course endowed by CSL Behring; MS and KN have received research funding from Bioverativ Inc./Sanofi S.A.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Yoshimura, T., Horiuchi, K., Shimonishi, N. et al. Modified expi293 cell culture system using piggyBac transposon enables efficient production of human FVIII. Int J Hematol 117, 56–67 (2023). https://doi.org/10.1007/s12185-022-03468-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-022-03468-9