Abstract
Climate regionalization is an important but often under-emphasized step in studies of climate variability. While most investigations of regional climate make at least an implicit attempt to focus on a study region or sub-regions that are climatically coherent in some respect, rigorous climate regionalization––in which the study area is divided on the basis of the most relevant climate metrics and at a resolution most appropriate to the data and the scientific question––has the potential to enhance the precision and explanatory power of climate studies in many cases. To facilitate the application of rigorous regionalization for climate studies, we introduce an improved hierarchical clustering method, describe a new open-source R package designed specifically for climate regionalization, and offer concise suggestions for performing appropriate regionalization. This paper describes the regionalization algorithms and presents a demonstration application in which the R package is used to regionalize Africa on the basis of interannual precipitation variability. Both the proposed methodology and the R package can be used for a broad range of applications and over different areas of the globe.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Climate regionalization is the process of dividing an area into smaller regions that are homogeneous with respect to a specified climatic characteristic. Regionalization is a fundamental exercise in climate studies because it enables us to distinguish between the mechanisms responsible for spatio-temporal variability specific to each region (e.g., Dezfuli and Nicholson 2013; Nicholson and Dezfuli 2013). Methodologically, climate regionalization is typically achieved using some form of objective multivariate statistical technique. Cluster Analysis (CA) of various types has been applied widely for this purpose (e.g., Burn 1989; Gong and Richman 1995; Ramachandra Rao and Srinivas 2006; Isik and Singh 2008; Dezfuli 2011). CA methods are divided into two main categories: hierarchical and nonhierarchical (or flat) methods (Jain et al. 1999), and hierarchical clustering in turn includes two different approaches: agglomerative (or bottom-up) and divisive (or top-down).
Each clustering technique has its own advantages and shortcomings, and there is no clear consensus in the literature regarding the best-performing method (Manning et al. 2008). Recent studies, however, have identified a number of advantages of agglomerative hierarchical clustering (AHC) relative to other methods. First, AHC offers an easily understandable cluster definition process that successively merges the most similar members (or small-size clusters). The divisive hierarchical approach is harder to trace, as it splits clusters using a flat clustering algorithm (Cimiano et al. 2004; Manning et al. 2008). Second, AHC methods are more deterministic, informative and predictable than the highly variable nonhierarchical methods that return unstructured set of clusters, and “often converge to a local optimum of poor quality” (Manning et al. 2008). Third, AHC methods facilitate validating clusters (Fovell and Fovell 1993; Dezfuli 2011), which is perhaps the most challenging part of the cluster analysis (Jain and Dubes 1988). This issue is more difficult to address in nonhierarchical methods that require a prespecified number of clusters.
AHC methods are not without their limitations. For example, all commonly used AHC algorithms are unidirectional: once individual members are merged into a region they cannot be reassigned. AHC algorithms can also be negatively affected by noise in the input data––a problem that is not negligible in most climate datasets. However, previous studies have shown that the problem of noise can be significantly reduced when CA methods are used in conjunction with the principal component analysis (PCA, Baeriswyl and Rebetez 1997; Busuioc et al. 2001; Argüeso et al. 2011; Dezfuli 2011). In this approach, PCA is applied to raw data, and the leading PCs that together explain a large fraction of the variance are retained and used as the inputs of cluster model. The PCs are sometimes rotated, using orthogonal or oblique methods, in order to better identify the dominant modes of variability (e.g., Munoz-Diaz and Rodrigo 2004; Rogers and McHugh 2002; Dezfuli 2011). The choice of rotation method and number of PCs retained are known to affect the stability of clustering results (White et al. 1991; Comrie and Glenn 1998).
For climate regionalization, criteria used to evaluate cluster validity may vary with the analysis objectives. Here, we adopt a set of criteria suggested by Dezfuli (2011) that a satisfactory level is reached primarily when the regions are homogeneous and geographically contiguous, the size of regions is consistent with problem-specific size constraints (e.g., landscape structure, data coverage and density, known climate phenomena), and the total number of regions is consistent with the inherent physical properties of interest. We should emphasize that climate regionalization is a combined physical-statistical problem, so that the optimum solution involves some subjective decisions such as examining the contiguity or geographical characteristics of regions. The objective criteria, however, are often met by simultaneously minimizing the inter-regional correlations (i.e., correlations between clusters) and maximizing the intra-regional correlations (i.e., correlations between the mean of each cluster and its members).
In summary, there is a diversity of climate regionalization techniques in the literature, and these techniques are sensitive to conceptual approach, clustering algorithm, data processing, and validation criteria. The density of the clustering literature and the lack of easily accessible, climate-oriented clustering software tools presents a barrier to the utilization of objective regionalization in the climate science community and makes it difficult to compare across studies. This is our motivation for developing a flexible and clearly documented software tool designed for climate regionalization, which we have implemented as an open-source R package for hierarchical cluster-based climate regionalization (“HiClimR”). The remainder of this paper describes the theoretical basis for original functionalities in HiClimR (Section 2), design and implementation including a summary of the available features (Section 3), presents a demonstration application in which HiClimR is used to regionalize Africa on the basis of interannual precipitation variability (Section 4), and offers brief discussion of the uses of the package (Section 5).
Theoretical basis
The HiClimR package is based on statistical theory and software tools that are well established in the literature. Most relevantly, the package is built on the foundation of the efficient code of the “hclust” function in the “stats” library of the R project for statistical computing (Team 2012). This function includes seven AHC methods: Ward’s minimum variance, single linkage, complete linkage, average linkage, Mcquitty’s, Median, and centroid methods. Among these, Ward’s (Ward 1963; Murtagh 1983) and average linkage (Sokal 1958; Murtagh 1983) methods have been most frequently applied to climate analyses (El-Hamdouchi and Willett 1989; Fovell and Fovell 1993; Dezfuli 2011; Legendre and Legendre 2012).
We have added an eighth AHC method (called “regional linkage”) to the set of available methods in hclust. Like the other clustering methods in hclust, the regional linkage method is generally applicable to a wide range of clustering problems, but we present it here in the context of a spatio-temporal analysis, in which N spatial elements (e.g., weather stations) are divided into k regions, given that each element has a time series of length M. The regional linkage method is theoretically similar to the existing average linkage method, but it offers practical advantages of computational speed, a built-in objective tree cutting method based on inter-regional correlations (described in Section 3), and the ability to isolate noisy data during the clustering process. These advantages are achieved by using the mean time series and standard deviation of regions as input variables for the cluster update function. This differs from the average linkage method, for example, in which the update function is based on correlation between individual spatial elements.
In the regional linkage update method, similarity in temporal variability between two timeseries x and y is quantified using Pearson’s correlation r between the spatially averaged timeseries of each region:
where x i and y i are the spatial mean value of the variable of interest (e.g., precipitation) in two different regions (or elements) at time i, and \( \overline{x} \) are \( \overline{y} \) the temporal mean values for each region, and σ x and σ y are the standard deviations of the time series x and y, respectively. The time series mean \( \overline{x} \) is
The variance σ 2 x can be written in the form
The dissimilarity measure, between two regions based on their mean timeseries x and y is calculated as the Pearson correlation distance,
It can be shown that this distance is equivalent to the squared Euclidean distance if the data are standardized, which is the recommended metric for Ward’s method that minimizes the error sum of squares (Ward 1963; Murtagh 1983).
Given a set of N spatial elements, an AHC algorithm can be summarized generically as follows:
-
Compute N × N distance matrix (dissimilarities)
-
Assign each station to one cluster
-
i.
Find the most similar pair of clusters and merge them
-
ii.
Update distances between the new cluster and others
-
i.
-
Repeat steps i and ii until all clusters are merged
Clustering methods have different update formulae (step ii). Regional linkage modifies the average linkage algorithm, which has the following update formulae:
where n x and n y are the number of members (stations) used to calculate time series x and y. d x,z and d y,z are Pearson correlation distances between the two original time series x and y and a third time series, z, and d x ∪ y,z is the Pearson correlation distance between the time series \( x{\displaystyle \cup }y=\frac{n_xx+{n}_yy}{n_x+{n}_y} \), representative of a newly merged region, and z.Regional linkage modifies the average linkage update formulae by incorporating the standard deviation of the timeseries of the merged region x ∪ y as shown below
where σ x ∪ y is the standard deviation of the new region’s mean timeseries which can be calculated in terms of the standard deviations of the individual regions’ means as follows
Equations (6) and (7) can be directly derived from Eqs. (1) to (5). It is clear that standard deviation of the new region, in Eq. (7), is a function of the correlation between the individual regions and their standard deviations before merging. It is equal to the average of their standard deviations if and only if the correlation between the two merged regions is 100 %. In this special case, the regional linkage method is reduced to the classic average linkage clustering method. It is also reduced to the classic centroid linkage method if the data are standardized. Note that the range of possible values for correlation is between −1 and 1 and the dissimilarity measure has a range between 0 and 2. The correlation distance can be divided by 2 to make a standard range between 0 and 1, but this has no effect on the regionalization results except for the dendrogram height which is more interpretable when using the correlation distance with regional linkage method as the maximum inter-regional correlation.
The merging history of this method is based on the inter-regional correlation between the temporal means of the regions. Regions with strongly correlated means are successively merged. This guaranties the homogeneity of each region, where homogeneity is defined as strength of correlation between the regional mean of that region and its members (stations). At the end of clustering process, the optimum cut-level of the tree diagram, which illustrates the arrangement of the clusters, is determined by minimizing the inter-regional correlations. An advantage of the regional linkage method is that it allows us to find the optimum number of clusters objectively by imposing a significance level threshold on the maximum acceptable value of inter-regional correlations. At each merging step, the highly correlated regions (maximum inter-regional correlation) are merged first. The statistical history of maximum inter-regional correlations is typically the merging criterion. It is a measure of separation or contiguity, and it is used as an objective measure to determine the tree cut (to find the “optimal” number of regions at a certain confidence level). Additionally, for validation purposes, detailed information on the inter-regional and intra-regional correlations for each selected region, together with cluster sizes, can be computed at any merging step. The average intra-regional correlation measures overall homogeneity, while the cluster size provides useful information for size constraints such as minimum cluster size.
Note that the physical meaning of regions returned by any AHC algorithm depends entirely on the nature of the time series data. A regionalization based on weekly or monthly data, for example, may be dominated by differences in the seasonal cycle across the analysis area, while a regionalization based on deseasonalized, annual, or repeated month or season data (e.g., “July only” or “winter only” time series) will capture differences in interannual variability.
Design and implementation
The clustering algorithm described above is contained in the R package HiClimR described in the flowchart in Fig. 1. The core function, also called HiClimR, performs AHC using any of the eight clustering algorithms listed above. The required input is an N row by M column matrix of ‘double’ values: N objects (spatial points or stations) to be clustered by M observations (temporal points or years). While we describe N and M in terms of typical climate datasets, any data matrix can be used as input (described in the manual). x is the input N × M data matrix, xc is the coarsened N 0 × M data matrix where N 0 ≤ N (N 0 = N only if lonStep = 1 and lonStep = 1), x m is the masked and filtered N 1 × M 1 data matrix where N 1 ≤ N 0 (N 1 = N 0 only if the number of masked stations/points is zero) and M 1 ≤ M (M 1 = M only if no columns are removed due to missing values), and x 1 is the reconstructed N 1 × M 1 data matrix if PCA is performed. Zero-variance rows (e.g., stations with zero variability) and/or missing values (e.g., years with missing observations) are allowed, as they can be removed by preprocessor functions (recommended) or will be removed automatically during clustering.
Detailed flowchart for the package as executed by the HiClimR function. Cyan blocks represent helper functions, green is input data or parameters, yellow indicates agglomeration Fortran code, and purple shows graphics options. For multi-variate clustering (MVC), the input data are a list of matrices (one matrix for each variable with the same number of rows to be clustered; the number of columns may vary per variable). The blue dashed boxes involve a loop for all variables to apply mean and/or variance thresholds, detrending, and/or standardization per variable before weighing the preprocessed variables and binding them by columns in one matrix for clustering
In addition, the package includes six helper functions (cyan blocks in Fig. 1) that can be used as free-standing routines or can be called internally by the HiClimR function to perform initialization, masking, preprocessing, and postprocessing, including validation and visualization. The validClimR helper function is used to select the “optimal” number of clusters objectively based on a specified significance level and to return information and summary statistics for the clusters. The regional linkage method is automatically supported in the objective selection of number of clusters, since its update formulae––Eqs. (6) and (7) ––are based on minimizing inter-regional correlations (i.e., the history of maximum inter-regional correlation is computed directly at each merging step). Other methods can utilize the objective tree cut either by calling the validClimR function with a user-specified range for the number of clusters or by using a hybrid hierarchical-regional clustering feature in the package.
Several features have been implemented to facilitate spatio-temporal analysis applications as well as cluster validation. These include options for preprocessing and postprocessing (Wilks 2011) as well as efficient code execution for large datasets. The ability to perform multi-variate clustering (MVC) and hybrid hierarchical-regional clustering introduced in section 3.1 and 3.2, respectively. Section 3.3 describes the available features for preprocessing, while the postpocessing features are presented in Section 3.4. Section 3.5 describes code performance.
Multi-variate clustering
In many climate regionalization applications, the clustering may involve multiple variables. For example, traditional climate zones are often defined by temperature and precipitation along with other characteristics. Multi-variate clustering (MVC) can be performed by providing HiClimR with a list of matrices (one matrix for each variable) rather than a single variable matrix. These matrices should have the same number of rows (objects or stations to be clustered) while the number of columns may vary per variable (e.g., different temporal periods or record lengths). The preprocessing options are then separately applied for each variable including filtering all variables before preprocessing, detrending and standardization of each variable, and applying weight for the preprocessed variables. Standardization is strongly recommended since variables may have different magnitudes. The correlation distance for MVC represents the (weighted) average of distances between all variables.
Hybrid clustering
This feature allows the user to apply any of the available clustering algorithms to generate the AHC dendrogram and then invoke the regional linkage method as a second step for objective tree cut. The upper part of the tree at a user-specified number of clusters will be reconstructed. By default, when the hybrid option is requested the first merging cost (the loss of overall homogeneity at each merging step) larger than the mean merging cost for the entire tree will be used. For hybrid clustering, the updated upper part of the tree will be used for cluster validation.
Preprocessing
Gridding and geographic masking
For gridded data, two gridding functions are offered to assist in data management. The grid2D helper function generates longitude (lon) and latitude (lat) matrices for gridded data, and the coarseR function can be used to thin large datasets using user-specified skip values for longitude and latitude (lonSkip and latSkip). coarseR is useful when applying regionalization on machines with limited memory resources and when performing initial tests or sensitivity studies.
Geographic masking capabilities are also included in the package, as there are many cases in which a user may want to focus on an area that is a mask-defined subset of the full dataset. For instance, the NASA Tropical Rainfall Measuring Mission (TRMM) data covers ocean and land, while a researcher might be interested in the precipitation variability only over land, a country, or a list of countries (e.g., Nile Basin countries). This masking capability is supported by the helper function geogMask, which can preprocess input data matrix within the HiClimR function if the geogMask logical parameter is set to TRUE as shown in Fig. 1. Alternatively, geogMask can be run as a preprocessor, and the output can be supplied to HiClimR as the gMask argument. This saves computational time when repeating the analysis. geogMask requires the longitude and latitude vectors together with a string (or array of strings) to specify continent name(s), region name(s), or country ISO3 character code(s) via either continent, region, or country parameters. Valid geogMask parameter values can be obtained by running geogMask(). World mask data are based on the Humanitarian Information Unit (HIU) Large Scale International Boundaries (LSIB) dataset (https://hiu.state.gov/data/).
Data thresholds
Optional thresholds can be applied to the observation mean (meanThresh) and/or variance (varThresh) in the HiClimR function to mask zero- and near-zero-variance data. Observations with mean/variance less than or equal to meanThresh/varThresh will be removed. The default is to only mask zero-variance data (meanThresh = NULL and varThresh = 0). The user can increase the variance threshold and/or set a value for the mean threshold. The masked data by thresholds and/or geographic masking is checked again for the correct dimensions before proceeding to the next processing step (Fig. 1).
Detrending and standardization
The HiClimR function uses an optional logical parameter detrend for removing a linear trend from the data. This is important when variation rather than secular change is of interest (e.g., interannual variability). Another logical optional parameter, standardize, can be turned on/off in the HiClimR function to standardize the data before clustering. When data are standardized, clustering algorithms are applied to the mean of equally-weighted elements within each cluster (cluster mean = mean of standardized variables within the cluster). Otherwise, the mean of the raw data will be used (cluster mean = mean of raw variables within the cluster). The variance of the mean is updated at each agglomeration step.
PCA
Principal component analysis (PCA) can be conducted to filter the data before clustering. The nPC parameter in the HiClimR function represents the number of PCs to be retained. If nPC=NULL, then the raw data will be used for clustering. Otherwise, the data will be filtered and reconstructed using nPC PCs obtained from PCA based on singular value decomposition (SVD). The eigenvalues, explained variance, and accumulated variance will be returned to inform the choice of the most appropriate number of PCs. The detrend and/or standardize options will be applied, if requested, before PCA. The preprocessed data are returned in the output so that the user can easily apply HiClimR preprocessing tools for other applications.
Postprocessing
A number of options are available to return additional processing information, statistical summaries, and plots from the HiClimR function. A logical parameter plot can be turned on to display the dendrogram tree immediately after processing completes. The validClimR function validates clustering results on the basis of cluster means, sizes, intra- and inter-cluster correlations, and overall statistical properties, and can be invoked as a flag on HiClimR or as an independent call. Statistical information and validation indices can be computed based on either the raw data or PCA-filtered data (if PCA preprocessing is applied), as controlled by the rawStat parameter. An optional parameter can be used to validate clustering for a selected number of clusters k. If k=NULL, the default, objective cutting of the tree to select the optimal number of clusters will be applied based on a user-specified significance level (alpha parameter). HiClimR and validClimR call the fastCor function to compute the correlation matrix efficiently, and the minSigCor function to estimate the “cut level”—defined as the minimum significant correlation for a given sample size (number of observations or temporal points in a timeseries) at a specified confidence level. Maps of climate regions can be produced for gridded data using the plot parameter.
In the regional linkage method, noisy spatial elements are isolated or placed in their own very small-size clusters since they do not correlate well with any other elements. They can be excluded from validation indices by setting a value for the minimum size of clusters (parameter minSize) greater than one. The excluded clusters are identified in the output of validClimR in the clustFlag component, which is assigned a value of one for valid clusters and zero for excluded clusters. The sum of clustFlag elements represents the selected number of clusters.
Performance
The clustering code is available in both R and Fortran languages. The R code is easier to modify when the user needs to customize the code for his or her own application/development. The Fortran code is a modification of “hclust” function in the “stats” library of the R project for statistical computing (Team 2012), in which we have included an optimized algorithm to deal with only the upper/lower triangular-half of the symmetric dissimilarity matrix instead of the old algorithm that uses the full matrix in the merging steps. For high-resolution gridded data, the function coarseR enables the user to coarsen data in any spatial dimension: longitude, latitude, or both. This can be useful for very large datasets and on older computers. The fastCor function computes the correlation matrix by calling the cross product function in the Basic Linear Algebra Subroutines (BLAS) library used by R. A significant performance improvement can be achieved when building R on 64-bit machines with an optimized BLAS library, such as ATLAS, OpenBLAS, or the commercial Intel® Math Kernel Library. It also uses a memory-efficient algorithm that allows for splitting the data matrix and computes only the upper-triangular half of the correlation matrix. This almost halves memory use, which can be very important for big data with very large number of objects (stations of observations).
Demonstration
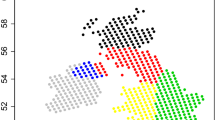
To demonstrate the functionality of HClimR we apply the package to regionalize Africa on the basis of interannual variability in precipitation. This is a simple demonstration for using the package with some of the available features. The current version of HiClimR makes the extension to multi-variate applications straightforward with more options to preprocess each variable separately. The data matrix for each variable controls the nature of clustering problem: rows represent objects or stations to be clustered while columns are for observations that define a specific climatic metric. For instance, Fig. 2a shows regionalization of Africa based on annual cycle (columns are 12 observations for the mean rainfall of each month) while regions in Fig. 2b are based on interannual variability of annual rainfall (columns are observation for year-to-year variations). Only precipitation data and Ward’s clustering with 12 regions are used for simplicity, but the extension to multi-variate clustering using multiple climate variables and/or other clustering methods is a straightforward application of HiClimR.
Regionalization of Africa based on: (a) Annual Cycle and (b) Annual Mean via Ward’s clustering with 12 regions. Only precipitation data (1949–1989) is used for simplicity, but the extension to multi-variate clustering using multiple climate variables and/or other clustering methods is a straightforward application
The input dataset is the University of East Anglia Climatic Research Unit (CRU) TS (timeseries) precipitation dataset version 3.2 (Harris et al. 2013). CRU TS 3.21 data (1901–2012) are monthly gridded precipitation with 0.5° resolution. The dataset used in this demonstration case is included in the HClimR package, and the core commands to repeat the analysis are listed in the package manual. The “Set3” color palette from package RColorBrewer (Neuwirth 2011) was used for colPalette (an optional argument of HiClimR to customize colors in region maps). As regions can change by month and by season, we present only the regionalization for interannual variability in January precipitation. Additional regionalizations can be performed for other months, groups of months, or total annual precipitation.
Method intercomparison
Figure 3 shows a comparison of three AHC methods: regional linkage (Fig. 3a), average linkage (Fig. 3b), Ward’s (Fig. 3c), and the popular nonhierarchical k-means method (Fig. 3d). The regions are created for the same preprocessing options and a statistically “optimal” number of clusters (k = 9) obtained from the validClimR function at 99 % confidence level (alpha = 0.01 and minimum significant correlation = 0.398) and minimum cluster size of 100 –– which is equivalent to an area of 5° × 5°. In this particular application, the regional linkage method strongly outperforms Ward’s and k-means methods with regard to inter-regional correlation (i.e., it provides insignificant inter-regional correlations less than 0.398 while the other methods tend to divide South Africa in to two regions with relatively a significant inter-regional correlation of ~0.5 or more) and slightly outperforms the average linkage method. Differences in homogeneity are negligible across methods, though Ward’s and k-means methods do provide slightly higher intra-regional correlation values. As described above, the regional linkage method isolates and masks noisy regions. This can be seen in parts of Western Equatorial Africa (WEA), which is consistent with the fact that this region suffers from extremely limited data availability and is characterized by intrinsically complex patterns of rainfall variability (Dezfuli 2011; Nicholson et al. 2012).
Sensitivity analysis
Figure 4 shows the effect that detrending and standardizing input data has on the regions produced by the regional linkage method. Figure panels are for regionalization with both detrending and standardization (Fig. 4a), no preprocessing (Fig. 4b), only detrending (Fig. 4c), and only standardization (Fig. 4d). There are differences between all four maps, especially in WEA and Southern Africa. However, the spatial patterns in East Equatorial Africa (EEA) are very similar for all approaches, implying that EEA has a small or spatially consistent trend in January, and that the amount of the precipitation in this month is relatively uniform across the region. The choice of the best set of detrending and standardization options depends on the purpose of the regionalization––e.g., focus on interannual variability vs. focus on multi-decadal variability (e.g., climate change response), or focus on total amount of precipitation vs. focus on common variabilities across precipitation gradients.
Figure 5 shows the sensitivity of regionalization results to geographic masking and PCA. It is clear that masking the data for a study area of interest before clustering does affect the results. Figure 5a shows the reference case where geogMask function was applied for Africa together with detrending and/or standardizing the raw data. Figure 5b is the same as Fig. 5a but without geographic masking. This causes of parts of southern Europe and southwest Asia to be included in the analysis, which influences the clustering algorithm in a way that alters regionalization in central and southern Africa. For other applications––for example, when the region of interest is surrounded by regions with very different variability, or when the climate dataset includes observations over ocean as well as land––the influence of geographic masking will be even more dramatic. Figure 5c shows the regions generated by the regional linkage method when PCA is applied on the reference case and 27 PCs (~90 % of the total variance) are used to reconstruct a filtered data before clustering. Figure 5d shows regions when PCA is applied as in Fig. 3d but without detrending or standardization. Again, the choice of pre-processing options has a noticeable influence on regionalization results. The analyst must consider whether the noise-reducing effect of PCA has a beneficial impact on the regionalization in any given application, as well as whether detrending and standardization are justified for the given scientific purpose of the regionalization. There is no objective “right” answer for these preprocessing decisions, since the value of the regionalization is a function of both objective performance metrics and the purpose of the exercise.
Discussion
This paper has introduced an R package designed to support objective climate regionalization using hierarchical cluster analysis. The package includes Ward’s method and average linkage method clustering algorithms, which have been widely applied to climate regionalization in the past, a number of other methods (single linkage, complete linkage, Mcquitty’s, Median, and centroid) that may be of interest to users applying the package to other clustering problems, and a new, modified clustering algorithm that is designed specifically for climate regionalization—the “regional linkage” method. The regional linkage method is a modification of the average linkage method that minimizes inter-regional correlations between region means. It also provides the ability to identify noisy elements for quality control and to perform an objective tree cut based on correlation significance.
In addition to the core clustering algorithms, the HiClimR package includes several preprocessing and postprocessing features to facilitate climate regionalization and related applications.
Availability and requirements
A stable release of the package is available through the Comprehensive R Archive Network at http://CRAN.R-project.org/package=HiClimR. This release includes sample data used in the test case presented in this paper. In the future we plan to expand the package to include additional clustering algorithms and processing options, as informed by user experience.
The package requires R, which is a free software environment for statistical computing and graphics. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS. The hardware requirements depend on the problem size, mainly the memory required for very large data sets.
References
Argüeso D, Hidalgo-Muñoz J M, Gámiz-Fortis S R, Esteban-Parra M J, Dudhia J, Castro-Díez Y (2011) Evaluation of Wrf parameterizations for climate studies over southern spain using a multistep regionalization. J Clim 24
Baeriswyl P-A, Rebetez M (1997) Regionalization of precipitation in Switzerland by means of principal component analysis. Theor Appl Climatol 58:31–41
Burn DH (1989) Cluster analysis as applied to regional flood frequency. J Water Res Plan Manag 115:567–582
Busuioc A, Chen D, Hellström C (2001) Temporal and spatial variability of precipitation in Sweden and its link with the large-scale atmospheric circulation. Tellus A 53:348–367
Cimiano P, Hotho A, Staab S (2004) Comparing Conceptual, Divise and Agglomerative Clustering for Learning Taxonomies from Text. Proceedings of the 16th Eureopean Conference on Artificial Intelligence, Ecai’2004, Including Prestigious Applicants Of Intelligent Systems, Pais 2004
Comrie AC, Glenn EC (1998) Principal components-based regionalization of precipitation regimes across the southwest United States and Northern Mexico, with an application to monsoon precipitation variability. Clim Res 10:201–215
Dezfuli AK (2011) Spatio-temporal variability of seasonal rainfall in western equatorial africa. Theor Appl Climatol 104:57–69
Dezfuli, A K, Nicholson S E (2013) The relationship of rainfall variability in western equatorial africa to the tropical oceans and atmospheric circulation. Part Ii: The boreal autumn. J Clim 26
El-Hamdouchi A, Willett P (1989) Comparison of hierarchic agglomerative clustering methods for document retrieval. Comput J 32:220–227
Fovell RG, Fovell M-YC (1993) Climate zones of the conterminous united states defined using cluster analysis. J Clim 6:2103–2135
Gong X, Richman MB (1995) On the application of cluster analysis to growing season precipitation data in north america east of the rockies. J Clim 8:897–931
Harris I, Jones P, Osborn T, Lister D (2013) Updated high-resolution grids of monthly climatic observations–the Cru Ts3. 10 dataset. Int J Climatol
Isik S, Singh VP (2008) Hydrologic regionalization of watersheds in turkey. J Hydrol Eng 13:824–834
Jain A K, Dubes R C (1988) Algorithms For Clustering Data. Prentice-Hall, Inc.
Jain AK, Murty MN, Flynn PJ (1999) Data clustering: a review. Acm Comput Surv (Csur) 31:264–323
Legendre P, Legendre L (2012) Numerical Ecology. Elsevier
Manning CD, Raghavan P, Schütze H (2008) Introduction to Information Retrieval. Cambridge University Press, Cambridge
Munoz-Diaz D, Rodrigo F S (2004) Spatio-Temporal Patterns of Seasonal Rainfall In Spain (1912–2000) Using Cluster and Principal Component Analysis: Comparison. Annales Geophysicae. Copernicus Gmbh, 1435–1448
Murtagh F (1983) A survey of recent advances in hierarchical clustering algorithms. Comput J 26:354–359
Neuwirth E (2011) Rcolorbrewer: Colorbrewer Palettes. R Package Version, 1.0-5
Nicholson S E, Dezfuli A K (2013) The relationship of rainfall variability in western equatorial africa to the tropical oceans and atmospheric circulation. Part i: the boreal spring. J Clim 26
Nicholson SE, Klotter D, Dezfuli AK (2012) Spatial reconstruction of semi-quantitative precipitation fields over africa during the nineteenth century from documentary evidence and gauge data. Quat Res 78:13–23
Ramachandra Rao A, Srinivas V (2006) Regionalization of watersheds by hybrid-cluster analysis. J Hydrol 318:37–56
Rogers J, Mchugh M (2002) On the separability of the north Atlantic oscillation and arctic oscillation. Clim Dyn 19:599–608
Sokal RR (1958) A statistical method for evaluating systematic relationships. Univ Kans Sci Bull 38:1409–1438
Team R C (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2012
Ward JH Jr (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
White D, Richman M, Yarnal B (1991) Climate regionalization and rotation of principal components. Int J Climatol 11:1–25
Wilks D S (2011) Statistical Methods in the Atmospheric Sciences. Academic Press
Acknowledgments
This study was supported by the Department of Earth and Planetary Sciences, The Johns Hopkins University, and NASA Applied Sciences grant NNX09AT61G.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: H. A. Babaie
Rights and permissions
About this article
Cite this article
Badr, H.S., Zaitchik, B.F. & Dezfuli, A.K. A tool for hierarchical climate regionalization. Earth Sci Inform 8, 949–958 (2015). https://doi.org/10.1007/s12145-015-0221-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12145-015-0221-7