Abstract
In this study, a simple and rapid high-throughput method for the detection of horse meat in processed food products is described. Specific loop-mediated isothermal amplification (LAMP) primers were designed to target the mitochondrial genome of horse (Equus caballus). No cross-reactions were observed for beef, pork, and chicken. Sensitivity tests showed reliable detection of 0.1 ng of extracted horse DNA. Spiking experiments were performed to show that the assay is capable of detecting 0.1 % horse meat in prepared model sausages, independent from their cooking time. Additionally, five different commercial horse meat products were analyzed to ensure the robustness of the assay when applied to varying food matrices. All experiments were performed on a heating block followed by visual detection using an intercalating dye. Results were confirmed by real-time fluorescence monitoring using a thermal cycler and compared to a previously published real-time PCR assay. In conclusion, this method is a good candidate for the simple and efficient testing of horse meat in food-products in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2012 and 2013, the European Commission performed official controls in several European Union member states, revealing the addition of horse meat in pre-packaged food products labelled as 100 % beef. Followed by these discoveries, the meat adulteration scandal was sparked in January 2013, especially in the UK, Ireland, and Germany, where most adulterated products were found (European Commission 2013). Although horse meat is considered a delicacy in some countries, the European Commission raised concerns about these findings. Since labelling requirements were violated, the fraudulent nature of this matter can be assumed. Apart from that, the consumption of non-declared horse meat might also pose a health risk to consumers. Phenylbutazone is a widely used veterinary drug whose use is only allowed on non-food-producing animals. The use of phenylbutazone has to be excluded clearly for any meat species going into the food chain (European Commission 2010). Although several protein-based (enzyme-linked immunosorbent assay, ELISA) and DNA-based (polymerase chain reaction, PCR) test systems for meat species identification exist, these methods are time consuming and require costly equipment, e.g., thermal cyclers, and trained personnel to perform these reactions (Walker et al. 2013). Rapid high-throughput methods are needed to facilitate controls to ensure proper labelling in the future and in compliance with the law.
In 2000, an alternative DNA amplification method termed loop-mediated isothermal amplification (LAMP) was developed (Notomi et al. 2000). Three primer sets bind to six distinct regions of the target sequence. Through the use of a Bst polymerase with strand displacement activity (Aliotta et al. 1996) a dumbbell-like structure is formed which then undergoes cyclic amplification producing various-sized DNA strands with alternately inverted repeats of the target sequence (Notomi et al. 2000; Nagamine et al. 2002). Since a denaturation step is not required, the assay can be carried out under isothermal conditions. A thermal cycler is not needed to perform the amplification reaction, thus making this method highly cost- and time-efficient as well as field-applicable and easy-to-perform. LAMP products can be detected with the naked eye through the use of an intercalating dye such as SYBR Green I (Iwamoto et al. 2003). Although the first developed LAMP assay by Notomi et al. (2000) targeted the HB virus, LAMP as well as isothermal methods in general are becoming more and more interesting for food analysis, e.g., for the detection of allergens and genetically modified organisms (GMOs) and for meat species identification (Ahmed et al. 2010; Zahradnik et al. 2014a, b).
In this study, we report the develeopment of a loop-mediated isothermal amplification assay for the detection of horse meat in meat products targeting part of the mitochondrial genome which was demonstrated to be specific for horse in earlier publications (Köppel et al. 2008, 2009). Several LAMP primer sets were evaluated according to their specificity and sensitivity and compared with the results of a previously published real-time PCR assay. To test the capability of the assay to detect horse meat in highly complex food matrices, model sausages with defined horse meat contents were prepared and analyzed. Additionally, five different commercial food products, containing horse meat, were tested. The LAMP assay performed similar to PCR regarding the detection of horse meat in food products but can be performed much faster with an analysis time of around 30 min. The use of a simple heating block and the visual detection using SYBR Green I make this assay much more time- and cost-efficient compared to PCR and could prove to be a valuable tool for the detection of horse meat in pre-packaged food products.
Material and Methods
Meat Material and Samples
Horse meat and five horse meat products (two differently spiced hard cured sausages, meat loaf, blood pudding, and knackwurst) were obtained from a local butcher, specialized on the commerce of horse meat. Other meat species (beef, pork, chicken) and ingredients for the production of model sausages were obtained in local supermarkets.
Preparation of Model Sausages
For the preparation of model sausages, in a meat mixture containing beef (35 %), pork (35 %), bacon (20 %), salt (2 %), pepper (0.7 %), worcestercause (0.7 %), vinegar (0.7 %), and garlic (0.7 %) as well as various herbs and spices (5,2 %), each ingredient was added in (w/w). The mixture was homogenized thoroughly with fixed portions of horse meat in concentrations of 10, 5, 1, 0.5, 0.1 and 0 % using a meat cutter and filled into sheep gut. The sausages were then cooked for 40 min.
DNA Extraction
All samples were extracted using a previously published, slightly modified DNA extraction protocol for plant tissue (Amani et al. 2011). Each sample was extracted three times. Two hundred milligrams of each sample were incubated with extraction buffer, followed by addition of 500 μL chloroform/isoamylalcohol (24:1). After centrifugation (15,000 × g) the supernatant was added to 400 μL isopropanol (precooled at −20 °C) and incubated at −20 °C for 30 min. After another centrifugation step, the liquid phase was removed with a pipette and the pellet was dissolved in 100 μL × 1 TE buffer (10 mM Tris, 1 mM EDTA, pH 8). DNA concentration and purity were measured photometrically using the NanoVue Plus (VWR, Radnor, USA). Extracted DNA samples were stored at −20 °C until further use.
Primers and Probes
All oligonucleotides were synthesized by Eurofins MWG (Ebersberg, Germany). Real-time PCR primers and probe were used as published by Köppel et al. (2008) and Köppel et al. (2009). Seven LAMP primer sets were designed using the Primer Explorer V4 software with standard settings (Eiken Chemical Co., Ltd.; Tokyo, Japan) on basis of the complete sequence of the Equus caballus mitochondrial DNA, GenBank acc. no. X79547. The LAMP primer sequences, used for the final assay, target the NADH dehydrogenase at the position [3353–3565 bp] and are given in Table 1.
Real-Time PCR Reaction
One microliter diluted DNA extracts (1:10) were added to 14 μL of reaction mix containing Kapa™ Probe® Fast (Peqlab, Erlangen, Germany), primers, and probe (in a final concentration of 0.3 μmol/L each). Amplification reactions were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, New York, USA) according to the following thermal cycling protocol: initial steps of 2 min at 95 °C, followed by 60 cycles of 15 sec at 95 °C and 1 min at 62 °C. Amplification reactions were carried out in five replicates. To prove the presence of amplifyable DNA, all DNA extracts were amplified using a previously published 12S rRNA universal primerset (Kitano et al. 2007) (Supporting Information, Table S1). One microliter diluted DNA extracts (1:10) were added to 14 μL of reaction mix containing Kapa™ SYBR® (Peqlab, Erlangen, Germany) and primers (in a final concentration of 0.3 μmol/L each).
LAMP Reactions
LAMP reactions were carried out in a total reaction volume of 25 μL containing 0.8 μM each FIP and BIP, 0.2 μM each F3 and B3, 0.4 μM each LoopF and LoopB, 1.4 μM of each dNTP (Peqlab, Erlangen, Germany), 0.8 M betaine solution (Sigma-Aldrich, St. Louis, USA), 8 U Bst DNA polymerase (New England Biolabs, Ipswich, USA), 20 mM Tris-HCl (pH 8.5), 10 mM KCl, 10 mM (NH4)2SO4, 3 mM MgSO4, 0.1 % Triton X-100 (Sigma-Aldrich, St. Louis, USA), 1 μL diluted (1:10) DNA extract, and 1 μL Syto® 9 Green Fluorescent Nucleic Acid Stain (Life Technologies, Carlsbad, USA) (Monis et al. 2005) for detection via real-time fluorescence monitoring. Reactions were run in the thermal cycler at 67 °C for 90 min. Amplification reactions were carried out in five replicates. Alternatively, reactions were incubated on an Eppendorf Thermomixer Plus (Eppendorf, Hamburg, Germany) at 67 °C for 90 min. For visual detection, 1 μL × 1 SYBR Green® I was added to each reaction tube after incubation. In order to prove that the signals measured via LAMP are a product of DNA amplification, a 1.5 % agarose gel electrophoresis with × 1 TAE buffer (40 mM Tris acetate, 1 mM EDTA, pH 8,2) was performed (Supporting Information, Fig. S1).
Analytical Sensitivity and Specificity of the Horse Meat LAMP Assay
To investigate the analysis sensitivity of the presented assay, horse meat DNA in a concentration of 10 ng/μL was serially diluted (1:10) with beef DNA in a constant concentration of 10 ng/μL and analyzed with PCR, using a previously published primer set for the detection of horse meat (Köppel et al. 2009), and LAMP via real-time fluorescence monitoring via Syto® 9 Green Fluorescent Nucleic Acid Stain using the same conditions as mentioned above.
To ensure the specific detection of horse meat, DNA extracts of horse, beef, pork, chicken, goat, mutton, deer, roe, rabbit, boar, turkey, and donkey with a concentration of 40 ng/μL, respectively, were introduced as template into real-time LAMP reactions, under the same reaction conditions.
Limit of Detection for Horse Meat
Model sausages with fixed concentrations of horse meat (10, 5, 1, 0.5, 0.1, and 0 %, respectively) were analyzed with real-time PCR and LAMP in five replicates each. LAMP reactions were performed on two different amplification platforms, real-time fluorescence monitoring via Syto® 9 using a thermal cycler and amplification on a heating block with subsequent detection via SYBR Green® I, using the same conditions as mentioned above. Positive (PPV) and negative predicted values (NPV) were calculated as follows: PPV = (number of true positives) / (number of true positives + number of false positives) × 100; NPV = (number of true negatives) / (number of true negatives + number of false negatives) × 100.
Analysis of Commercial Horse Meat Products
To evaluate the performance of the LAMP assay when analyzing commercial horse meat products containing high amounts of fat, salt, and spices, DNA from five different meat products was extracted and introduced as template (diluted 1:4, except for blood pudding which was used undiluted) into PCR and LAMP reactions, using the same conditions as mentioned above, and subsequent staining of the LAMP products via SYBR Green® I.
Results
Analytical Sensitivity and Specificity
The detection probability was 100 % for 0.1 ng/μL horse meat in 10 ng/μL beef DNA background with a detection time of 21 min (±1.6 min). Non-horse meat samples and no template controls did not result in any positive amplification signals (Table 2). Specificity tests for horse and donkey resulted in positive signals (ten out of ten replicates). Testing the specificity with DNA extracted from pork, beef, chicken, beef, mutton, goat, deer, roe, boar, rabbit, and turkey meat resulted in negative signals (zero out of ten replicates) (Table 3).
Limit of Detection (LOD) of the LAMP Assay
In order to establish a limit of detection (LOD), based on real food matrices, the prepared model sausages with different concentration of horse meat (10, 5, 1, 0.5, 0.1, and 0 %, respectively) were analyzed via LAMP. The LAMP reactions were performed on two different amplification platforms, real-time fluorescence monitoring via Syto® 9 using a thermal cycler (n = 5), and amplification on a heating block with subsequent detection via SYBR Green® I (n = 2). The time of detection reached from 26.4 min (±0.5 min) for 0.1 % to 13.7 min (±0.8 min) for 10 %, respectively (Fig. 1). No amplification signals were observed for the model sausages containing no horse meat at all and for the non-template controls. The visual detection via SYBR Green® I gave similar results (Fig. 2).
Analysis of Commercial Products
To evaluate the performance of the LAMP assay when analyzing commercial horse meat products containing high amounts of fat, salt, and spices, DNA from five different meat products was extracted and introduced as template into PCR and LAMP reactions. Table 4 shows DNA concentrations in extracts for each of the products and the results obtained from real-time PCR and real-time LAMP experiments. All samples were analyzed in five replicates and reliably detected, except for blood pudding. Similar to the analysis of the model sausages, performing the LAMP assay on a thermal cycler and on a heating block gave the same results. Figure 3 shows the results for the visual detection via staining with SYBR Green I.
Discussion
The 2013 meat adulteration scandal drastically raised the awareness of the authorities and the public regarding the authenticity of meat and meat products. The increasing demand for versatile and high-quality products poses a challenge not only for controlling agencies but also for food industry and retail companies. Due to increased analysis efforts and increasing sample numbers, rapid test system that allow a simple on-site detection of specific analytes are required.
The authentication of meat species is more or less limited to DNA-based test systems due to the high specificity provided by PCR-based methods. However, conventional PCR methods are laborious and time consuming and cannot be transferred into a more field-applicable approach. Since LAMP is capable of amplifying a DNA target under isothermal conditions and the amplification products can be visually detected, this method offers a great advantage for the rapid on-site testing of food products and has been successfully applied for the detection of allergens, genetically modified organisms (GMOs), and food-borne pathogens (Zahradnik et al. 2014a; Lee et al. 2009; Yamazaki et al. 2008). However, to date, only a few publications have utilized the LAMP method for meat species authentication, like the detection of ostrich meat (Abdulmawjood et al. 2014) and the detection of meat species via an electrochemical DNA sensor (Ahmed et al. 2010).
The here-presented LAMP assay for the detection of horse meat is capable of reliably detecting 0.1 % horse meat in prepared model sausages in less than 20 min. The calculated PPV and NPV were 100 % for the analyzed model sausages. These results underline the findings of previous publications that LAMP is much less susceptible to known PCR inhibitors such as fats and salts (Kaneko et al. 2007). Specificity tests were able to show that positive amplifications signals are obtained only for horse (E. caballus) and donkey (Equus asinus), which were both found in food products during the 2013 meat adulteration scandal (Walker et al. 2013), but no other more or less frequently consumed meat species in Europe (turkey, chicken, pork, beef, deer, roe, boar, rabbit, mutton, and goat). The high specificity is provided by the use of three primer sets, all of which have to bind perfectly in order to amplify the target sequence. The analytical sensitivity of the assay was found to be 100 pg/μL with a detection probability of 100 %. Although lower sensitivities for LAMP assays for the detection of meat have been reported (Abdulmawjood et al. 2014), the addition of horse meat to meat products is an issue of food adulteration and not of trace analysis. Therefore, the analytical sensitivity obtained in this publication is sufficient in order to detect horse meat in meat products when admixed by a butcher or producer to increase the financial gain. The analysis of commercial horse meat products resulted in the reliable detection of all products except for blood pudding. Although three DNA extractions were performed, the photometrically obtained yields were <5 ng/μL; no horse meat was detected in the blood pudding sample with PCR or LAMP. This result might have been caused by the fact that blood pudding mainly consists of coagulated blood which on its part consists of >90 % erythrocytes which do not contain nuclei or mitochondria. Hence, extracted DNA will originate from thrombocytes and leucocytes which represent only a minor part of the composition of mammalian blood (Bowen 1963) and of genomic DNA extracted from spices and bacon, which are part of the typical Austrian recipe for blood pudding. All other commercial horse meat products (meat loaf, knackwurst, two different types of hard cured sausages) were detected with 100 % probability, in less than 25 min.
Conclusion
The loop-mediated isothermal amplification assay was shown to be a specific and sensitive method for the detection of horse meat in food products. Although it is not suitable for the quantification of trace levels, it represents an accurate and robust technique that can be performed in less than 1 h without special equipment. With a reaction time of less than 30 min, it is a valuable tool for the rapid control of food samples for horse meat. Furthermore, the naked eye visualization using SYBR Green I eliminates the need for a UV transilluminator and allows the on-site analysis of food samples. This work contributes to a future approach on food analysis by meeting the demands for quick and easy-to-perform analytical methods.
References
Abdulmawjood A, Grabowski N, Fohler S, Kittler S, Nagengast H, Klein G (2014) Development of loop-mediated isothermal amplification (LAMP) assay for rapid and sensitive identification of ostrich meat. PLoS One 9(6):1–6
Ahmed MU, Hasan Q, Hossain MM, Saito M, Tamiya E (2010) Meat species identification based on the loop mediated isothermal amplification and electrochemical DNA sensor. Food Control 21(5):599–605
Aliotta JM, Pelletier JJ, Ware JL, Moran LS, Brenner JS, Kong H (1996) Thermostable Bst DNA polymerase I lacks a 3’ → 5’ proofreading exonuclease activity. Genet Anal: Biomol Eng 12(5–6):185–195
Amani J, Kazemi R, Abbasi AR, Salmanian AH (2011) A simple and rapid leaf genomic DNA extraction method for polymerase chain reaction analysis. Iran J Biotechnol 9(1):69–71
Bowen HJM (1963) The elementary composition of mammalian blood. Wantage, Berkshire, United Kingdom
European Commission (2010) Off J Eur Union 15:1
European Commission (2013) Off J Eur Union 48:1
Iwamoto T, Sonobe T, Hayashi K (2003) Loop-mediated isothermal amplification for direct detection Mycobacterium tuberculosis Complex, M. avium, and M. intracellulare in Sputum Samples. J Clin Microbiol 41(6):2616–2622
Kaneko H, Kawana T, Fukushima E, Suzutani T (2007) Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods 70:499–501
Kitano T, Umetsu K, Tian W, Osawa M (2007) Two universal primer sets for species identification among vertebrates. Int J Legal Med 121:423–427
Köppel R, Ruf J, Zimmerli F, Breitenmoser A (2008) Multiple real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur Food Res Technol 227:1199–1203
Köppel R, Zimmerli F, Breitenmoser A (2009) Heptaplex real-time PCR for the identification and quantification of DNA from beef, pork, chicken, turkey, horse meat, sheep (mutton) and goat. Eur Food Res Technol 230:125–133
Lee D, La Mura M, Allnutt TR, Powell W (2009) Detection of genetically modified organisms (GMOs) using isothermal amplification of target DNA sequences. BMC Biotechnol 9:7
Monis PT, Giglio S, Saint CP (2005) Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concetration on amplification and DNA melting curve analysis. Anal Biochem 340(1):24–34
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16:223–229
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):I–VII
Walker MJ, Burns M, Burns DT (2013) Horse meat in beef products—species substitution 2013. J Assoc Publ Anal 41:67–106
Yamazaki W, Ishibashi M, Kawahara R, Inoue K (2008) Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol 8:163
Zahradnik C, Martzy R, Mach RL, Krska R, Farnleitner AH, Brunner K (2014a) Detection of the food allergen celery via loop-mediated isothermal amplification technique. Anal Bioanal Chem 406(27):6827–6833
Zahradnik C, Kolm C, Martzy R, Mach RL, Krska R, Farnleitner AH, Brunner K (2014b) Detection of the P35S promotor in transgenic maize via various isothermal amplification techniques: a practical approach. Anal Bioanal Chem [Epub ahead of print]
Acknowledgments
This work was financially supported by the Federal Country Lower Austria in cooperation with the European Regional Development Fund (ERDF).
Conflict of interest
Celine Zahradnik declares that she has no conflict of interest. Roland Martzy declares that he has no conflict of interest. Robert L. Mach declares that he has no conflict of interest. Rudolf Krska declares that he has no conflict of interest. Andreas H. Farnleitner declares that he has no conflict of interest. Kurt Brunner declares that he has no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
The presence of amplifyable DNA was proven via real-time PCR using a universal primer set for vertebrates targeting parts of the 12S rRNA, as previously published (Kitano et al. 2007), n = 10. (DOC 11 kb)
Figure S1
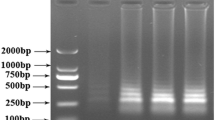
Agarose gel electrophoresis of the amplification products, resulting in a pattern typical for the various-sized LAMP products. Lane 1-10 - horse meat [10 ng/μL], lanes 11-15 - beef [10 ng/μL], lanes 16-20 - non-template control (NTC). (DOC 255 kb)
Rights and permissions
About this article
Cite this article
Zahradnik, C., Martzy, R., Mach, R.L. et al. Loop-Mediated Isothermal Amplification (LAMP) for the Detection of Horse Meat in Meat and Processed Meat Products. Food Anal. Methods 8, 1576–1581 (2015). https://doi.org/10.1007/s12161-014-0072-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-014-0072-8