Abstract
Background
Breast cancer (BC) remains a significant global health challenge, contributing substantially to cancer-related deaths worldwide. Its prevalence and associated death rates remain alarmingly high, highlighting the persistent public health burden. The objective of this study was to systematically examine the involvement of SUSD3 (Sushi Domain-Containing 3) in BC, highlighting its crucial role in the pathogenesis and progression of this disease.
Methods
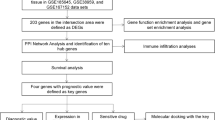
BC-related gene microarray data, along with corresponding clinicopathological information, were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. Leveraging TIMER and HPA databases, we conducted comparative analyses to evaluate SUSD3 expression in BC. We then analyzed the association between SUSD3 and clinical traits, as well as the prognostic value of SUSD3. SUSD3-related differential expression genes (DEGs) were sent for analysis utilizing GO, KEGG, and GSEA. We utilized SUSD3 mRNA expression to assess immune cells’ scores in BC tissues calculated by single-sample enrichment analyses based on “CIBERSORT” R package. Drug sensitivity analysis was used to screen potential drugs sensitive to SUSD3. R software was used for statistical analyses and graphical representation of the data.
Results
Our findings confirmed a significant upregulation of SUSD3 expression in BC, which correlated with a favorable prognosis. Clinical correlation analysis further emphasized the strong association between SUSD3 expression and key clinical parameters like estrogen receptor (ER) status, progesterone receptor (PR) status, stage, and T classification in breast cancer. Univariate and multivariate Cox regression analyses showed that SUSD3 could be used as an independent prognostic factor for BC. Differentially expressed genes (DEGs) co-expressed with SUSD3 were significantly associated with various biological processes, such as the cell cycle, DNA replication, p53 signaling pathway, cancer-related pathways, and Wnt signaling pathway, as indicated by gene set enrichment analysis (GSEA). Furthermore, our analysis demonstrated that SUSD3 generally exhibited negative associations with immune modulators. Drug sensitivity analysis revealed positive correlations between SUSD3 and the efficacy of Fulvestrant, Raloxifene, and Fluphenazine.
Conclusion
The research emphasizes the significance of SUSD3 as a potential marker for BC, providing insights into the underlying molecular mechanisms implicated in tumorigenesis. SUSD3 holds promise in helping the classification of breast cancer pathological groups, predicting prognosis, and facilitating targeted therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) represents a prevalent malignancy characterized by substantial mortality and morbidity [1]. Notably, China ranks highest globally in both BC incidence and related fatalities, accounting for 17.6% of BC cases and 15.6% of BC-related deaths worldwide [2]. Despite this burden, there is a shortage of dependable biomarkers for early screening and diagnosis of BC. The advent of high-throughput sequencing has particularly facilitated the identification of an expanding array of genes serving as potential markers for early-stage BC. However, there is a critical need to identify key driver genes capable of influencing the immune microenvironment of BC and its progression. Currently, there is a growing emphasis on identifying central genes associated with BC through the analysis of RNA-seq data sourced from databases such as The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO). This endeavor is promising as it helps advance the diagnosis, prognosis, and targeted therapy of BC.
Sushi domains, also referred to as complement control protein modules or short consensus repeats, represent evolutionarily conserved protein domains present in numerous complement and adhesion proteins [3]. Sushi domain-containing proteins typically localize to the outer membrane and play roles in nutrient binding and signaling. SUSD3 (Sushi Domain-Containing 3) is notable as a cell surface protein, possessing extracellular, transmembrane, and cytoplasmic domains [4]. Prior research has highlighted a correlation between increased expression levels of SUSD3 and BC prognosis [5, 6]. However, the precise functions of SUSD3 remain elusive. Therefore, the objective of this study was to assess the prognostic implications and underlying mechanisms of SUSD3 in BC.
Bioinformatic analysis of sequencing data sourced from common databases such as TCGA and GEO enables the interpretation of gene regulatory pathways and disease networks. In this study, we conducted a comparative analysis of differentially expressed genes (DEGs) in patients using breast cancer gene (BRCA) samples from the TCGA database. The expression profiles of these genes were correlated with the clinical data of the patients. Specifically, the RNA expression analysis encompassed 1029 tumor samples and 112 non-tumor samples from the TCGA dataset. To further assess the potential biological functions of SUSD3 in BC, bioinformatics tools were systematically used to validate molecular mechanisms through Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. In addition, we assessed the prognostic significance of SUSD3 in breast cancer and comprehensively assessed its potential clinical use based on analyses of TCGA and GEO datasets. Our findings indicated SUSD3 is highly expressed in BC and associated with a good prognosis, which provide valuable insights into the potential molecular mechanisms influencing BC prognosis.
Materials and methods
Data collecting and screening
RNA-seq data and clinical information for BC were retrieved from the TCGA database. After excluding patients with incomplete clinical information, a dataset comprising 1029 samples from patients diagnosed with BC and 112 samples from normal individuals were obtained. Detailed information on the 1029 patients with BC is provided in Table 1.
Differential expression analysis of SUSD3 in BC
The TIMER database comprehensively encompasses genetic information from a vast array of samples, totaling 10,897 samples, primarily derived from the TCGA database [7]. This repository was leveraged to conduct a pan-cancer assessment of SUSD3 expression. Subsequently, expression profiles of SUSD3 were extracted and scrutinized across 1029 BC samples, 112 normal samples, along with 112 paired samples, using the “Limma” R package [8].
Correlation analysis between SUSD3 and clinical traits
Clinical data from patients with BC including parameters such as T/N stage, stage, PR/ER/Her-2 status, and age, were integrated with SUSD3 expression for comprehensive analysis. Logistic regression analysis was conducted using R (Version 4.3.2) to assess the relationship between clinical data for BC individuals and SUSD3 expressions. A significance level of p < 0.05 was considered statistically significant.
Independent prognostic analysis
In our investigation of the impact of SUSD3 expression on the prognosis of patients with BC, we used a comprehensive approach. Initially, univariate Cox regression analysis was applied to quantify the correlation between SUSD3 expression levels and patient survival. Subsequently, multivariate analysis was conducted to determine whether SUSD3 serves as an independent prognostic factor for survival in these patients. The Cox data of the patients were statistically analyzed using the survival package within the R software, using both univariate and multivariate methods to identify independent prognostic indicators. The results were visualized using forest plots, with the significance threshold set at p < 0.05.
Validation using the Gene Expression Omnibus (GEO) databases
To ensure the accuracy of the results obtained from the TCGA group, they were validated using the GSE42568 dataset from the GEO database to assess SUSD3 expression levels. This dataset comprised 104 BRCA samples and 17 adjacent normal tissue samples. As the GSE42568 group lacked clinical information, the GSE86166 dataset, which includes 280 breast cancer samples, was used to validate the correlation between SUSD3 expression and clinical traits, as well as to assess the prognostic significance of SUSD3. Detailed information on the 280 patients with BC is provided in Table 2.
Functional enrichment analysis
To interpret the underlying biological processes and pathways associated with SUSD3 in BC, DEGs between the SUSD3 high and SUSD3 low groups were identified using the Limma package, applying criteria of an adjusted p value < 0.05 and a |logFC|> 2. In addition, the “ClusterProfiler” R package was used to perform and visualize KEGG and GO analysis, encompassing molecular function (MF), biological process (BP), and cellular component (CC) [9]. Enrichment to significant pathways was determined based on an adjusted p value < 0.05.
GSEA
The GSEA software, obtained from the GSEA website, is a powerful platform for conducting gene set enrichment analysis. The initial molecular signature database encompasses 1325 gene sets across various categories like biological pathways, chromosomal locations, upstream cis-patterns, drug therapy responses, and expression profiles from microarray datasets. Also, users have the flexibility to create additional gene sets through genetic and chemical perturbations, computational analysis of genomic information, and other biological annotations. GSEA results provide a computational enrichment score (ES), indicating the extent to which a gene set S is overrepresented in the extreme (top or bottom) of the overall sorted list L.
To facilitate result interpretation, the ES is normalized to account for genome size, yielding a normalized enrichment score (NES). To control for false positives, the false discovery rate (FDR) corresponding to each NES is calculated. Patient data were divided into high- and low-SUSD3 gene expression groups based on the median level in our study. These TCGA data were formatted into text files and imported into the GSEA software.
Subsequently, the grouping and expression matrix files were uploaded into GSEA 4.3.2 lineage enrichment analysis. The resulting data were exported to analyze significant functional and pathway differences between the high- and low-SUSD3 expression groups. The genomic alignment was repeated 1000 times for each analysis. Meanwhile, p values, FDR values, ES values, and NES values from GSEA were analyzed with expression differences being considered significant if they met the criteria of p < 0.05 and FDR < 25% in the gene enrichment analysis.
Immune cells infiltration analysis
SUSD3 relative scores across 28 types of immune cells in humans were quantitatively assessed using single-sample gene set enrichment analysis (ssGSEA), facilitated by the R package “GSVA” [10]. In addition, the quantification of 22 distinct infiltrating immune cell types in lung cancer was conducted using the R package “CIBERSORT”. The significance threshold for both analyses was set at a p value < 0.05.
Therapeutic analysis
We acquired expression data from the Cancer Cell Line Encyclopedia (CCLE) and drug sensitivity data from the CellMiner database [11]. This enabled us to comprehensively investigate the relationship between SUSD3 expression and drug responses in tumor cell lines.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 9.0 and R (version 4.3.3), with the implementation of ggpubr, ggplot2, and limma packages for data analysis and visualization. For paired samples, the Wilcoxon signed-rank test was used to assess SUSD3 expression, while for unpaired samples, the Wilcoxon rank sum test was used. The relationship between clinical features and SUSD3 expression was thoroughly examined using the limma package for statistical analysis and ggpubr for graphing. Univariate and multivariate analyses of Cox proportional hazard regression models were performed to identify independent prognostic factors like gender, age, stage T, N, stage, ER, PR, Her-2, and SUSD3 expression, in assessing the risk of death. The CIBERSORT algorithm was applied to predict the abundance of 22 infiltrating immune cells in the tumor microenvironment (TME) using the BC gene expression profile obtained from the TCGA database. The results were visualized using the limma package, ggplot2 package, and ggpubr package. The Wilcoxon test was used to compare stromal/immune scores among different clinicopathological groups, with a significance threshold set at p < 0.05.
Results
The levels of SUSD3 mRNA in breast cancer and other cancers
To assess the differences in SUSD3 expression between tumor and normal tissues, we analyzed SUSD3 mRNA levels across various tumor types and corresponding controls using data from TIMER. Along with BC, we observed significant increases in SUSD3 mRNA levels in several other cancer types, like esophageal cancer (ESCA), head and neck cancer (HNSC), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), rectum adenocarcinoma (READ), and stomach adenocarcinoma (STAD). Conversely, SUSD3 mRNA levels were significantly lower in cholangiocarcinoma (CHOL), kidney clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), prostate adenocarcinoma (PRAD), and uterine corpus endometrial carcinoma (UCEC) when compared to the corresponding normal tissues (Fig. 1A). Subsequently, we further analyzed SUSD3 mRNA expression levels in 1020 patients with BC and 112 normal patients with complete clinical data. The findings revealed a significant increase in the expression level of SUSD3 in breast cancer samples compared to the normal control group (Fig. 1B). In paired samples, the analysis demonstrated that SUSD3 levels within the tumor group were consistently higher than those observed in the corresponding normal group (Fig. 1C). Also, GEO data revealed that SUSD3 was highly expressed in BC (Fig. 1D).
There was a notable increase in SUSD3 expression in the BC samples. A The Timer online tool was used to measure SUSD3 expression in various tumor tissues found in the TCGA database. B, C The TCGA dataset indicated an increase in SUSD3 expression in BC. D Based on the GEO dataset, the expression of SUSD3 increased in BC. E, F Using immunohistochemistry (IHC) data, SUSD3 protein expression was examined in both normal and tumor tissues in BC
We conducted an analysis of SUSD3 protein expression in both normal and tumor tissues of breast cancer using IHC data. Our investigation revealed contrasting SUSD3 staining patterns: SUSD3 staining was observed in glandular cells and myoepithelial cells in normal tissues, whereas weak SUSD3 staining was evident in tumor tissues (Fig. 1E, F).
SUSD3 expression relates to BC patients’ clinicopathological characteristics based on TCGA and GEO data
Further investigation into the relationship between SUSD3 gene expression and clinicopathological variables in patients diagnosed with BC was deemed necessary. The association between SUSD3 gene expression and clinicopathological variables in these patients was assessed using the Wilcoxon signed-rank test and logistic regression analysis. These patients were divided into two groups based on their SUSD3 expression levels, and relevant parameters were analyzed alongside dichotomized SUSD3 expression (high/low) using thermographic analysis. The findings revealed statistically significant associations between PR, ER, and Her-2 status and SUSD3 expression (p < 0.001) in both TCGA and GEO datasets (Fig. 2A, B).
The expression of SUSD3 correlates with the clinicopathological characteristics of patients with BC. A, B Thermal imaging analysis of clinical parameters in patients with BC divided by high and low expression of SUSD3, using data from TCGA and GEO datasets. C–I Analysis based on the TCGA dataset revealed a positive correlation between SUSD3 expression and parameters such as age, clinical stage, T stage, ER status, PR status, and Her-2 status, but not with N stage. J–N Analysis based on the GEO dataset revealed a positive correlation between SUSD3 expression and clinical histological grade, ER status, PR status, and Her-2 status, but not with clinical stage
Subsequently, SUSD3 mRNA expression was assessed across cohorts divided by age, clinical stage, T/N stages, and ER/PR/Her-2 status. The results demonstrated a significant association between SUSD3 overexpression and age (≤ 65 and > 65, p = 0.038), stage (stage I and stage II, p = 0.00083), T stage (T1 and T2, p = 0.0007), ER status (negative and positive, p < 2.22e−16), PR status (negative and positive, p < 2.22e−16), and Her-2 status (negative and positive, p = 4.6e−05) in the TCGA dataset (Fig. 2C–I). In the GEO dataset, SUSD3 mRNA expression was assessed across groups stratified by grade, clinical stage, and ER/PR/Her-2 status. The results demonstrated a significant association between SUSD3 overexpression and grade (grade 1 and grade 3, p = 2.9e−05), ER status (negative and positive, p < 2.22e−16), PR status (negative and positive, p < 2.22e-16), and Her-2 status (negative and positive, p = 9e-08) (Fig. 2J–N).
Potential significance of SUSD3 in BC prognosis
Based on the aforementioned findings, it can be inferred that the overexpression of SUSD3 may hold significant prognostic implications in BC. To substantiate this hypothesis, a comprehensive analysis and validation were conducted using multiple datasets. To assess the prognostic value of SUSD3 in patients with BC, Kaplan–Meier (KM) survival curves were constructed using the TCGA and GSE86166 datasets. In the TCGA-BRCA dataset, patients with high expression of SUSD3 exhibited a significantly lower risk of mortality (p = 0.003) compared to those with low expression (Fig. 3A). This trend was also observed in the GSE86166 group, where high SUSD3 expression was associated with a better OS outcome (p < 0.001) (Fig. 3B). Also, patients diagnosed with BC were grouped into two groups based on clinical features like age (age ≤ 65 and age > 65), clinical stage (Stage I–II and Stage III–IV), T stage (T 1–2 and T 3–4), N stage (N0 and N1–3), ER (positive and negative), PR (positive and negative), and Her-2 (positive and negative).
SUSD3 could be used as an independent prognostic factor for BC. A, B The TCGA and GEO datasets collectively indicate that patients diagnosed with BC having high-SUSD3 expression experience significantly longer overall survival compared to those with low-SUSD3 expression. C The calibration curve of the nomogram and the nomogram integrating SUSD3 along with other prognostic factors of BC, derived from TCGA data. D The calibration curve of the nomogram and the nomogram integrating SUSD3 along with other prognostic factors of BC, based on GEO data. E, F Univariate analysis and multivariate analysis demonstrating SUSD3 as an independent predictor of survival time in patients with BC, using TCGA data. G, H Univariate analysis and multivariate analysis displaying SUSD3 as an independent predictor of survival time in BC patients, utilizing GEO data
In the TCGA-BRCA dataset, patients with high SUSD3 expression had a better OS outcome regardless of whether they were younger than or older than 65 years (Fig. S1A). In patients with Stage III–IV BC, those with high-SUSD3 expression lived longer (Fig. S1B). Similarly, in patients with T3-4 or N1-N3, those with high-SUSD3 expression exhibited longer survival (Fig. S1C, D). Also, in patients with ER-positive status, those with high-SUSD3 expression had a better OS outcome (Fig. S1E).
However, for the clinical trait groups of PR or Her-2, SUSD3 expression level did not reveal significant differences in prognosis (Fig.S1F, G). In the GSE86166 dataset, patients with stage II–III and high-SUSD3 expression lived longer (Fig. S1H). Moreover, in patients with ER-positive, PR-positive, or Her-2-positive status, high-SUSD3 expression was associated with a better OS outcome (Fig. S1I–K).
Development of a prognostic model integrating SUSD3 expression and clinical factors
Using SUSD3 expression and other clinical parameters in the TCGA dataset, we developed a prediction model for OS, integrating SUSD3 as a biomarker for BC. A calibration curve assessed the performance of the nomogram for SUSD3 expression versus 1-year, 3-year, and 5-year survivals, and a nomogram was established to visually represent this prediction model (Fig. 3C). We validated this prognosis model using the GEO dataset (GSE86166) (Fig. 3D).
Meanwhile, univariate regression and multivariate analysis were used to assess whether SUSD3 could be an independent risk factor for overall survival in patients diagnosed with BC. In the TCGA dataset, univariate regression analysis revealed that SUSD3 gene expression was a low-risk factor for BC (HR 0.853; confidence interval, 0.781–0.933; p < 0.001) (Fig. 3E). In the subsequent multivariate analysis, SUSD3 mRNA expression emerged as an independent risk factor for overall survival in these patients (HR 0.824; confidence interval, 0.752–0.903; p < 0.001) (Fig. 3F). In the GEO dataset (GSE86166), univariate regression analysis indicated that SUSD3 gene expression was also a low-risk factor for BC (HR 0.744; confidence interval, 0.644–0.861; p < 0.001) (Fig. 3G). In the subsequent multivariate analysis, SUSD3 mRNA expression emerged as an independent risk factor for overall survival in these patients (HR 0.720; confidence interval, 0.616–0.840; p < 0.001) (Fig. 3H). Both univariate and multifactorial Cox regression analyses in the TCGA and GEO datasets consistently identified SUSD3 gene expression as a low-risk factor for BC.
Enrichment analyses of SUSD3‑related genes in BC through GO , KEGG and GSEA based on TCGA data
To conduct a comprehensive examination of the involvement of SUSD3 in BC, genes highly associated with SUSD3 (correlation coefficient > 0.6) were screened in BC using Pearson correlation analysis. The representative SUSD3-related genes are illustrated in Fig. 4A. ESR1, IL6ST, CA12, NINJ1, MAPT, C5AR2, PGR, and FGD3 exhibited a positive correlation with SUSD3 expression. Furthermore, genes highly associated with SUSD3 were identified in BC using differential expression analysis. The identified DEGs were then classified into high- and low-SUSD3 expression groups based on the median value of SUSD3 in TCGA data. To understand the functional relevance of the identified SUSD3-associated genes, GO and KEGG pathway analyses were conducted. GO enrichment analysis assessed the involvement of BP, CC, and MF, and revealed that SUSD3-related genes were primarily enriched in functional categories related to epidermis development and cell communication. Notably, these genes revealed significant enrichment for “skin development”, “epidermal cell differentiation”, “collagen trimer”, “cytokine activity”, and “endopeptidase activity”, indicating potential roles in cell–cell interactions (Fig. 4B).
Gene enrichment analysis to identify SUSD3-related pathways. A Genes highly correlated with SUSD3 (correlation coefficient > 0.6) were identified in BC through Pearson’s test method analysis. B GO enrichment analysis of DEGs related to SUSD3. C KEGG enrichment analysis of DEGs associated with SUSD3. D, E GSEA revealed signaling pathways associated with SUSD3 mRNA expression in BC
KEGG pathway analysis provided further insights, highlighting enrichment in various signaling pathways crucial for cell–cell communication and regulation. Genes associated with SUSD3 were significantly enriched in pathways such as neuroactive ligand-receptor interactions, IL-17 signaling pathway, protein digestion and absorption, and estrogen signaling pathway (Fig. 4C). To assess the potential regulatory mechanisms of the SUSD3 gene in BC, GSEA was performed using data from the TCGA database. The results revealed associations of SUSD3 mRNA expression with endocytosis, GPI anchor biosynthesis, peroxisome, SNARE interactions in vesicular transport, vasopressin-regulated water reabsorption, cell cycle, DNA replication, p53 signaling pathway, pathways in cancer, and Wnt signaling pathway (Fig. 4D, E).
Correlation between SUSD3 and immune cell infiltration
To analyze the association between SUSD3 and immune cell infiltration, we calculated the correlation between SUSD3 and 22 immune cells using CIBERSORT. The results revealed variations in the abundance of immune cell subtypes between low- and high-SUSD3 expression groups (Fig. 5A). Specifically, mast cells resting, monocytes, and dendritic cells resting exhibited increased abundance in the high-SUSD3 expression group, while T cells CD4 memory activated, macrophages M0, NK cells resting, and eosinophils were activated in the low-SUSD3 expression group (Fig. 5B). Analysis of the linear relationship between SUSD3 and immunological checkpoint genes revealed that SUSD3 generally had a negative regulatory relationship with immunological checkpoint genes (Fig. 5C). Considering that tumor mutational burden (TMB) plays a pivotal role in tumorigenesis and progression, we investigated the correlation between TMB and SUSD3 expression in BC and assessed the connection between SUSD3 expression and mutations. Our findings indicated a significant correlation between increased SUSD3 expression in BC and decreased TMB (Fig. 5D). Furthermore, patients diagnosed with BC were divided into low- and high-SUSD3 expression groups based on their SUSD3 expression levels. Compared with the low expression group of SUSD3, the high expression group of SUSD3 had a lower TMB (Fig. 5E). Figure 5F–G reveals that genes, including PIK3CA, TP53, TTN, and CDH1, were significantly different in the high and low-SUSD3 expression groups.
SUSD3 may participate in the immune regulation and TMB. A The correlation between SUSD3 and 22 immune cell types in 1020 BC samples was computed using CIBERSORT. B A lollipop chart was generated to visualize the immune cells most strongly associated with SUSD3 expression. C The correlation between SUSD3 and immune checkpoint genes was analyzed. D, E An inverse relationship between SUSD3 expression and TMB in BC was observed, indicating that increased SUSD3 expression correlates with decreased TMB. F, G Waterfall plots depicting gene mutations in BC samples divided by low and high expression of SUSD3 were generated
Correlation analysis between SUSD3 expression and drug sensitivity
The relationship between SUSD3 expression and drug sensitivity was analyzed using CCLE data. The sensitivity of the SUSD3 gene to commonly used anti-tumor drugs was investigated through the CellMiner database. The relationship between gene expression and drug sensitivity was calculated, revealing a significant association of SUSD3 expression with the sensitivity of 12 drugs (Fig. 6). Notably, SUSD3 was positively associated with Fulvestrant, Raloxifene, Fluphenazine, Isotretinoin, Belinostat, Hydroxyurea, Umbralisib, LDK-378, Dromostanolone Propionate, 6-Thioguanine, Etoposide, and LEE-011. Some of these drugs, which exhibit sensitivity to SUSD3 expression levels, have not yet undergone extensive clinical testing. Their potential as promising candidates for future therapeutic interventions warrants further assessment.
Discussion
Assessing biomarkers and molecular targets is crucial for the early diagnosis and effective treatment of BC, a prevalent malignancy among women. Through comprehensive bioinformatics analyses of data from TCGA and GEO databases, we identified genes associated with the pathogenesis and clinical prognosis of BC. Our investigation has revealed a significant upregulation of SUSD3 in BC, raising compelling questions about its specific role in the development and progression of this particular cancer subtype. The increased expression of SUSD3 in BC, compared to other tumors, indicates a unique function within BC biology. This discovery prompts further assessment to determine whether the role of SUSD3 in BC differs from its functions in other cancer types. Understanding this distinction is crucial for unraveling the complexities of BC and holds the potential to uncover new therapeutic avenues tailored to its specific characteristics.
In the present study, the prognostic significance of SUSD3 expression in BC provides new insights into patient stratification and treatment approaches. Our investigation has revealed a strong association between high-SUSD3 expression and enhanced survival outcomes. Specifically, high-SUSD3 expression is correlated with a favorable prognosis for patients diagnosed with BC. However, this contrasts with findings in other diseases, such as acute myeloid leukemia (AML), where high-SUSD3 expression is associated with a poor prognosis [12]. This disparity may be related to the differing malignancy levels of these tumors, with BC generally being less malignant than myeloid leukemia.
Furthermore, the high expression of SUSD3 in BC may enhance the efficacy of various therapeutic drugs, thereby enhancing drug sensitivity and therapeutic outcomes, potentially extending patient survival. Therefore, increased SUSD3 expression in BC tissues may inhibit the malignant progression of cancer cells and prolong patient survival. Positioning SUSD3 as a promising prognostic biomarker highlights its significance in therapeutic decision-making and its potential to guide the development of targeted therapies. Recognizing this correlation underscores the importance of considering SUSD3 expression in treatment planning and highlights its potential as a guiding factor in the development of personalized treatment strategies.
In a thorough examination of the impact of SUSD3 on clinicopathological parameters in patients diagnosed with BC using data from both the TCGA and GEO datasets, our findings indicate that SUSD3 serves as an independent prognostic factor influencing survival in these patients. Integrating SUSD3 expression levels into established prognostic models has the potential to enhance the precision of survival predictions, providing valuable insights for personalized treatment planning. The high expression of SUSD3 in BC not only signifies its potential as a prognostic marker but also indicates its involvement in tumorigenesis and early progression. Notably, across different clinical traits, patients with high-SUSD3 expression consistently exhibited better prognoses compared to those with low-SUSD3 expression. This underscores the significant clinical value of SUSD3 in guiding prognosis. Therefore, investigating the role of SUSD3 in tumor development and its potential as a biomarker for early detection could provide crucial insights into the temporal changes in SUSD3 expression from the early to late stages of BC. Such insights could pave the way for the development of new therapeutic strategies and enhance the overall management of these patients.
Co-expressed genes typically share similarities in their functions, providing insights into their collective impact on BC. In our analysis, we assessed the correlation between SUSD3 and its co-expressed genes to gain a broader understanding of their involvement in BC. The findings indicated a robust and positive correlation between SUSD3 and several genes, including ESR1, IL6ST, CA12, NINJ1, MAPT, C5AR2, PGR, and FGD3. Both ESR1 and PGR serve as established biomarkers for BC and are clinically significant in guiding diagnosis and treatment strategies [13,14,15,16,17]. Given that aromatase inhibitors (AI) represent the standard endocrine therapy for postmenopausal BC, there is a pressing need for predictive biomarkers to identify potential responders to AI treatment more effectively [18,19,20].
Our analysis revealed significant differences in SUSD3 expression between ER-positive and ER-negative groups, as well as PR-positive and PR-negative groups. Moreover, SUSD3 demonstrated strong correlations with ER and PR, with correlation coefficients of 0.6 and 0.66, respectively. In addition, drug sensitivity analysis indicated that SUSD3 exhibits high sensitivity to Fulvestrant and Raloxifene. These findings indicate that SUSD3 may serve as a novel predictor of response to endocrine therapy and potentially represent a therapeutic target when combined with ER or PR.
To comprehensively assess the functional pathways associated with SUSD3, we conducted enrichment analyses encompassing GO, KEGG, and GSEA. Our KEGG analysis revealed enrichment of SUSD3 in several pathways like neuroactive ligand–receptor interaction, IL-17 signaling pathway, protein digestion and absorption, and estrogen signaling pathway. Concurrently, GO analysis highlighted the involvement of SUSD3 in epidermis development, skin development, epidermal cell differentiation, and endopeptidase activity. Also, GSEA analysis revealed a positive correlation between SUSD3 expression and pathways such as endocytosis, GPI anchor biosynthesis, peroxisome, snare interactions in vesicular transport, and vasopressin-regulated water reabsorption. Conversely, low-SUSD3 expression exhibited tight associations with pathways related to cell cycle, DNA replication, the p53 signaling pathway, pathways in cancer, and the Wnt signaling pathway. These findings indicate that dysregulation of these pathways may significantly contribute to the progression and development of breast cancer.
The TME encompasses the non-cancerous cellular elements and molecular components surrounding the tumor. These elements, along with their interactions with tumor cells, are essential in influencing tumor initiation, progression, metastasis, and response to therapies. Given its significance, the tumor microenvironment has emerged as a promising therapeutic target in cancer research and clinical practice [21]. Increasing evidence underscores the crucial role of immune infiltrates in shaping both the prognosis and response to systemic treatments in patients with BC [22,23,24,25]. Therefore, investigating the involvement of SUSD3 within the TME, particularly its interactions with immune cells and immune checkpoints, presents opportunities for developing immunomodulatory strategies. Our study used CIBERSORT analysis, revealing that high-SUSD3 expression in BC correlates with mast cells resting, monocytes, and dendritic cells resting, while low-SUSD3 expression is associated with T cells CD4 memory activated, macrophages M0, NK cells resting, and eosinophils. Notably, the activation of CD4+ T cells, known for their regulatory role in cytolytic mechanisms and the enhancement of B cell and CD8+ T cell responses, indicates intricate dynamics in BC progression [26].
Although tumor mutation burden (TMB) has been closely associated with the efficacy of immune checkpoint inhibitors (ICIs), with high tumor mutation burden (TMB-H) proposed as a predictive biomarker for ICI response, its predictive value has not been consistent across all cancer types, including breast cancer, prostate cancer, and glioma [27,28,29]. In our study, we observed a significant correlation between increased SUSD3 expression and decreased TMB in patients with BC. These findings indicate that SUSD3 may not be adequate for guiding breast cancer immunotherapy. Further research is warranted to elucidate the complex interplay between SUSD3 expression, TMB, and the response to immunotherapeutic interventions in breast cancer.
In conclusion, this study enhances our understanding of the involvement of SUSD3 in BC, providing insights into its role in disease pathogenesis and potential therapeutic targets. By leveraging data from public databases, we have provided a comprehensive assessment of BC molecular biology, laying the groundwork for further experimental investigations.
However, it is important to acknowledge certain limitations. First, reliance on publicly available databases and published literature may introduce biases, and the quality of data could affect the robustness of our findings. Second, the precision of database resources and the choice of statistical methodologies could influence result interpretation. However, the consistency of our results across multiple datasets strengthens the reliability of our conclusions.
Data availability statement
All data supporting the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- BC:
-
Breast cancer
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GSEA:
-
Gene Set Enrichment Analysis
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660. (Epub 2021 Feb 4, PMID: 33538338).
Global Burden of Disease 2019 Cancer Collaboration, Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of disease study 2019. JAMA Oncol. 2022;8(3):420–44. https://doi.org/10.1001/jamaoncol.2021.6987.
Gaboriaud C, Rossi V, Bally I, Arlaud GJ, Fontecilla-Camps JC. Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle. EMBO J. 2000;19(8):1755–65. https://doi.org/10.1093/emboj/19.8.1755.
Yu Z, Jiang E, Wang X, Shi Y, Shangguan AJ, Zhang L, et al. Sushi domain-containing protein 3: a potential target for breast cancer. Cell Biochem Biophys. 2015;72(2):321–4. https://doi.org/10.1007/s12013-014-0480-9. (PMID: 25556073).
Moy I, Todorović V, Dubash AD, Coon JS, Parker JB, Buranapramest M, et al. Estrogen-dependent sushi domain containing 3 regulates cytoskeleton organization and migration in breast cancer cells. Oncogene. 2015;34(3):323–33. https://doi.org/10.1038/onc.2013.553. (Epub 2014 Jan 13; PMID: 24413080; PMCID: PMC4096609).
Aushev VN, Lee E, Zhu J, Gopalakrishnan K, Li Q, Teitelbaum SL, et al. Novel predictors of breast cancer survival derived from miRNA activity analysis. Clin Cancer Res. 2018;24(3):581–91. https://doi.org/10.1158/1078-0432.CCR-17-0996. (Epub 2017 Nov 14; PMID: 29138345; PMCID: PMC6103440).
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. https://doi.org/10.1158/0008-5472.CAN-17-0307. (PMID:29092952; PMCID:PMC6042652).
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. https://doi.org/10.1093/nar/gkv007. (Epub 2015 Jan 20; PMID: 25605792; PMCID: PMC4402510).
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2(3):100141. https://doi.org/10.1016/j.xinn.2021.100141. (PMID: 34557778; PMCID: PMC8454663).
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;16(14):7. https://doi.org/10.1186/1471-2105-14-7. (PMID:23323831; PMCID:PMC3618321).
Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. Cell miner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72(14):3499–511. https://doi.org/10.1158/0008-5472.CAN-12-1370. (PMID:22802077; PMCID:PMC3399763).
Cheng Y, Yang X, Wang Y, Li Q, Chen W, Dai R, et al. Multiple machine-learning tools identifying prognostic biomarkers for acute myeloid leukemia. BMC Med Inform Decis Mak. 2024;24(1):2. https://doi.org/10.1186/s12911-023-02408-9. (PMID:38167056; PMCID:PMC10759623).
Badr NM, Zaakouk M, Zhang Q, Kearns D, Kong A, Shaaban AM. Concordance between ER, PR, Ki67, and HER2-low expression in breast cancer by MammaTyper RT-qPCR and immunohistochemistry: implications for the practising pathologist. Histopathology. 2024. https://doi.org/10.1111/his.151932. (Epub ahead of print; PMID: 38651302).
Dailey GP, Rabiola CA, Lei G, Wei J, Yang XY, Wang T, et al. Vaccines targeting ESR1 activating mutations elicit anti-tumor immune responses and suppress estrogen signaling in therapy resistant ER+ breast cancer. Hum Vaccines Immunother. 2024;20(1):2309693. https://doi.org/10.1080/21645515.2024.2309693. (Epub 2024 Feb 8; PMID: 38330990; PMCID: PMC10857653).
Ng CW, Tsang YTM, Gershenson DM, Wong KK. The prognostic value of MEK pathway-associated estrogen receptor signaling activity for female cancers. Br J Cancer. 2024;130(11):1875–84. https://doi.org/10.1038/s41416-024-02668-w. (Epub 2024 Apr 6; PMID: 38582811; PMCID: PMC11130254).
Przewoźny S, Rogaliński J, de Mezer M, Markowska A, Markowska J, Żurawski J. Estrogen receptor (ER) and progesterone receptor (PgR) expression in endometrial cancer—an immunohistochemical assessment. Diagnostics (Basel). 2024;14(3):322. https://doi.org/10.3390/diagnostics14030322. (PMID:38337838; PMCID:PMC10855223).
Yang Z, Chen H, Yin S, Mo H, Chai F, Luo P, et al. PGR-KITLG signaling drives a tumor-mast cell regulatory feedback to modulate apoptosis of breast cancer cells. Cancer Lett. 2024;589:216795. https://doi.org/10.1016/j.canlet.2024.2167956. (Epub 2024 Mar 30; PMID: 38556106).
Bahçacı U, Atasavun Uysal S, Erdogan İyigün Z, Ordu Ç, Soybir GR, Ozmen V. Progressive relaxation training in patients with breast cancer receiving aromatase inhibitor therapy-randomized controlled trial. PLoS ONE. 2024;19(4): e0301020. https://doi.org/10.1371/journal.pone.0301020. (PMID:38635763; PMCID:PMC11025930).
Guglielmi G, Del Re M, Gol LS, Bengala C, Danesi R, Fogli S. Pharmacological insights on novel oral selective estrogen receptor degraders in breast cancer. Eur J Pharmacol. 2024;969:176424. https://doi.org/10.1016/j.ejphar.2024.176424. (Epub 2024 Feb 23; PMID: 38402929).
Mansour B, Ngo C, Schlemmer D, Robidou P, Blondel J, Marin C, et al. Simultaneous quantification of four hormone therapy drugs by LC–MS/MS: clinical applications in breast cancer patients. J Pharm Biomed Anal. 2024;242:116032. https://doi.org/10.1016/j.jpba.2024.116032. (Epub 2024 Feb 12; PMID: 38367520).
Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221:107753. https://doi.org/10.1016/j.pharmthera.2020.107753. (Epub 2020 Nov 28; PMID: 33259885; PMCID: PMC8084948).
Dieci MV, Miglietta F, Guarneri V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells. 2021;10(2):223. https://doi.org/10.3390/cells10020223. (PMID:33498711; PMCID:PMC7911608).
Nalio Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, Bromley CP, Bugatti M, Núñez NG, et al. Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer. Cell. 2022;185(7):1189-1207.e25. https://doi.org/10.1016/j.cell.2022.02.021. (Epub 2022 Mar 23 PMID: 35325594).
Virassamy B, Caramia F, Savas P, Sant S, Wang J, Christo SN, et al. Intratumoral CD8+ T cells with a tissue-resident memory phenotype mediate local immunity and immune checkpoint responses in breast cancer. Cancer Cell. 2023;41(3):585-601.e8. https://doi.org/10.1016/j.ccell.2023.01.004. (Epub 2023 Feb 23; PMID: 36827978).
Zhao Y, Liu Z, Liu G, Zhang Y, Liu S, Gan D, et al. Neutrophils resist ferroptosis and promote breast cancer metastasis through aconitate decarboxylase 1. Cell Metab. 2023;35(10):1688-1703.e10. https://doi.org/10.1016/j.cmet.2023.09.004. (PMID:37793345; PMCID:PMC10558089).
Zhai Y, Liu X, Huang Z, Zhang J, Stalin A, Tan Y, et al. Data mining combines bioinformatics discover immunoinfiltration-related gene SERPINE1 as a biomarker for diagnosis and prognosis of stomach adenocarcinoma. Sci Rep. 2023;13(1):1373. https://doi.org/10.1038/s41598-023-28234-7. (PMID:36697459; PMCID:PMC9876925).
Snyder A, McGranahan N. The importance of ancestry to understanding tumor mutation burden in cancer. Cancer Cell. 2022;40(10):1076–8. https://doi.org/10.1016/j.ccell.2022.09.004. (Epub 2022 Sep 29; PMID: 36179683).
Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39(2):154–73. https://doi.org/10.1016/j.ccell.2020.10.001. (Epub 2020 Oct 29; PMID: 33125859; PMCID: PMC7878292).
McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32(5):661–72. https://doi.org/10.1016/j.annonc.2021.02.006. (Epub 2021 Mar 15; PMID: 33736924; PMCID: PMC8053682).
Acknowledgements
The authors gratefully acknowledge the Timer, TCGA, GEO, and HPA database for providing a platform to upload and access valuable datasets.
Funding
This study was supported by the National Natural Science Foundation of China (no. 81572916).
Author information
Authors and Affiliations
Contributions
Conception and design of the research: Yan-Ming Dong, Guo-Qiang Bao. Acquisition of data: Yan-Ming Dong. Analysis and interpretation of the data: Yan-Ming Dong. Statistical analysis: Yan-Ming Dong. Obtaining financing: Guo-Qiang Bao. Writing of the manuscript: Yan-Ming Dong. Critical revision of the manuscript for intellectual content: Guo-Qiang Bao. All authors read and approved the final draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval and Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12094_2024_3641_MOESM1_ESM.jpg
Supplementary file1 Figure S1: Clinical significance of SUSD3 gene expression in the prognosis of BC. A–G. Analysis of TCGA data reveals the prognostic significance of SUSD3 expression under various clinical traits, including age, stage, T stage, N stage, ER, PR, and Her-2 status. H–K. Similarly, the GEO dataset also demonstrates the prognostic value of SUSD3 expression under different clinical traits, encompassing Stage, ER, PR, and Her-2 status (JPG 1163 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dong, YM., Bao, GQ. Characterization of SUSD3 as a novel prognostic biomarker and therapeutic target for breast cancer. Clin Transl Oncol (2024). https://doi.org/10.1007/s12094-024-03641-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12094-024-03641-y