Abstract
Two full-length cDNAs encoding NAM, ATAF, and CUC (NAC)-family transcription factors (TFs) were isolated from two different cultivars of Panicum virgatum L. (switchgrass) and named PvNAC1 and PvNAC2. Phylogenetic analysis of PvNAC1 and PvNAC2 grouped them with NAC proteins involved in senescence in annual plant species. Transcript profiling revealed that both PvNAC1 and PvNAC2 are induced during leaf senescence. Expression of a PvNAC1-green fluorescent protein (GFP) fusion in plant cells revealed a nuclear location of the protein, consistent with a role in transcriptional regulation. Expression of PvNAC1 in an Arabidopsis nap stay-green mutant suppressed its senescence defect. Expression of PvNAC1 in wild-type Arabidopsis triggered early leaf senescence and remobilization. Transcriptome analysis implicated leaf protein degradation and nitrogen recycling enzymes in NAC-dependent seed protein increase in Arabidopsis. Overexpression of pvNAC2 in switchgrass resulted in increased aboveground biomass associated with increased transcript levels of key nitrogen metabolism genes in leaves and nitrate and ammonium transporter genes in roots. The results indicate that NAC TFs play conserved roles in leaf senescence in the plant kingdom not only in annual monocots and dicots but also in perennial plants such as switchgrass. PvNAC1 and PvNAC2 hold promise for improving nutrient use efficiency in switchgrass through genetic manipulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Switchgrass (Panicum virgatum L.) is a perennial plant with water-efficient C4 photosynthesis that is native to the North American tall grass prairies and has been targeted for development as a bio-energy crop in the USA. Switchgrass has higher biomass yield than many annual crops and can be grown on marginal soils, which avoids competition with food crops for prime arable land [1–3]. There are two switchgrass ecotype classes, “upland” and “lowland,” which refer to latitude. Average annual biomass yields of switchgrass in the USA were estimated to be 12.9 and 8.7 tonnes/hectare for lowland and upland ecotypes, respectively [4].
Successful development of switchgrass for biofuel production will depend, in part, on agricultural sustainability including nutrient conservation in the soil-plant system [1, 5]. Like other perennial grasses, switchgrass plants remobilize a portion of some nutrients, including N, P, and K, from shoots to roots during yearly senescence [5]. Nonetheless, substantial amounts of soil nutrients are removed with the harvested shoot biomass. For example, total N removed with biomass varied from 31 to 63 kg N ha−1 year−1 in a one-cut fall harvest after senescence, compared to 90 to 144 kg ha−1 year−1 for a two-cut system with the first harvest prior to senescence, over 5 years of measurements [6]. Such nutrient withdrawal rates inevitably deplete soil N and necessitate the addition of fertilizer N to maintain switchgrass productivity. Synthesis and application of fertilizer N are energy-intensive and economically and environmentally costly. High N content in harvested biomass can be an additional liability because it yields NOx compounds upon oxidation, which are potent atmospheric pollutants [7]. Therefore, decreasing the nutrient concentration in harvested switchgrass, with the exception of C, H, and O that are converted to biofuels, will help to conserve soil nutrients, reduce the need for fertilizers, and, thereby, reduce the cost and environmental impact of switchgrass production.
Senescence is the final stage of plant development, leading to the death of specific organs or the whole plant [8]. Leaf senescence is a regulated process and is important for remobilization of nutrients from leaves to other plant parts. For annual plants including many crop species, leaf senescence mobilizes nutrients for seed production. Small-grain cereals like rice, wheat, and barley mobilize up to 90 % of the nitrogen in vegetative organs to the grain [9, 10]. In perennial grasses like switchgrass, nutrients in leaves are remobilized during yearly senescence not only to the inflorescence for seed production but also to the root system for storage and later re-use. There is natural variation among switchgrass accessions for nitrogen remobilization efficiency, with a range of 20–61 % remobilization from shoots recorded in one set of experiments [5]. Nothing is known about the genetic regulation of nutrient remobilization in switchgrass and other perennial grasses at present. Such knowledge and the corresponding regulatory genes could pave the way to improved cultivars with increased nutrient remobilization and, consequently, increased nutrient use efficiency for biomass production.
Transcription factor (TF) genes have been implicated in the control of leaf senescence and nutrient remobilization to seeds in annual plant species. A NAC TF gene, NAM-B1, from wild emmer wheat was associated with accelerated senescence and increased grain zinc, iron, and protein content [11]. Repression of four homologs of NAM-B1 in a hexaploid wheat, via RNA interference (RNAi), delayed plant senescence by more than 3 weeks and reduced grain protein, zinc, and iron content by more than 30 %, while flag leaves of RNAi plants retained more residual nitrogen, zinc, and iron than wild-type controls [11]. Rice OsNAC5 was found to be induced earlier and to a greater extent in flag leaves and panicles of rice cultivars with higher seed protein content, indicating that OsNAC5 may be involved in amino acid remobilization from green tissues to seeds [12]. Although there have been some microarray-based transcriptomic studies of senescence in Arabidopsis [13–15], Populus [16], wheat [17], and barley [18], little is known about processes that are regulated directly by NAC TFs. Recently, the effect of downregulation of TtNAM-B1 on wheat flag-leaf gene expression during senescence (12 days after anthesis) was investigated using RNA-seq technology [19]. The list of putative TtNAM-B1-regulated genes included transporters, hormone-regulated genes, and other transcription factors.
Here, we report the identification and characterization of two NAC TF genes, PvNAC1 and PvNAC2, from two different switchgrass cultivars, Summer VS16 and Alamo AP13, respectively. Phylogenetic analysis of PvNAC1 and PvNAC2 grouped them with NAC proteins involved in senescence in annual plant species. PvNAC1 and PvNAC2 transcript levels increased in switchgrass leaves during natural and artificial, dark-induced senescence. Expression of PvNAC1 in an Arabidopsis stay-green mutant, atnap, suppressed the defective-senescence phenotype of the mutant. Expression of PvNAC1 in wild-type Arabidopsis promoted leaf senescence and nitrogen remobilization. Transcriptome analysis linked PvNAC1 expression in Arabidopsis to induction of protein and nitrogen-recycling genes and leaf protein degradation. Overexpression of pvNAC2 in switchgrass increased nitrogen use efficiency and biomass production in switchgrass.
Results
Cloning and Phylogenetic Analysis of PvNAC1 and PvNAC2
Fragments of several putative NAC TF cDNA were amplified from switchgrass cultivar Summer VS16 by 5′-Rapid Amplification of cDNA Ends (RACE) and 3′-RACE (see experimental procedures). The N-terminal portions of deduced putative NAC proteins were aligned with typical NAC TFs from other plant species, resulting in the identification of a switchgrass NAC protein with five characteristic N-terminal DNA binding motifs (A–E) that we called PvNAC1 (Fig. S1) [20, 21]. The full-length PvNAC1 coding sequence was found to be 1,104 bp (GenBank accession no. JN673545). Comparison of PvNAC1 cDNA and genomic sequences, obtained via PCR-amplification, revealed two introns in the PvNAC1 gene. Another NAC TF cDNA, with a coding sequence of 1113 bp, was amplified from switchgrass cultivar Alamo AP13 and named PvNAC2. The coding sequences of PvNAC1 and PvNAC2 share 95.2 % identity and their deduced proteins share 92 % identity, indicating that they may have similar functions in the two cultivars.
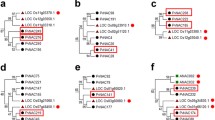
Phylogenetic analysis of various plant NAC TFs grouped PvNAC1 and PvNAC2 with AtNAP, TtNAM-B1, and OsNAC5, each of which have been implicated in senescence and/or nutrient remobilization [22, 11, 12] (Fig. 1). At the amino acid level, the complete PvNAC1 protein shares 48.1 % identity with AtNAP, 37.9 % with TtNAM-B1, and 34.4 % with OsNAC5.
Phylogenetic analysis of PvNAC1 and PvNAC2 with representative rice NACs as well as other NACs with known or indicative functions. The branch within the dashed box includes PvNAC1 and PvNAC2 (indicated by solid arrows) and NAC TFs related to senescence in Arabidopsis (AtNAP) [22] and nutrient remobilization in wheat (TtNAM-B1) [11] and rice (ONAC009/OsNAC5) [12] (indicated by empty arrows). Confidence levels are shown as percentages
Expression of PvNAC1 and PvNAC2 in Leaves Is Induced During Natural Senescence and by Prolonged Darkness
Natural senescence in grasses begins in the oldest, most basal leaves of vegetative plants and proceeds to younger, higher leaves of the panicle as plants mature and transition to reproductive development [23]. Transcript levels of PvNAC1 in bottom (B), intermediate (I), and top (T) leaves of young (Y) and middle-aged (M) and old (O) tillers of cultivar Summer were quantified by quantitative real-time (RT)-PCR (qRT-PCR) (Fig. 2a). PvNAC1 transcript levels in bottom leaves were lowest in young tillers (YB), higher (19-fold) in late vegetative tillers (MB), and highest (91-fold) in reproductive tillers (OB; Fig. 2b). Within old, reproductive tillers, PvNAC1 transcript levels were lowest in young leaves at the top of tillers (OT), intermediate in intermediate leaves (OI), and highest in the oldest, bottom leaves (OB; Fig. 2c). In other words, PvNAC1 expression increased with plant development and leaf age. Notably, PvNAC1 transcript levels in leaves increased prior to visible signs of senescence (leaf yellowing). Finally, leaf PvNAC1 transcript levels increased in response to prolonged darkness, which induced senescence/yellowing of detached leaves (Fig. 2d). PvNAC2 expression in Alamo AP13 showed similar responses to leaf aging and dark incubation (Fig. S2). Taken together, these results are consistent with a possible role of PvNAC1 and PvNAC2 in the regulation of leaf senescence.
Expression of PvNAC1 in leaves of switchgrass genotype VS16. a Young (Y), middle-aged (M), and old (O) tillers and leaves at different positions (T, top; I, intermediate; and B, bottom). b Relative transcript levels of PvNAC1 in the bottom leaves of young, middle-aged, and old tillers (YB, MB, and OB, respectively), determined by qRT-PCR. c Relative transcript levels of PvNAC1 in the top, intermediate, and bottom leaves of old tillers (OT, OI, and OB). d Relative transcript levels of PvNAC1 in intermediate leaves detached from middle-aged tillers and incubated at room temperature on wet paper towel in continuous darkness for 0, 3, 6, and 9 days. All transcript levels are expressed relative to that of the switchgrass Ubiquitin gene in each sample. Error bars indicate SD of three biological replicates. Different letters above columns in each figure indicate significant differences based on Tukey’s test (p < 0.05)
PvNAC1 Is Localized in the Nucleus
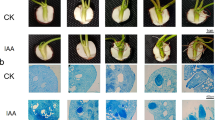
To determine the subcellular location of PvNAC1, a translational fusion of PvNAC1 and green fluorescent protein (GFP) was constructed under the control of the constitutive CaMV-35S promoter, p35S (Fig. 3a). In particle-bombarded onion epidermal cells expressing the PvNAC1-GFP fusion protein, GFP fluorescence was detected only in nuclei (Fig. 3b–d), whereas in cells bombarded with p35S-GFP, green fluorescence was detected in multiple compartments including nuclei, plasma membrane, and cytoplasm (Fig. 3e–g). Nuclear localization of PvNAC1 is consistent with a possible role of the protein in regulation of gene transcription.
Localization of PvNAC1 protein in plant cells. a Schematic representation of the PvNAC1-GFP fusion construct in vector pAVA121. b–d Nuclear localization of p35S::PvNAC1-GFP fusion protein detected in onion epidermal cells. e–g Subcellular localization of p35S::GFP as a control. b, e GFP fluorescence. c, f White-light micrographs. d, g Overlay of b and c and e and f, respectively
Complementation of an Arabidopsis Atnap Mutant with PvNAC1
PvNAC1 and PvNAC2 are on the same branch of the phylogenetic tree as AtNAP (Fig. 1). To test the function of PvNAC1 in planta, an Arabidopsis atnap T-DNA insertion mutant was isolated (Fig. S3ab), and homozygous mutant plants were transformed with constructs that contained PvNAC1 or GUS (negative control) coding sequence under the control of 2 kb of the Arabidopsis AtNAP promoter sequence. Fully expanded, non-senescing leaves of wild type (WT) and atnap mutant plants were excised and incubated on a wet filter paper in total darkness at 22 °C for 5 days, which resulted in yellowing of WT leaves but not of atnap leaves, which remained green (Fig. 4a). In the same assay, leaves of atnap plants transformed with PvNAC1 turned yellow, like the WT, while those transformed with GUS remained green (Fig. 4a). The chlorophyll concentration of WT and PvNAC1-expressing atnap were not significantly different (approx. 300 ng/g FW), while those of atnap and GUS-expressing atnap were more than threefold higher than the WT (approx. 1100 ng/g FW; Fig. 4b). Thus, PvNAC1 restored the WT phenotype to atnap, indicating that the switchgrass protein PvNAC1 can substitute for AtNAP and trigger leaf senescence.
PvNAC1 suppression of the stay-green phenotype of the Arabidopsis nap mutant. a Leaves were detached from wild type (WT), atnap and atnap transformed with PvNAC1 (two independent lines) or the GUS gene (as a negative control), and incubated in complete darkness for 5 days prior to photography. b Chlorophyll concentration in different leaves after 5 days of dark treatment, with abbreviations as in (a). Values are means ± SD (n = 4). Asterisk indicates significant difference at p < 0.01 compared with WT, using the Student’s t test
Expression of PvNAC1 Triggers Leaf Senescence and Nitrogen Translocation
PvNAC1 was cloned into vector pER8 under the control of an estrogen-inducible transactivator [24] and transformed into Arabidopsis wild type. Detached leaves of WT, pER-PvNAC1, and pER-GFP (negative control) transgenic plants were incubated on filter paper wetted with estradiol (EST) solution and kept under constant light for 5 days, which resulted in senescence (yellowing) of pER-PvNAC1 leaves but not those of WT or pER-GFP (Fig. S4).
Estradiol treatment of whole plants had no effect on pER-GFP or WT controls but resulted in accelerated senescence (yellowing) of mature leaves on pER-PvNAC1 transgenic plants (Fig. 5a). Interestingly, young leaves of pER-PvNAC1 plants remained green after estradiol treatment. DMSO solution without estradiol had no effect on any of the plant genotypes. Mature leaves of estradiol-treated pER-PvNAC1 plants had 53 % less chlorophyll (Fig. S5a) and 24 % less total nitrogen than DMSO-treated controls, after 5 days of treatment (Fig. 5b). PvNAC1 was found to be expressed only in estradiol-treated pER-PvNAC1 transgenic leaves (Fig. S5b). Thus, expression of PvNAC1 was sufficient to trigger leaf senescence in whole plants, resulting in chlorophyll degradation and nitrogen translocation out of leaves. However, transient expression of PvNAC1 for 5 days did not increase nitrogen concentration in mature seeds (Fig. S5c).
Phenotypic effect of PvNAC1 expression on Arabidopsis. a Phenotype of whole plants of WT, pER-GFP, and pER-PvNAC1 transgenics after 5 days of treatment with 100 μM estradiol (EST, in 0.1 % DMSO) and control (0.1 % DMSO). Inflorescences were removed to reveal the leaf phenotypes. b Leaf nitrogen concentration in DMSO- or EST-treated mature leaves. Values are means ± SD (n = 4). Asterisk indicates significant difference at p < 0.01 of EST-treated pER-PvNAC1 compared with all other lines, using the Student’s t test
Role of PvNAC1 in Nitrogen Remobilization During Senescence
To understand how PvNAC1 mediates leaf senescence and nitrogen remobilization when expressed in Arabidopsis, we carried out transcriptome analysis of pER-PvNAC1 transgenic Arabidopsis leaves, following estradiol-induced PvNAC1 expression for 3, 6, 12, 72, and 120 h. PvNAC1 transcript level peaked after 6 h of estradiol treatment, then declined gradually (Fig. S6a). Transcript levels of the senescence marker gene, SAG12, increased while chlorophyll concentration decreased significantly within 72 h of estradiol treatment, compared to the DMSO-treated control (Fig. S6bc). Six hours after the commencement of estradiol treatment, at the peak of PvNAC1 expression (Fig. S6a), transcript levels of 248 genes had increased by at least twofold, and transcripts of 18 genes decreased at least twofold compared to the DMSO control (Table S1). After 72 h of PvNAC1 expression, there were 921 genes upregulated and 1,151 genes downregulated by a factor of at least two (Table S1). To explore the biological processes regulated directly or indirectly by PvNAC1, MapMan analysis (Pathways: Regulation_overview) [25] was performed on transcriptome data. After 6 h of estradiol treatment, a total of 99 genes with at least twofold change in transcript level were assigned to multiple processes, but especially regulation of transcription with 22 genes, 19 of which were induced and 3 repressed; receptor kinase signaling with 18 genes all being induced; calcium signaling with 17 of 18 genes induced; hormone metabolism and signaling with 13 of 16 genes induced; protein degradation with 11 of 13 genes induced; and posttranslational protein modification with five of six genes induced (Fig. 6a). Transcript levels of 569 genes with a change of at least twofold by 72 h of PvNAC1 expression, compared to DMSO treatment control, were assigned to a variety of regulatory processes (Fig. 6b). Transcriptional regulators constituted the largest functional class of genes regulated by 6 and 72 h of PvNAC1 expression. Interestingly, induced genes involved in protein degradation moved from the fifth most numerous set of regulated genes at 6 h to the second most numerous set by 72 h, perhaps reflecting a shift towards cellular deconstruction by that time. Consistent with this idea, chlorophyll concentration of plant leaves decreased by 12 % after 72 h of PvNAC1 expression (Fig. S6c). In contrast to the protein degradation genes, the relative importance of genes for receptor kinase signaling and calcium signaling among those regulated following PvNAC1 expression was higher at 6 h than at 72 h, indicating that signaling pathways played more central roles early after PvNAC1 induction.
MapMan analysis of nitrogen and GABA (4-aminobutyrate) metabolism identified genes encoding two cytosolic glutamine synthetases (GS1: GLN1.1 and 1.3), a ferredoxin-dependent glutamate synthase (Fd-GOGAT) and a glutamate dehydrogenase (GDH2) that were upregulated following 72 h of PvNAC1 induction. These enzymes have been implicated in N-recycling during leaf senescence in the past [26]. Further, qRT-PCR confirmed the role of PvNAC1 in inducing expression of these genes (Fig. S7). Except Fd-GOGAT, which showed a 1.7-fold increase in expression, the expression levels of the other three genes were enhanced more than twofold by PvNAC1 expression for 72 h.
Overexpression of PvNAC2 in Switchgrass Increases Aboveground Biomass and Nitrogen Content
An efficient switchgrass transformation protocol has been developed for cultivar Alamo [27] but not for cultivar Summer. Therefore, we transformed Alamo PvNAC2, driven by the CaMV 35S promoter, into the Alamo cultivar to investigate its physiological role in switchgrass. Together with a transgenic negative control (Tc), three independent transgenic switchgrass lines (T20, T42, T54) were selected for physiological analysis based on their high levels of pvNAC2 expression (Fig. S8a). Split tillers (vegetative clones) of different lines were grown continuously in a greenhouse for 6 months prior to harvesting of whole shoots. No accelerated yellowing of whole shoots was observed in transgenic plants during this 6-month period.
Inflorescences of T20 and T42, leaves of T54, and stems of all three transgenic lines exhibited significantly higher dry biomass than the control, resulting in the significantly higher total aboveground biomass (sum of leaves, stems, and inflorescences) in all lines, i.e., 87, 85, and 103 g/plant in T20, T42, and T54, respectively, compared to 60 g/plant in the control (Figs. 7a and S8b). In contrast, there were no statistical differences in biomass of underground organs (crown and roots) between transgenic and control plants (Figs. 7a and S8b). Nitrogen concentration of T42 leaves was significantly lower than that of Tc, while nitrogen concentration of all three transgenic stems was significantly higher. No significant difference was found for nitrogen concentration in inflorescences, crowns, and roots between transgenics and control (Fig. 7b). The nitrogen content was calculated by multiplying nitrogen concentration by biomass. The inflorescences of T20 and T54 and stems of all three transgenic lines showed significant higher nitrogen content than the control, while the nitrogen contents in leaves of transgenic and control plants were statistically similar (Fig. 7c). For underground organs, nitrogen content in T20 and T54 root was higher than that of control (Fig. 7c). We further calculated the aboveground and underground nitrogen content (Fig. S8c). All three transgenic lines showed significantly higher nitrogen contents in aboveground organs (sum of inflorescences, leaves, and stems), but only T54 showed significantly higher nitrogen content in underground organs.
Overexpression of PvNAC2 in switchgrass. a Biomass yield. b Nitrogen concentration. c Nitrogen content in inflorescences, leaves, stems, crowns, and roots in control and transgenic plants after 6 months of growth in greenhouse. d Relative transcript levels of putative nitrogen metabolism genes (PvGS1, GOGAT, and GDH) in leaves at intermediate position on mature tillers and genes of nitrate transporter (PvNRT1, NRT2) and ammonium transporter (PvAMT2) in roots of three independent transgenic lines (T20, T42, and T54), compared to those of control (Tc). Values are means ± SD of three biological replicates. Asterisk indicates significant difference at p < 0.05 of transgenic lines compared with control, using the Student’s t test
To uncover possible mechanisms of increased biomass and nitrogen content in the shoots of PvNAC2 overexpressor lines, we measured transcript levels of the nitrogen metabolism genes PvGS1, PvGOGAT, and PvGDH in leaves at an intermediate position on mature tillers from transgenic and control plants (c.f. Fig. 2a). Transcript levels of PvGDH, but not PvGS1 or PvGOGAT, were consistently higher in all three PvNAC2 overexpressor lines than in the control (Fig. 7d). Transcript levels of the nitrate and ammonium transporter genes, PvNRT1, PvNRT2, and PvAMT2 in roots, were also measured and found to be significantly higher in PvNAC2 overexpressor lines than in the control (Fig. 7d).
Discussion
We have identified two TFs of switchgrass, PvNAC1 and PvNAC2, that appear to be positive regulators of leaf senescence and associated with nitrogen use efficiency, based on the following observations: the proteins are closely related to NAC TFs of other species that are known to be involved in leaf senescence and nutrient remobilization; expression of PvNAC1 and PvNAC2 increased with leaf age and was induced by prolonged darkness, concomitant with leaf senescence; PvNAC1 protein was localized to the nucleus; PvNAC1 suppressed the stay-green phenotype of the Arabidopsis atnap mutant; inducible expression of PvNAC1 in wild-type Arabidopsis triggered senescence and nitrogen remobilization in mature leaves; and constitutive expression of pvNAC2 in switchgrass increased aboveground biomass, which was associated with upregulation of genes for nitrogen metabolism in leaves and nitrate and ammonium transport in roots.
Previous work revealed functional conservation among related NAC TFs of different annual species, including Arabidopsis, rice, and kidney bean. For instance, rice and kidney bean homologs of AtNAP suppressed the stay-green mutant phenotype of Arabidopsis atnap [22]. Using the same approach, we showed that PvNAC1 from switchgrass can substitute for AtNAP in Arabidopsis (Fig. 4). Thus, we demonstrated that conservation of NAC function in senescence extends beyond annual plants into perennial plants like switchgrass.
We found that ectopic expression of PvNAC1 in wild-type Arabidopsis was sufficient to trigger controlled senescence in mature leaves of intact plants (Fig. 5). This implies that PvNAC1 and its homologs (such as TtNAM-B1) act early in the senescence developmental program [11]. In contrast, no senescence (yellowing) was observed in young leaves of estradiol-treated pER-PvNAC1 transgenic Arabidopsis plants (Fig. 5), which may reflect an absence in young leaves of additional regulatory factors that are required for senescence or the presence of factors that interfere with PvNAC1 activity.
In contrast to Arabidopsis, switchgrass plants grown in our greenhouse and growth cabinets do not undergo whole-shoot senescence and as they do in nature. Instead, they continually produce new tillers while mature tillers mostly remain green. Overexpression of PvNAC2 in switchgrass did not trigger premature shoot senescence in plants grown in the greenhouse, perhaps for the same reason that PvNAC1 overexpression in young leaves of Arabidopsis did not trigger senescence there (see previous paragraph). Taken together, our experiments with Arabidopsis and switchgrass indicate that developmental and environmental signals influence NAC proteins, possibly via other transcription factors, to determine where and when senescence occurs in plant shoots.
Development of estradiol-inducible pER-PvNAC1 transgenic lines of Arabidopsis allowed us to identify biological processes and potential gene targets of NAC TF activity. A relatively large number of TF genes responded rapidly (at 6 h) to PvNAC1 induction (Fig. 7a and Table S1), which indicates that PvNAC1 may be a master positive regulator of other TFs that in turn regulate the many processes associated with senescence. Among the latter are 105 genes involved in protein degradation, which represented the second largest group regulated by 72 h following PvNAC1 expression (Fig. 7b). Sixty-four protein degradation-related genes were upregulated at least twofold 72 h after PvNAC1 induction, nearly half of which (30) were also induced during natural senescence in Arabidopsis [13]. Natural senescence occurs more slowly and in a different developmental context than was the case in our experiments with vegetative plants.
Although PvNAC1 transcript levels in pER-PvNAC1 transgenic lines of Arabidopsis peaked after 6 h of estradiol treatment, there were no significant increases in transcript levels of genes for nitrogen metabolism, compared to the DMSO control, at that stage (Table S1). However, after 72 h of PvNAC1 expression, four genes involved in ammonium recycling and metabolism, i.e., GLN1.1, GLN1.3, Fd-GOGAT, and GDH2, exhibited significantly higher transcript levels (Fig. S7). GLN1.1 and GLN1.3 encode isoforms of cytosolic glutamine synthetase (GS1). GS1 has been implicated in nitrogen remobilization during leaf senescence [26]. For example, maize mutants lacking specific GS1 isoforms accumulated large amounts of amino acids and ammonia in leaves below ears due to a dysfunction in nitrogen export [28]. Ferredoxin-dependent glutamate synthase (Fd-GOGAT) is known to play an important role in leaf photorespiratory ammonium assimilation [29]. Induction of Fd-GOGAT during PvNAC1-induced senescence in Arabidopsis leaves implicates it in nitrogen remobilization during this developmental process. Glutamate dehydrogenase (GDH) plays a central role liberating ammonium from amino acids following hydrolysis of protein reserves in seeds during germination or proteins in other organs during senescence [29]. The Arabidopsis GDH2 gene was induced substantially 72 h after the onset of PvNAC1 expression (Fig. S7). A similar level of induction of GDH2 was found during natural senescence of Arabidopsis [13]. In view of the delay between PvNAC1 induction and the accumulation of transcripts of genes involved in nitrogen mobilization, it is unlikely that PvNAC1 regulates these genes directly. However, some of the TFs induced within 6 h of PvNAC1 expression may regulate nitrogen metabolism genes. In any case, the results presented here shine light on the network of genes and processes that respond to expression of senescence-related NAC TF genes.
Nitrogen use efficiency (NUE) in plants is determined by how well they take up inorganic nitrogen from the soil, assimilate nitrate and ammonium, and recycle organic nitrogen to harvested organs [30]. Although transient expression of PvNAC1 in pER-PvNAC1 Arabidopsis plants at the bolting stage of early reproductive development resulted in nutrient remobilization from older leaves, as reflected by 24 % less nitrogen and 53 % less chlorophyll (Figs. 5b and S6a), no increase in seed nitrogen concentration was found (Fig. S5c). Presumably, other sink tissues such as expanding leaves or the inflorescence competed with seeds for the remobilized leaf nitrogen. Alternatively, some nitrogen may have been lost from plants as gaseous nitrogen, such as NH3 [31, 32].
NUE is an important target for switchgrass breeding as high NUE will help to minimize the need for fertilizer N, which will reduce biomass and biofuel production costs [1], and the environmental costs of fertilizer loss from agricultural systems. Although efficient uptake of N compounds from the soil to maximize biomass production will be an important part of NUE in switchgrass, high N content of the harvested biomass will not be, in contrast to crop and forage species where the objective is generally high protein (N) in food and feed. Remobilization of N from aboveground to belowground organs in switchgrass is a natural process that occurs during shoot senescence in autumn, which helps to conserve nitrogen in the plant for regrowth of the shoot in spring. There is substantial variation in N remobilization efficiency during shoot senescence between switchgrass ecotypes, which could be harnessed in breeding programs to conserve N [5]. The results presented here point to roles of PvNAC1 and PvNAC2 in leaf senescence in switchgrass and to the possibility of using these TFs to improve plant NUE. Overexpression of PvNAC2 in switchgrass increased aboveground biomass and nitrogen content under greenhouse conditions (Fig. S8bc). Stems of all three PvNAC2 overexpressor lines exhibited significantly higher biomass, nitrogen concentration, and content (Fig. 7abc). Because stems serve as a conduit for N movement between organs, the higher N concentration in stems of the PvNAC2 overexpressors compared to the control may have reflected an increase in N uptake from the soil or N remobilization from mature leaves. In fact, we found evidence for both, as discussed below.
Transient overexpression of PvNAC1 in Arabidopsis leaves and constitutive overexpression of PvNAC2 in switchgrass led to increased transcript levels of GS1 and GDH (Figs. S7 and 7d), which also occurs during natural and dark-induced senescence [13, 15]. Remobilization of N compounds from leaves to other organs, mediated by GS1, GDH, and other enzymes, may explain the drop in N concentration in leaves expressing PvNACs (Figs. 5b and 7b). In the case of switchgrass constitutively expressing PvNAC2, the greater N content of shoots (Figs. 7c and S8c) indicates that more ammonium and/or nitrate was taken up from the soil by the roots and transported aboveground. Consistent with this, we found higher transcript levels of PvNRT1, PvNRT2, and PvAMT2 in roots of the PvNAC2 transgenics (Fig. 7e). Increased nitrogen remobilization from leaves and/or increased N uptake by roots may explain the increased aboveground biomass of these plants compared to control (Figs. 7a and S8b).
In summary, we have identified and characterized two NAC TFs from two different ecotypes of switchgrass that have similar roles as positive regulators of senescence. Our work demonstrates functional conservation among closely related NAC genes in annual and perennial plants. PvNAC1 and PvNAC2 show promise as tools to control senescence and nitrogen remobilization in shoots and, indirectly, nitrogen uptake in roots. Thus, they are interesting targets for improvement of NUE in switchgrass in the future.
Materials and Methods
Plant Materials and Growth Conditions
Wild-type switchgrass (P. virgatum L.) cultivars Summer (genotype VS16) and Alamo (genotype AP13) were used for gene cloning. Wild-type and transgenic switchgrass plants (Alamo background) were propagated asexually by tiller-splitting and grown in a greenhouse under standard conditions (temperature range, 25–29 °C) with a 16-h day from 0600 to 2200 hours. Single tiller splits (all of similar size) of transgenic and control plants were grown in 1-gal pots with Metro-Mix 830 soil for 6 months. Plants were watered every week using tap water and fertilized monthly with 250 ml of 0.1 % Peters Professional Peat–Lite 20-10-20 fertilizer solution (Scotts, USA) in which nitrogen was supplied as 5.72 mM NH4NO3 and 2.86 mM KNO3 with a ratio of nitrate to ammonium of 60:40.
Arabidopsis ecotype Columbia-0 was used as wild type. An atnap (At1g69490) T-DNA insertion mutant (SALK_005010) was obtained from the Arabidopsis Biological Resource Center (Ohio State University). Homozygous mutant plants lines were confirmed as described previously [33]. Arabidopsis were grown in soil (Metro-Mix 350, Sun Gro Horticulture, Canada) in a controlled culture room at 23 °C with relative humidity of 55 % under long day (16-h light and 8-h dark) or 24-h light condition with white light illumination (120 μmol photons m−2 s−).
Isolation of Putative Switchgrass NAC cDNAs
Total RNA was extracted from switchgrass senescing leaves with yellow tips. cDNA was synthesized from 2 μg purified total RNA and used as template for 5′- and 3′-RACE (FirstChoice RLM-RACE Kit, Ambion, USA). The two nested degenerate primers for 5′-RACE were 5′-ACTCGTGCATGATCCAGTYKGT-3′ (outer primer) and 5′-KCGGGSWGAAGAAGTACCACTC-3′ (inner primer). The primers for 3′-RACE were 5′-TCGACCTCTACAAGTTCGAYCC-3′ (outer primer) and 5′-GCGAGMAGGAGTGGTACTTCTT-3′ (inner primer).
Sequence Alignment and Phylogenetic Analysis
RACE products were sequenced and the N-terminal portion of putative NAC protein sequences were aligned with several known NAC sequences using ClustalW of MegAlign (DNASTAR, Madison, USA) to confirm the presence of the conserved motifs of the NAC family. The full-length sequences of representative rice and Arabidopsis NAC proteins were first aligned with PvNAC1 and PvNAC2 using ClustalW with the default parameter values. The alignment results were then used to produce the neighbor-joining phylogenetic tree using MEGA5 [34]. The bootstrap method, with 100 replications, was used to provide confidence levels (reported as a percentage) of branch points on the phylogenetic tree.
Expression Profiling of PvNAC1 and PvNAC2 in Switchgrass
Switchgrass tillers at the developmental stages, vegetative 3, elongation 5, and reproductive 4 [35], were classified as young, medium, and old tillers, respectively. Transcript levels of PvNAC1 were measured by qRT-PCR in the first leaves (numbered from bottom to top) on different tillers and in leaves at different positions (top, intermediate, and bottom) on old tillers. Intermediate leaves detached from middle-aged tillers were incubated on wet filter paper in continuous darkness to artificially induce senescence prior to qRT-PCR analysis. Transcript levels of PvNAC1 were expressed relative to the transcript level of switchgrass Ubiquitin (GenBank accession JQ425118) in each sample.
Subcellular Localization
The open reading frame of PvNAC1 was PCR-amplified with gene-specific primers containing SalI and PciI enzyme sites and cloned into vector pAVA121 cut with XhoI and NcoI [36]. The resulting 35S:PvNAC1-GFP construct or the 35S:GFP control vector was transiently expressed in onion (Allium cepa) epidermal cells using particle bombardment [37]. Subcellular localization of the GFP signal was determined using a confocal laser scanning microscope (TCS SP2 AOBS; Leica).
Plasmid Constructs and Plant Transformation
Approximately 2.0 kb of AtNAP (At1g69490) promoter sequence (P NAP ) was amplified as described previously [22] to replace CaMV-35S promoter, via the PstI and NcoI sites of pCAMBIA3301 which contains GUS reporter gene driven by CaMV-35S promoter (35S-GUS) (http://www.cambia.org), thus resulting in P NAP -GUS construct.
P NAP -NAC1
The open reading frame (ORF) of PvNAC1 was amplified by PCR using a pair of gene-specific primers, 5′-(BglII) TTAGATCTATGGCGGTAAGCTCTGC-3′ (forward) and 5′-(BstEII) TAGGTCACCCTAGTGTTTTTTTCTTTCATATTTGAATTTG-3′ (reverse), and cloned into the P NAP -GUS construct via the BglII and BstEII restriction sites to replace GUS sequences.
pER-NAC1
The PvNAC1 ORF was cloned via AscI and SpeI sites into an estradiol-inducible vector pER8 [24] and named pER-NAC1. The pER8 vector containing GFP as reporter gene (pER-GFP) was used as a negative control vector.
35S-NAC2
The ORF of PvNAC2 was cloned into pCAMBIA1305 (http://www.cambia.org) via the BglII and BstEII restriction sites to replace GUS sequences.
Following the floral dip method [38], Arabidopsis homozygous mutant atnap was transformed with Agrobacterium tumefaciens strain C58 containing P NAP -GUS or P NAP -NAC1 for complementation testing (COM); Arabidopsis wild type (Col-0) was transformed with pER-GFP or pER-NAC1 for inducible overexpression (IOE).
Switchgrass cultivar Alamo was transformed with A. tumefaciens strain EHA105 containing 35S-NAC2 or 35S-GUS, following an established protocol [27].
Dark Incubation of Detached Arabidopsis Leaves and Estrogen Treatment on Whole Plants
Leaves (number 5 or 6) from 4-week-old plants of P NAP -NAC1 complementation transgenic, WT, and atnap Arabidopsis were excised and placed on filter papers moisturized with distilled H2O in Petri dishes with adaxial side facing up. The plates were kept in darkness at 22 °C for 5–7 days.
Estradiol treatments were conducted as previously described [24]. The detached leaves of Arabidopsis plants of pER-NAC1 transgenics and wild type were placed on filter paper moisturized with 100 μm EST (in 0.1 % DMSO) or 0.1 % DMSO solution in Petri dishes with adaxial side facing up. The plates were kept under 120 μmol white light at 22 °C for 5–7 days.
Three-week-old pER-PvNAC1 and pER-GFP transgenic as well as WT plants which were grown in soil under continuous light (120 μmol m−2 s−1) were sprayed with 100 μM EST (in 0.1 % DMSO) or 0.1 % DMSO solution every 3 h to ensure continuous wetting of foliage area. The sprayed plants were kept under a transparent plastic dome for 120 h. The phenotype of plants was observed, and leaves were harvested for physiological and molecular analysis.
Measurement of Chlorophyll Concentration
Arabidopsis leaves were ground to powder in liquid nitrogen. Chlorophyll was extracted from powdered samples with 80 % acetone, and chlorophyll concentration was determined by measuring absorption at 663 and 646 nm [39].
Measurement of Nitrogen Concentration
Leaves or seeds from inducible overexpression transgenic Arabidopsis plants were dried at 37 °C for at least 3 days, then ground into a fine powder using a SPEX SamplePrep 6870 Freezer/Mill (Metuchen, New Jersey, USA).
After 6 months of growth, whole plants of 35S-NAC2 transgenic switchgrass lines and a Tc were harvested. Aboveground biomass was separated into inflorescence (including seeds), leaves, and stems (including sheaths), while underground biomass was separated into roots and crowns (including rhizomes). Samples were dried at 65 °C for at least 2 days, weighed, and then ground into fine powder.
The concentration of total nitrogen in each sample was analyzed by Servi-Tech Laboratories (Amarillo, Texas, USA) using a combustion method [40] and expressed in percent (mg/100 mg). Nitrogen content in switchgrass samples was calculated by multiplying nitrogen concentration by biomass.
BLAST Search of Switchgrass Nitrogen Metabolism/Uptake Genes
The coding regions of maize genes for glutamine synthetase, GS1.4 (GenBank accession X65929), glutamate dehydrogenase, GDH1 (NM_001111831), glutamate synthase, GOGAT (NM_001112223), low-affinity nitrate transporter, NRT1.2 (NM_001112455), high-affinity nitrate transporter, NRT2.2 (NM_001279422), and ammonium transporter 2, AMT2 (NM_001154311), were used to search a Switchgrass Unique Transcript Database (http://switchgrassgenomics.noble.org). The top query results with complete coding regions were selected as switchgrass homologs of GS1, GDH, GOGAT, NRT1, NRT2, and AMT2 (Unitranscript IDs are listed in Table S3). Oligonucleotide primers for quantitative real-time PCR were designed based on conserved sequences in the switchgrass and maize homologs.
qRT-PCR
Transcript levels of PvNAC1, PvNAC2, and putative GS1.4, GDH1, and GOGAT in switchgrass leaves and PvNAC1 in transgenic Arabidopsis plants were determined with a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). As described previously [41], each 5-μL reaction included 2 μL primers (0.5 μM each primer), 2.5 μL SYBR Green Master Mix reagent (Applied Biosystems), and 0.5 μL 1:10 diluted cDNA from the reverse transcription step. The qRT-PCR data were analyzed using SDS 2.2.1 software (Applied Biosystems). The relative transcript levels were normalized to those of the constitutively expressed housekeeping gene, Arabidopsis UBQ10 (At4g05320) or switchgrass UBQ (GenBank accession JQ425118), using the equation 2−ΔCt, where Ct is the threshold cycle for each gene in every sample [42]. qRT-PCR was also conducted to confirm the microarray results on transcript changes of nitrogen metabolism genes due to PvNAC1 expression. The qRT-PCR primers are listed in Table S4.
Microarray Analysis
Total RNA samples from estradiol (100 μM, in 0.1 % DMSO)-treated IOE leaves (pER-NAC1 transgenic) were subjected to Affymetrix microarray analysis. To exclude genes that respond to treatment of DMSO or estradiol, RNAs from estradiol-sprayed pER-GFP leaves were used as the control. RNA was isolated with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol, then purified and concentrated using the RNeasy MinElute Cleanup Kit (Qiagen). Probe labeling, hybridization, and scanning were conducted by the Genomics/Microarray Facility at the Noble Foundation, according to the manufacturer’s instructions for the GeneChip 3′ IVT Express Kit (Affymetrix).
References
Parrish DJ, Fike JH (2005) The biology and agronomy of switchgrass for biofuels. Crit Rev Plant Sci 24:423–459
Ghimire S, Charlton N, Craven K (2009) The mycorrhizal fungus, Sebacina vermifera, enhances seed germination and biomass production in switchgrass (Panicum virgatum L). BioEnergy Res 2:51–58
Hallam A, Anderson IC, Buxton DR (2001) Comparative economic analysis of perennial, annual, and intercrops for biomass production. Biomass Bioenergy 21:407–424
Wullschleger SD, Davis EB, Borsuk ME, Gunderson CA, Lynd LR (2010) Biomass production in switchgrass across the United States: database description and determinants of yield. Agron J 102:1158–1168
Yang J, Worley E, Wang M, Lahner B, Salt D, Saha M, Udvardi M (2009) Natural variation for nutrient use and remobilization efficiencies in switchgrass. BioEnergy Res 2:257–266
Reynolds JH, Walker CL, Kirchner MJ (2000) Nitrogen removal in switchgrass biomass under two harvest systems. Biomass Bioenergy 19:281–286
Lemus R, Parrish D, Abaye O (2008) Nitrogen-use dynamics in switchgrass grown for biomass. Bioenerg Res 1:153–162
Lim PO, Kim HJ, Gil Nam H (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136
Gregersen PL, Holm PB, Krupinska K (2008) Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol 10:37–49
Kichey T, Hirel B, Heumez E, Dubois F, Le Gouis J (2007) In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Res 102:22–32
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301
Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, Grusak MA, Fett JP (2009) Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230:985–1002
Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585
Lin J-F, Wu S-H (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39:612–628
van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge U-I, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141:776–792
Andersson A, Keskitalo J, Sjodin A, Bhalerao R, Sterky F, Wissel K, Tandre K, Aspeborg H, Moyle R, Ohmiya Y, Bhalerao R, Brunner A, Gustafsson P, Karlsson J, Lundeberg J, Nilsson O, Sandberg G, Strauss S, Sundberg B, Uhlen M, Jansson S, Nilsson P (2004) A transcriptional timetable of autumn senescence. Genome Biol 5:R24
Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotech J 5:192–206
Jukanti AK, Heidlebaugh NM, Parrott DL, Fischer IA, McInnerney K, Fischer AM (2008) Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol 177:333–349
Cantu D, Pearce S, Distelfeld A, Christiansen M, Uauy C, Akhunov E, Fahima T, Dubcovsky J (2011) Effect of the down-regulation of the high Grain Protein Content (GPC) genes on the wheat transcriptome during monocarpic senescence. BMC Genomics 12:492
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280:547–563
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Sadras V (2000) Profiles of leaf senescence during reproductive growth of sunflower and maize. Ann Botany 85:187–195
Zuo J, Niu Q-W, Chua N-H (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24:265–273
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Masclaux-Daubresse C, Reisdorf-Cren M, Orsel M (2008) Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol 10(S1):23–36
Xi Y, Fu C, Ge Y, Nandakumar R, Hisano H, Bouton J, Wang Z-Y (2009) Agrobacterium-mediated transformation of switchgrass and inheritance of the transgenes. Bioenerg Res 2:275–283
Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, Tercé-Laforgue T, Quilleré I, Coque M, Gallais A, Gonzalez-Moro M-B, Bethencourt L, Habash DZ, Lea PJ, Charcosset A, Perez P, Murigneux A, Sakakibara H, Edwards KJ, Hirel B (2006) Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18:3252–3274
Lea PJ, Ireland RJ (1999) The enzymes of glutamine, glutamate, asparagine, aspartate metabolism. In: Singh BK (ed) Plant amino acid: biochemistry and biotechnology. Marcel Dekker, New York, pp 49–110
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Botany 105:1141–1157
Morgan JA, Parton WJ (1989) Characteristics of ammonia volatilization from spring wheat. Crop Sci 29:726–731
Parton WJ, Morgan JA, Altenhofen JM, Harper LA (1988) Ammonia volatilization from spring wheat plants. Agron J 80:419–425
Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27:521–549
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Moore KJ, Moser LE, Vogel KP, Waller SS, Johnson BE, Pedersen JF (1991) Describing and quantifying growth stages of perennial forage grasses. Agron J 83:1073–1077
von Arnim AG, Deng X-W, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221:35–43
Varagona MJ, Schmidt RJ, Raikhel NV (1992) Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4:1213–1227
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Porra R (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73:149–156
Horneck DA, Miller RO (1998) Determination of total nitrogen in plant tissue. In: Kalra YP (ed) Handbook of reference methods for plant analysis. vol CRC Press, Boca Raton, Boston, London, New York, Washington DC pp 75–83
Modolo L, Blount J, Achnine L, Naoumkina M, Wang X, Dixon R (2007) A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol 64:499–518
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Acknowledgments
We thank Prof. Arvid Boe (Plant Science Department, South Dakota State University) for generously providing seeds of switchgrass cultivar Summer and Dr. Albrecht von Arnim (Department of BCMB, University of Tennessee) for the gift pAVA121 vector. We also thank Jianfei Yun for the help with collecting switchgrass samples and caring for Arabidopsis plants. This work was supported by the Office of Biological and Environmental Research of the US Department of Energy via the BioEnergy Science Center (BESC) (grant number DE-PS02-06ER64304) and by the Samuel Roberts Noble Foundation.
Conflict of Interest
The authors have no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, J., Worley, E., Torres-Jerez, I. et al. PvNAC1 and PvNAC2 Are Associated with Leaf Senescence and Nitrogen Use Efficiency in Switchgrass. Bioenerg. Res. 8, 868–880 (2015). https://doi.org/10.1007/s12155-014-9566-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-014-9566-x