Abstract
Objective
Our aim is to validate the process steps implemented by the French CATI platform to assess amyloid status, obtained from 18F-Florbetapir PET scans, in a cohort of 318 cognitively normal subjects participating in the INSIGHT-preAD study. Our objective was to develop a method with partial volume effect correction (PVEC) on untransformed PET images, using an automated pipeline (“RACHEL”) adapted to large series of patients and including quality checks of results.
Methods
We compared RACHEL using different options (with and without PVEC, different sets of regions of interest), to two other methods validated in the literature, referred as the “AVID” and “CAEN” methods. A standard uptake value ratio (SUVR) was obtained with the different methods for participants to another French study, IMAP, including 26 normal elderly controls (NEC), 11 patients with mild cognitive impairment (MCI) and 16 patients with Alzheimer’s disease (AD). We determined two cutoffs for RACHEL method by linear correlation with the other methods and applied them to the INSIGHT-preAD subjects.
Results
RACHEL including PVEC and a combination of the whole cerebellum and the pons as a reference region allowed the best discrimination between NEC and AD participants. A strong linear correlation was found between RACHEL and the other two methods and yielded the two cutoffs of 0.79 and 0.88. According to the more conservative threshold, 19.8% of the INSIGHT-preAD subjects would be considered amyloid positive, and 27.7% according to the more liberal threshold.
Conclusions
With our method, we clearly discriminated between NEC with negative amyloid status and patients with clinical AD. Using a linear correlation with other validated cutoffs, we could infer our own positivity thresholds and apply them to an independent population. This method might be useful to the community, especially when the optimal cutoff could not be obtained from a population of healthy young adults or from correlation with post-mortem results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
There is growing evidence that the pathophysiological process of Alzheimer’s disease (AD) begins several decades before the clinical diagnosis of AD [1]. Within the continuum of the disease, the symptomatic stages (including prodromal and dementia stages) are preceded by a preclinical phase characterized by normal cognitive functioning in the presence of pathophysiological biomarkers of AD that can now be assessed in vivo [2].
CATI (Centre d’acquisition et traitement des images, cati-neuroimaging.com) is the French platform that was funded in 2010 with the aim of supporting multicentre clinical trials involving neuroimaging [3]. The main project devoted to the CATI was to handle the multimodality imaging aspect of MEMENTO (http://clinicaltrials.gov/show/ NCT01926249), a large French multicentric study aiming at better understanding the natural history of AD, and to develop post-processing quantification tools for MRI and PET biomarkers.
The objective of this paper is to present and validate the process steps implemented by the CATI to assess amyloid status, obtained from Florbetapir PET scans, in participants of the INSIGHT-preAD study, an ancillary study of MEMENTO. It is a mono-centric French cohort at the Pitié-Salpêtrière Hospital in Paris including 318 cognitively normal elderly individuals, with subjective memory complaints (SMC) and with defined brain amyloid status. The objective of the INSIGHT-preAD study is to determine the best biomarker/biomarker combination or the best algorithm for predicting the secondary occurrence of prodromal AD or cognitive decline in the subjects with positive PET amyloid imaging.
Amyloid burden is usually quantified by calculating a mean standard uptake value ratio (SUVR) between a set of cortical regions of interest (ROIs) most affected by amyloid pathology, and a reference region [4]. Various options exist for the choice of cortical and reference ROIs, as well as for the analysis method. Quantification is often done in Montreal Neurological Institute (MNI) space, using a PET or MRI template. Our aim was to develop a method including partial volume effect correction (PVEC) on untransformed PET images, to reduce possible quantification biases related to spatial normalization or co-registration, using an automated pipeline, “RACHEL”, adapted to a large series of patients and including a quality check of the results.
To validate the quantification processing developed by the CATI, we compared it to two reference methods validated in the literature, using data from another French study: the “Imagerie Multimodale de la maladie d’Alzheimer à un stade Précoce” (IMAP) study (Caen, France) [5, 6]. These data included normal amyloid-negative elderly controls and AD patients, allowing comparisons of the discriminating power of the different methods.
Finally, as the SUVR positivity threshold for amyloid status depends not only on the radiotracer but also the quantification method, we had to determine the threshold for the method chosen by the CATI.
Materials and methods
Subjects
INSIGHT-preAD study group
INSIGHT-preAD is an on going and mono-centric cohort study from the Salpêtrière Hospital, Paris, France, which started on 25th May 2013. The cohort includes cognitively normal individuals, over 70 years, with subjective memory complaints (SMC) but normal cognitive and memory scores according to the Mini Mental State Examination (MMSE ≥ 27), Clinical Dementia Rating (CDR = 0) and Free and Cued Selective Reminding Test (Total Recall ≥ 41). Demographic, cognitive, functional, ApoE status, genomics, MRI (anatomical, diffusion, resting state-fMRI, arterial spin labeling sequences), FDG-PET imaging, EEG recordings with resting state and ERP, were performed at baseline with optional Actigraphy and CSF investigations. All subjects participate in follow-up with neuropsychological assessment every 6 months; EEG, Actigraphy every year; blood samplings for research on biomarkers, MRI, FDG-PET and amyloid-PET scans every 2 years. The demographic data are presented in Table 1. The main characteristics of the INSIGHT-preAD cohort are: a mean age of 76 (± 3.5) with a mean MMSE score of 28.7 (± 0.96), a predominance of females (63.2%) and a high level of education. Sixty-two subjects (19.5%) were APOE-ε4 carriers.
IMAP study group
To validate our methods of quantification, we applied them to Florbetapir amyloid PET images collected in Caen for IMAP (PI: Gael Chételat & Vincent de la Sayette): 26 amyloid-negative normal elderly controls, 11 patients with mild cognitive impairment (MCI) and 16 patients with AD. The IMAP study’s objectives and the participants have been previously described [5, 6]. The IMAP study was approved by a regional ethics committee (Comité de Protection des Personnes Nord-Ouest III) and is registered with http://clinicaltrials.gov (number NCT01638949). All participants gave written informed consent to the study prior to the investigation.
MRI and PET acquisitions
INSIGHT-preAD study group
In Pitié-Salpêtrière centre, as in all nuclear medicine centres participating in the MEMENTO study, the CATI determined PET acquisition parameters using Jaszczack’s and 3D-Hoffman’s phantoms measurements prior to patient studies, with the aim of harmonizing the contrast recovery while preserving, if possible, the spatial resolution. The methods and results for this optimization have been detailed in a previous paper [7].
All amyloid PET scans were acquired in a single session on a Philips Gemini GXL CT-PET scanner 50 (± 5) min after the injection of approximately 370 MBq (333–407 MBq) of 18F-Florbetapir (AVID radiopharmaceuticals). PET acquisition consisted of 3 × 5-min frames, in a 128 × 128 acquisition matrix, with a voxel size of 2 × 2 × 2 mm3. Images were then reconstructed using the iterative LOR-RAMLA algorithm (10 iterations). Reduction of noise modulated the relaxation parameter lambda, which was set at 0.7. All corrections (attenuation, scatter and random coincidence) were integrated in the reconstruction.
MRI scans were acquired on a Siemens Verio 3T scanner using a 3D TurboFLASH sequence (orientation sagittal; repetition time 2300 ms; echo time 2.98 ms; inversion time 900 ms; flip angle 9°; 176 slices; slice thickness 1 mm; field of view 256 × 240 mm2; matrix 256 × 240; bandwidth 240 Hz/Px).
MRI and PET-CT data were securely forwarded to the CATI via the Imagys Cloud platform (http://www.keosys.com). A quality check was then performed by the CATI team: frames were realigned, averaged and visually inspected for possible artefacts, such as subject motion, mismatch between CT and emission scans and attenuation correction artefacts. Automated software allowed for the extraction of information from the DICOM headers, which were checked for compliance with acquisition and reconstruction protocols.
IMAP study group
For the IMAP subjects from Caen, MRI scans were acquired on a Philips Achieva 3T scanner using a 3D fast-field echo sequence (sagittal; repetition time 20 ms; echo time 4.6 ms; flip angle 10°; 180 slices; slice thickness 1 mm; field of view 256 × 256 mm2; matrix 256 × 256). PET scans were acquired on a GE Health-care Discovery RX VCT 64 PET-CT device (resolution 3.76 × 3.76 × 4.9 mm; voxel size 1.95 × 1.95 × 3.27 mm3). A 20-min PET scan was acquired 50 min after intravenous injection of 4 MBq/kg of Florbetapir.
Florbetapir PET image processing
Three methods of ROI-based analysis were compared. The first was the semi-automated quantitative analysis (cortical-to-cerebellar SUVR) used by Clark et al. [8, 9] to compare Florbetapir results to the presence of neuritic amyloid plaques on autopsy. The description and materials (in particular, template and ROI masks) were kindly provided by the AVID Company. This method is referred to as the “AVID method” in the paper. The second method was developed in INSERM U1077 research laboratory in Caen, and will be referred to as the “CAEN method”. The third method is the method developed by the CATI and will be referred to as “RACHEL software”.
AVID method
This method has been largely used in the literature and will be briefly described here. All images were normalized to a PET amyloid template in MNI space using statistical parametric mapping (SPM) software (Wellcome Trust Centre for Neuroimaging, London, UK; http://www.fil.ion.ucl.ac.uk/spm/), and cortical-to-cerebellum SUVR were calculated from the average activity in 6 small target cortical ROIs (average of right and left: frontal, temporal, parietal, anterior cingulate, posterior cingulate and precuneus), using the whole cerebellum as a reference region [10].
CAEN method
IMAP data were analysed in Caen using a method described in previous publications from the group [5, 6, 11, 12]. Briefly, MRI 3D T1-weighted images were segmented into grey and white matter using the VBM5 toolbox (http://dbm.neuro.unijena.de/vbm/vbm5-for-spm5/) and spatially normalized into the MNI space. Florbetapir PET images were co-registered onto the corresponding MRI, corrected for partial volume effects using PMOD software (http://www.pmod.com), and spatially normalized into the MNI space using the deformation parameters obtained from the normalization of the MRI using the VBM5.1 toolbox (http://dbm.neuro.uni-jena.de) implemented in SPM5 software. The mean Florbetapir value in the cerebellum grey matter was extracted for each subject from the normalized PET images. Each Florbetapir image was then divided by its corresponding mean cerebellar value, resulting in Florbetapir-PET SUVR data. The global neocortical Florbetapir-PET SUVR value was then computed for each subject from the Florbetapir-PET SUVr data using a neocortex mask encompassing three right and three left ROIs: one including frontal, temporal, and anterior cingulate cortex, another one including precuneus and posterior cingulate cortex, the last one in parietal cortex).
RACHEL software
MRI 3D T1-weighted images were segmented and spatially normalized into the MNI space using the VBM8 package (http://dbm.neuro.uni-jena.de/vbm/) implemented in SPM8. Deformation fields and grey and white matter masks were generated. Structural MRI images were co-registered to Florbetapir-PET images using SPM8 with visual inspection to detect any co-registration errors. We created in MNI space a set of 6 right and 6 left cortical regions derived from AVID ones, but slightly larger (in particular, the precuneus). Using inverse deformation fields and matrix transformation, composite cortical ROIs and a reference region were placed in the individual native PET space. We then applied a PVEC algorithm, the RBV-sGTM method [13], that performs a region-based voxel-wise (RBV) correction of the entire image, using the anatomical parcellation of MRI scans and an accurate measure of the point spread function of the PET scanner (full width at half maximum: 8 mm). Parametric PET images were then created for each individual, by dividing each voxel with the mean activity extracted from the reference region. Finally, (SUVRs) were calculated by averaging the mean activity of all cortical ROIs in the individual PET native space.
All these steps were done with an automated pipeline developed with BrainVISA software (http://brainvisa.info/web/index.html) by the CATI engineers and were quality-checked by the CATI team. Figure 1 shows the different processing steps implemented in RACHEL. Figures 2 and 3 show the software interface used for the final quality checks.
We tested on IMAP images several sets of cortical regions (from the AVID, CAEN and CATI methods, and AAL atlas), scaled with different reference regions: white only, grey only or whole cerebellum, pons, or a combination of 2 or 3 of them. We also tested the effect of PVEC. As the discrimination of amyloid PET imaging between AD and amyloid-negative controls is perfect whatever the method, we used a bootstrap analysis to estimate confidence intervals of the difference between AD and amyloid-negative controls to classify the methods. Differences between the first quartile of AD group and the third quartile of controls group were calculated for all pipelines. The higher the difference is, the better the method is to discriminate both groups. One thousand bootstrap samples, with respect of the initial proportions between AD and controls size samples were then generated and quartile difference was evaluated for each bootstrap sample and each method. 95% confidence intervals for quartile difference were assessed on the distribution obtained by the bootstrap samples. Finally, to investigate the concordance between AVID, CAEN and RACHEL methods, we calculated the intraclass correlation coefficients.
SUVR threshold determination
Two SUVR cutoffs to determine the negative or positive amyloid status of the INSIGHT-preAD participants were extracted performing a linear correlation first between RACHEL and AVID SUVR, and second between RACHEL and CAEN SUVR, using PET images from the IMAP cohort.
The positivity threshold associated with the CAEN method was defined as the 90th percentile of the Florbetapir PET values estimated in an independent group of 26 healthy controls from the IMAP project (8 females; age 31 ± 8.4; education level 13.6 ± 2.5; MMSE 29.4 ± 0.64; 29% APOE 4 carriers), corresponding to a SUVR of 1.005 [5]. Thus, all subjects with an SUVR superior to 1.005 were considered positive.
The positivity threshold associated with the AVID method was defined as the confidence limit for the upper 5% of the SUVR distribution based on 2 groups of 10 and 11 healthy young controls (age interval 38–52 years) and corresponded to a value of 1.10 [10]. Thus, all subjects with an SUVR superior to 1.10 were considered positive.
Analysis of INSIGHT-preAD Florbetapir PET images
Finally, all Florbetapir PET images from the INSIGHT-preAD cohort were analysed with the “best” CATI method (defined as the method allowing the best discrimination between controls and AD patients). The number of positive and negative scans was calculated with the 2 cutoffs obtained as mentioned above, as well as the number of scans with SUVR values between the 2 thresholds (defined as “intermediate”). We also investigated a potential correlation between the SUVR and age or MMSE in the INSIGHT-preAD population (Pearson’s correlation test). Subgroups with negative, positive or intermediate amyloid status were compared for age, (ANOVA test), gender (χ 2 test), MMSE, FCSRT scores and level of education (Kruskal–Wallis test). Post hoc analyses were performed if pertinent, with Bonferroni correction for multiple corrections.
Results
SUVR calculations
The SUVR values for each IMAP group and different methods (AVID, CAEN and different ROI combinations of RACHEL) are presented in Fig. 4 and Table 2.
SUVRs obtained with a the AVID method, b the Caen method, c the CATI method without PVEC, d the CATI method with PVEC. For each method, three groups are plotted: patients with clinical Alzheimer’s disease (AD) or mild cognitive impairment (MCI) and elderly healthy controls (HCs). The CATI method with PVEC is the one allowing the best discrimination between HC and AD groups
The best discrimination between normal controls and AD patients was obtained with RACHEL using PVEC, the set of 6 right and left cortical ROIs created by CATI, and a combination of the whole cerebellum and pons as a reference region. The interval between the lowest SUVR value in AD patients and the highest SUVR value in amyloid-negative healthy controls was 0.30 units, while it was 0.08 for the AVID method and 0.22 with the CAEN method. As expected, MCI patients had a large range of negative and positive SUVR values. The results of bootstrap analysis are shown in Fig. 5. The quartile difference between the 2 groups was the smallest for AVID method and the largest for RACHEL and CAEN, with a slight superiority of CAEN method. However, the confidence interval at 95% was smaller for RACHEL method, suggesting that it is more robust. Intraclass correlation coefficient for RACHEL, CAEN and AVID methods was 0.84 [0.76–0.90], suggesting an excellent agreement between them.
Bootstrap analysis results (1000 samples). Differences values (represented by the points) between the first quartile Q 0.25 of AD group and the third quartile Q 0.75 of healthy controls group, and their 95% confidence intervals on the distribution obtained by the bootstrap samples (represented by the transversal bars), are shown for all tested methods. The abbreviations for the different methods are given in the legend of Table 2. The largest difference value and thus the best discrimination between AD and controls is observed for CATI_PVEC and CAEN. CATI_PVEC has a smaller confidence interval, suggesting a better robustness
Determination of positivity thresholds
A strong correlation was found between SUVRs calculated with RACHEL and AVID methods (p < 0.001; r = 0.90), and RACHEL and CAEN methods (p < 0.001; r = 0.92).
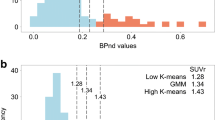
The threshold of 1.10 with the AVID method yielded, by linear interpolation, a value of 0.88 with RACHEL software. The threshold of 1.005 with the CAEN method yielded a value of 0.79 with RACHEL software. These results are shown in Fig. 6.
Analysis of Florbetapir scans in the INSIGHT-prAD population
The analysis of Florbetapir scans from the INSIGHT-preAD cohort with RACHEL and taking into account the 2 cutoffs yielded the following results:
-
Using the more conservative cutoff obtained from the AVID method, 63 subjects (19.8%) with an SUVR > 0.88 would be considered as amyloid positive.
-
Using the more liberal cutoff obtained from the CAEN method, 88 subjects (27.7%) with a SUVR > 0.79 would be considered amyloid positive.
-
25 subjects (7.9%) had an SUVR between 0.79 and 0.88.
Based on these results, we could define 3 groups: two “amyloid-negative” (n = 230) and “amyloid-positive” (n = 63) groups regardless of the chosen cutoff, and one “intermediate” group (n = 25) that could be considered amyloid positive or negative according to the choice of cutoff (liberal or conservative).
Within the whole population of the INSIGHT-preAD cohort, we found a significant correlation between SUVR and age (p = 0.0093, r = 0.15), but not with MMSE scores. Comparisons of age, gender, MMSE and sociocultural level between the 3 groups are presented in Table 1. Kruskal–Wallis test showed a significant difference between the 3 groups for age (p = 0.020) and MMSE scores (p = 0.033), but not for sociocultural level. Post hoc tests showed that only MMSE scores were significantly different between the amyloid-negative and -positive groups (p = 0.028), but this difference was no longer significant after adjusting for age.
Discussion
Choice of quantification method
In this work, we tested and validated a method of quantification of the amyloid load with 18F-Florbetapir PET images, with a partial volume effect correction, and a slightly different set of ROIs than the other methods. Our aim was to reduce quantification biases related to spatial normalization, co-registration and atrophy. We developed in-house software, RACHEL, allowing the amyloid load in grey matter of untransformed PET images to be quantified, with high throughput and a step-by-step quality check. We tested several variants of our method, compared them to two validated methods of the literature and showed that our approach was correct since it allowed the best discrimination between amyloid-negative elderly controls and clinical AD patients.
The AVID method uses a PET template for spatial normalization that is built from both negative and positive amyloid images. For some images, after normalization into MNI space, we observed a distortion of edges on upper brain slices. In addition, some subjects were clearly misclassified, according to visual assessment with the fusion to MRI images, because of cortical atrophy that yielded false results. As MRI acquisitions were available in the INSIGHT-preAD cohort, we chose to use an MRI template for normalization into MNI space and PVEC to address these two issues. We also performed the ROI analysis on native untransformed PET images to reduce potential bias effects of the processing steps, especially spatial normalization, on SUVR values. However, errors generated by the inverse transformation of ROIs, in particular from interpolation, cannot be completely avoided.
The CAEN method is close to our method, as it also relies on an MRI template and PVEC. But it uses PMOD software, which is not suited for processing large cohorts, but rather one subject at a time, and is, therefore, time consuming. We developed in-house software, RACHEL, that allows for iteratively processing an unlimited number of scans, with special attention to include a display of the results at each step for a quality check. Another difference between the CAEN and CATI methods is that PMOD only extracts grey matter probability maps from MRI images, while the CATI method also extracts white matter maps. We used the white matter maps on reference regions, such as the whole cerebellum or pons. We also plan to use them later to analyse the follow-up scans, as it has been suggested that a white matter reference would be best suited for the longitudinal evaluation of amyloid burden [14,15,16].
Finally, the method that allowed the best and robust discrimination between normal elderly amyloid-negative subjects and clinical AD patients was RACHEL software, using a set of cortical regions close to the ones from the AVID method, but larger in size: after inverse transformation of the ROIs from MNI to native space and applying a grey matter mask, the AVID ROIs would include too few voxels, especially in small regions as the precuneus or posterior cingulate cortex. Indeed, quantification in small ROIs is more sensitive to a slight mismatch with PET images, and is thus less robust. We also found that the best reference region associated with our method was the combination of the whole cerebellum and pons regions. The whole cerebellum is the reference region used in the AVID method, and the pons has been validated as the reference region for another amyloid ligand, the 18F-Flutemetamol [17].
Determination of positivity cutoffs for amyloid status
There are several ways of determining the amyloid positivity status of a given ligand and a given quantification method. One is to compare the amyloid PET SUVR to the presence of neuritic amyloid plaques on autopsy [8, 9], which is obviously not readily available. Another and perhaps better one is to measure the SUVR in a population of normal subjects with a very low probability of having amyloid plaques, such as young normal adults not carrying APOE4 ε4 [8]. The positivity cutoff can subsequently be inferred from the confidence limit (CI) for the upper 5% of the distribution, yielding a 1.10 cutoff for the AVID method [10]), or the 90th percentile of the values yielding a 1.005 cutoff for the CAEN method [5].
Another possibility is to obtain a positivity cutoff by linear correlation between the values obtained with their method and those obtained with a method of a reference with a known and validated cutoff. Several previous studies have shown that positivity cutoffs could be reliably converted between tracers and processing methods using the linear association across subjects [16, 18, 19]. Indeed, we found a strong linear relationship between RACHEL’s SUVR values and AVID’s or CAEN’s values and could infer a 0.88 cutoff value from AVID’s one and a 0.79 cutoff value from CAEN’s one. These cutoff values are lower than for the other methods, mainly because we added the pons (which is a hot region) to our reference region. A low cutoff value (0.62) has been previously validated with post-mortem results for the 18F-Flutemetamol, using pons only as reference region [17].
As we finally obtained 2 cutoff values, we leave to the INSIGHT-preAD investigators the choice between a more liberal or conservative threshold, according to their need for sensitivity or specificity in a given situation, as suggested previously by Jagust [20]. The conservative threshold would allow the INSIGHT-preAD results to be compared with those of other cohorts, for which AVID’s method and 1.10 cutoff was mostly used. However, a liberal cutoff could be justified in view of recent statistical and post-mortem data suggesting that existing thresholds are too high [21]. Another possibility is to consider the scans with values between the two cutoffs as “intermediate” scans. There has been accumulating evidence in the literature that elderly cognitively normal subjects may have ambiguous scans, reflecting both a lower number of ROIs with elevated amyloid uptake and lower SUVR values in these regions than the clearly positive cases [11, 22]. Clinical and neuroimaging follow-up will be necessary to assess the risk of conversion in these individuals [11].
Amyloid status in the INSIGHT-preAD cohort
According to a more conservative or liberal threshold, the percentage of amyloid-positive scans was 19.8 or 27.7% in the INSIGHT-preAD population, and 7.8% of the subjects could also be classified with an intermediate status. Percentages of positive amyloid scans between 20 and 30% are consistent with the previous ones reported in the literature in normal elderly controls [23, 24], and in particular in non-ApoE-ε4 normal controls [25]. Indeed, although INSIGHT-preAD participants had subjective memory complaints, they were recruited on the basis of their absence of objective cognitive deficits.
As previously observed in other cohorts, we found a significant correlation between amyloid burden and age in the cohort, but not with MMSE scores [25].
Conclusion
In this work, we evaluated a method of the quantification of amyloid burden, with partial volume effect correction and SUVRs calculated in ROIs of the native space of the untransformed PET images. Using this method, we could clearly discriminate between normal elderly controls with negative amyloid status and patients with clinical AD. For that purpose, we developed in-house software that allows an unlimited number of PET scans to be processed and includes quality checks of results. Using a linear correlation with other methods that yielded a validated cutoff of amyloid positivity, we could infer our own positivity thresholds and apply them to an independent population of 318 normal elderly subjects. This method might be useful to the community, especially when the optimal cutoff could not be obtained from a population of healthy young adults or from correlation with post-mortem results. It will be applied to the amyloid PET scans collected in the MEMENTO study.
References
Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–16.
Dubois B, Hampel H, Feldman HH, Scheltens P, Aisen P, Andrieu S, et al. Preclinical Alzheimer’s disease: definition, natural history, and diagnostic criteria. Alzheimers Dement. 2016;12(3):292–323.
Operto G, Chupin M, Batrancourt B, Habert MO, Colliot O, Benali H, et al. CATI: A large distributed infrastructure for the neuroimaging of cohorts. Neuroinformatics. 2016;14(3):253–64.
Wong DF, Rosenberg PB, Zhou Y, Kumar A, Raymont V, Ravert HT, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (Florbetapir [corrected] F 18). J Nucl Med. 2010;51(6):913–20.
Besson FL, La Joie R, Doeuvre L, Gaubert M, Mézenge F, Egret S, et al. Cognitive and brain profiles associated with current neuroimaging biomarkers of preclinical Alzheimer’s disease. J Neurosci. 2015;35(29):10402–11.
La Joie R, Landeau B, Perrotin A, Bejanin A, Egret S, Pélerin A, et al. Intrinsic connectivity identifies the hippocampus as a main crossroad between Alzheimer’s and semantic dementia-targeted networks. Neuron. 2014;81(6):1417–28.
Habert MO, Marie S, Bertin H, Reynal M, Martini JB, Diallo M, et al. Optimization of brain PET imaging for a multicentre trial: the French CATI experience. EJNMMI Phys. 2016;3(1):6.
Clark CM, Schneider JA, Bedell BJ, Beach TG, Bilker WB, Mintun MA, et al. Use of Florbetapir-PET for imaging beta-amyloid pathology. JAMA. 2011;305(3):275–83.
Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, et al. Cerebral PET with Florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-β plaques: a prospective cohort study. Lancet Neurol. 2012;11(8):669–78.
Joshi AD, Pontecorvo MJ, Clark CM, Carpenter AP, Jennings DL, Sadowsky CH, et al. Performance characteristics of amyloid PET with Florbetapir F 18 in patients with Alzheimer’s disease and cognitively normal subjects. J Nucl Med. 2012;53(3):378–84.
Chételat G, La Joie R, Villain N, Perrotin A, de La Sayette V, Eustache F, Vandenberghe R. Amyloid imaging in cognitively normal individuals, at-risk populations and preclinical Alzheimer’s disease. Neuroimage Clin. 2013;2:356–65.
La Joie R, Perrotin A, Barré L, Hommet C, Mézenge F, Ibazizene M, et al. Region-specific hierarchy between atrophy, hypometabolism, and β-amyloid (aβ) load in Alzheimer’s disease dementia. J Neurosci. 2012;32(46):16265–73.
Thomas BA, Erlandsson K, Modat M, Thurfjell L, Vandenberghe R, Ourselin S, Hutton BF. The importance of appropriate partial volume correction for PET quantification in Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2011;38(6):1104–19.
Brendel M, Högenauer M, Delker A, Sauerbeck J, Bartenstein P, Seibyl J, et al. Improved longitudinal [(18)F]-AV45 amyloid PET by white matter reference and VOI-based partial volume effect correction. Neuroimage. 2015;108:450–9.
Chen K, Roontiva A, Thiyyagura P, Lee W, Liu X, Ayutyanont N, et al. Improved power for characterizing longitudinal amyloid-β PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med. 2015;56(4):560–6.
Landau SM, Thomas BA, Thurfjell L, Schmidt M, Margolin R, Mintun M, et al. Amyloid PET imaging in Alzheimer’s disease: A comparison of three radiotracers. Eur J Nucl Med Mol Imaging. 2014;41(7):1398–407.
Thurfjell L, Lilja J, Lundqvist R, Buckley C, Smith A, Vandenberghe R, Sherwin P. Automated quantification of 18f-flutemetamol PET activity for categorizing scans as negative or positive for brain amyloid: concordance with visual image reads. J Nucl Med. 2014;55(10):1623–8.
Vandenberghe R, Van Laere K, Ivanoiu A, Salmon E, Bastin C, Triau E, et al. 18F-flutemetamol amyloid imaging in Alzheimer disease and mild cognitive impairment: a phase 2 trial. Ann Neurol. 2010;68(3):319–29.
Villemagne VL, Mulligan RS, Pejoska S, Ong K, Jones G, O’Keefe G, et al. Comparison of 11c-pib and 18f-florbetaben for aβ imaging in ageing and Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2012;39(6):983–9.
Jagust WJ. Amyloid imaging: Liberal or conservative? Let the data decide. Arch Neurol. 2011;68(11):1377–8.
Villeneuve S, Rabinovici GD, Cohn-Sheehy BI, Madison C, Ayakta N, Ghosh PM, et al. Existing Pittsburgh compound-b positron emission tomography thresholds are too high: statistical and pathological evaluation. Brain. 2015;138(Pt 7):2020–33.
Mormino EC, Brandel MG, Madison CM, Rabinovici GD, Marks S, Baker SL, Jagust WJ. Not quite PIB-positive, not quite PIB-negative: Slight PIB elevations in elderly normal control subjects are biologically relevant. Neuroimage. 2012;59(2):1152–60.
Pike KE, Savage G, Villemagne VL, Ng S, Moss SA, Maruff P, et al. Beta-amyloid imaging and memory in non-demented individuals: evidence for preclinical Alzheimer’s disease. Brain. 2007;130(Pt 11):2837–44.
Aizenstein HJ, Nebes RD, Saxton JA, Price JC, Mathis CA, Tsopelas ND, et al. Frequent amyloid deposition without significant cognitive impairment among the elderly. Arch Neurol. 2008;65(11):1509–17.
Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, et al. Amyloid imaging results from the Australian imaging, biomarkers and lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31(8):1275–83.
Acknowledgements
We sincerely thank Alain Giron for helping with the statistical analyses and Anne Bertrand for her input on the manuscript. INSIGHT-AD study group Audrain C, Bakardjian H, Benali H, Bertin, H, Boukadida L, Cacciamani F, Causse-Lemercier V, Cavedo E, Chiesa P, Colliot O, Dos Santos A, Dubois B, Durrleman S, Epelbaum S, Gagliardi G, Genthon R, Habert M-O, Hampel H, Jungalee N, Kas A, Lehericy S, Lamari F, Letondor C, Levy M, Lista S, Mochel F, Nyasse F, Poisson C, Potier MC, Revillon M, Rojkova K, Roy P, Santos-Andrade K, Santos A, Simon V, Sole M, Tandetnik C, Thiebaud De Schotten M.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Research involving human participants
A national ethics committee approved the INSIGHT study (ANSM 130134B-31), and all subjects gave written informed consent before their inclusion in the study. The IMAP study was approved by a regional ethics committee (Comité de Protection des Personnes Nord-Ouest III) and all participants gave written informed consent to the study prior to the investigation.
Funding
The INSIGHT-preAD study is supported by the IHU-A-ICM, Investissement d’Avenir from the French Ministry of Health, the French Foundation Plan-Alzheimer, Pfizer and AVID/Lilly companies. The French Foundation Plan-Alzheimer supports the CATI. Harald Hampel is supported by the AXA Research Fund, the Fondation Université Pierre et Marie Curie and the “Fondation pour la Recherche sur Alzheimer”, Paris, France. The research leading to these results has received funding from the program “Investissements d’avenir” ANR-10-IAIHU-06 (Agence Nationale de la Recherche-10-IA Institut Hospitalo-Universitaire-6).
Additional information
Members of INSIGHT-AD study group are listed in acknowledgements.
Rights and permissions
About this article
Cite this article
Habert, MO., Bertin, H., Labit, M. et al. Evaluation of amyloid status in a cohort of elderly individuals with memory complaints: validation of the method of quantification and determination of positivity thresholds. Ann Nucl Med 32, 75–86 (2018). https://doi.org/10.1007/s12149-017-1221-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-017-1221-0