Abstract
Background
Intrahepatic cholangiocarcinoma (ICC) is characterized by fibrous stroma and clinical behavior more aggressive than that of hepatocellular carcinoma (HCC). Scirrhous HCC is a subtype of HCC with fibrous stroma, frequently has partial cholangiocytic differentiation, and is more likely to have an aggressive behavior. This study explored the interaction of liver cancer cells with the extracellular matrix.
Methods and results
Liver cancer cells grown on collagen 1-coated plates showed upregulation of cholangiocytic marker expression but downregulation of hepatocytic marker expression. Three-dimensional sphere culture and Boyden chamber assay showed enhanced invasion and migration ability in collagen 1-conditioned liver cancer cells. Interaction with collagen 1 reduced liver cancer cell proliferation. RNA sequencing showed that in the liver cancer cells, collagen 1 upregulated cell cycle inhibitor expression and cell–matrix interaction, tumor migration, and angiogenesis pathways, but downregulated liver metabolic function pathways. Cholangiocytic differentiation and invasiveness induced by collagen 1 was mediated by the mitogen-activated protein kinase (MAPK) pathway, which was regulated by cell–matrix interaction-induced Src activation. Analysis of the Cancer Genome Atlas cohort showed that collagen 1 induced and suppressed genes were highly enriched in ICC and HCC, respectively. In HCC samples, collagen 1-regulated genes were strongly coexpressed and correlated with COL1A1 expression.
Conclusions
Liver cancer cell–matrix interaction induces cholangiocytic differentiation and switches liver cancer cells from a proliferative to an invasive phenotype through the Src/MAPK pathway, which may partly explain the differences in the behaviors of HCC and ICC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) are the two most common types of primary liver cancer. HCC typically occurs as a consequence of viral hepatitis, fatty liver disease, exposure to aflatoxin B1, and liver cirrhosis [1]. By contrast, the major risk factors for ICC are liver fluke infestation, hepatolithiasis, and biliary tract malformation. Accumulating evidence suggests that viral hepatitis and non-biliary cirrhosis are also risk factors for ICC, suggesting HCC and ICC may share some pathogeneses [2]. Approximately 2–5% primary liver tumors show both hepatocytic and cholangiocytic differentiations (i.e., combined hepatocellular–cholangiocarcinoma) [3]. The existence of such biphenotypic tumors indicates the plasticity of neoplastic cells toward differentiation into two cell lineages—similar to hepatic stem cells. Compared with HCC, ICC is a more aggressive, leading to more frequent extrahepatic metastasis and poorer outcome [4].
A vast majority of HCC tumors comprise polygonal cells with hepatocytic differentiation arranged in thickened trabeculae surrounded by vascular spaces. By contrast, ICC tumors are composed of cuboidal or columnar tumor cells arranged in a glandular pattern within a fibrous stroma. Fibrous stroma of variable proportion is also seen in HCC. If fibrous stroma occupies > 50% of the tumor area, the tumor is subtyped as scirrhous HCC [5]. Scirrhous HCCs are more likely to have portal vein invasion and early tumor recurrence, but their long-term survival outcomes are similar to those of non-scirrhous HCC [6, 7].
HCC with fibrous stroma tends to express the cholangiocytic marker keratin 19 (KRT19) and has a cholangiocarcinoma-like gene expression trait [8]. These KRT19-expressing tumors are associated with more aggressive tumor behavior, portal vein invasion, and poor prognosis [9, 10]. In a previous study, we found that the expression of the cholangiocytic transcription factor HNF-1β and the cholangiocyte-specific chromatin modification marker H3K36Me3 was strongly correlated with KRT19 expression and associated with early tumor recurrence and poorer long-term survival [11, 12]. Taken together, a group of HCCs has abundant fibrous stroma, demonstrate partial cholangiocytic differentiation, and is more likely to be aggressive.
Despite these clinical observations, the mechanisms underlying the association of HCC tumors with the fibrous stroma, cholangiocytic differentiation, and tumor invasiveness remain unknown. In this study, we found that contact with collagen 1 induced cholangiocytic marker expression but suppressed hepatocytic marker expression in liver cancer cells. Moreover, this interaction with collagen 1 reduced the proliferation of liver cancer cells but promoted their aggressive behavior. This phenotypic change occurred through Src/mitogen-activated protein kinase (MAPK)-mediated transcriptional activation of target genes. Our results may provide a mechanistic insight into the association of fibrous stroma, cholangiocytic differentiation, and tumor invasiveness in liver cancer and highlight the importance of tumor–matrix interaction in determining cancer cell phenotypes and behaviors.
Materials and methods
Cell culture and reagents
Liver cancer cell lines were purchased from Bioresource Collection and Research Center (Hsinchu, Taiwan). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Waltham, MA) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin, nonessential amino acids, and 1 mM sodium pyruvate. All cells were incubated at 37 °C in a water-saturated atmosphere of 5% CO2/95% air.
For the preparation of matrix-coated plates, rat tail collagen 1 (5 μg/cm2; Corning, Corning, NA) dissolved in 0.02 M acetic acid and fibronectin (1 μg/cm2; Gibco) dissolved in phosphate buffered saline (PBS) was coated onto cell culture plates and air-dried. PD98059 and dasatinib were purchased from Sellckchem (Houston, TX).
Western blotting
Protein samples (20–50 μg each) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electrotransferred onto nitrocellulose membranes (Amersham, Buckinghamshire, UK). The membranes were then incubated with appropriate primary and secondary antibodies at the optimum dilutions. Finally, the immunoreactive signals were detected using an Immobilon Crescendo Western HRP substrate (Millipore, Burlington, MA). The antibodies and dilutions used are listed in Supplementary Table 1.
Real-time reverse transcription polymerase chain reaction
Total RNA was isolated from cells using Trizol reagent (Life Technologies, Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. The SYBR green-based real-time reverse transcription (RT) polymerase chain reaction (PCR) was performed to determine the mRNA levels on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase, a housekeeping gene, was used as the control for RNA quantification. All the PCR primers are listed in Supplementary Table 2.
For 20-μL PCR reaction volume, we mixed 1 μL of complementary DNA template with 10 μL of 2 × Power SYBR PCR master mix (Applied Biosystems), 200 nM paired primers, and distilled water. The PCR amplification protocol included initial incubation at 50 °C for 2 min, followed by denaturing at 95 °C for 10 min and then 40 cycles of denaturing at 95 °C for 15 s and annealing at 60 °C for 1 min. The obtained melting curves were analyzed after each run to verify PCR reaction specificity.
Cell proliferation assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide (MTT) (Sigma, St. Louis, MO) colorimetric assay was used to measure the cell proliferation rate. We seeded 2000 cells into each well of a 96-well plate and incubated the plate at 37 °C in a humidified atmosphere with 5% CO2. At the appropriate interval, 2 mg/mL MTT in PBS was added, followed by incubation for 4 h. The resulting colored reaction product, MTT formazan, was extracted using dimethyl sulfoxide and the absorbance at 570 nm was measured.
Boyden chamber assay
Modified Boyden chambers with filter inserts (pore size = 8 µm) in 24-well plates (Nucleopore, Pleasanton, CA) were used for a transwell migration assay. Here, 5 × 104 Huh7 cells or 1 × 105 HepG2 cells in 100 µL of serum-free DMEM were placed in the upper chamber, and 0.5 mL of DMEM was placed in the lower chamber. After 24 h of incubation, the cells were fixed in 3.7% formaldehyde for 10 min and then stained with 0.05% crystal violet and 4′-6-diamidino-2-phenylindole in PBS for 15 min. The cells on the upper side of the filters were then removed using cotton-tipped swabs, and the filters were washed with PBS. The cells on the underside of the filters were viewed and counted under a fluorescent microscope. Each group was plated in triplicate in each experiment, and each experiment was repeated at least three times.
Three-dimensional spheroid culture
For three-dimensional (3D) spheroid culture, 2000 single cells were plated in 8-well chamber slides onto a basal layer of 5 mg/mL Matrigel (Corning). To study the effects of type I collagen on tumor invasion, collagen 1 was added, at a final concentration of 1.65 mg/mL, to the basal layer when indicated. The basal layer was overlaid with 2% Matrigel-growth medium mixture. All cultures were grown for 6–8 days and observed under a phase microscope.
RNA sequencing
Total RNA was extracted from cell lines and sequenced on a Hi-Seq system. RNA quality and quantity were assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA), respectively. Thereafter, the RNA was ligated to adapters for further amplification using a TruSeq Stranded mRNA Library Prep kit (Illumina, San Diego, CA). The obtained library was sequenced on an Illumina NovaSeq sequencer. After finishing the sequencing, we performed quality control for the raw RNA sequencing (RNA-seq) data using FastQC, and adapters were trimmed using BBDuk. All reads were mapped to GRCh37 reference using STAR. Next, we quantified gene expression through RSEM.
Differentially expressed genes were identified using the DESeq2 R package. Differential expression was defined as > twofold change and false discovery rate < 0.05. Over-Representative Analysis (ORA) on WEB-based GEneSeT AnaLysis Toolkit (http://www.webgestalt.org/) was used to decipher differentially enrichment pathways. The RNA-seq data were deposited in NCBI Genome Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number: GSE176270.
Assay of transposase-accessible chromatin using sequencing
An assay of transposase-accessible chromatin using sequencing (ATAC-seq) was performed as described previously [13]. In brief, nuclei isolated from 50,000 cells were used for transposition reaction with transposase using a Nextera DNA Library Prep Kit (Illumina). Column-purified DNA was amplified in 50-μL reactions with NEBNext High-Fidelity 2 × PCR Master Mix (New England Biolabs, Ipswich, MA) using primers with unique barcodes (Genomics, New Taipei City, Taiwan) to setup library. The library was then sequenced on an Illumina NovaSeq sequencer to obtain 150-bp paired-end reads.
ATAC-Seq data analysis
All sequencing reads from ATAC-seq were trimmed using cutadapt (v. 3.0) to improve mapping efficiency and reduce misalignment risk. Trimmed reads were aligned to the human genome (Genome Reference Consortium GRCh38) using BWA-mem (v. 0.7.17). Reads on autosomes excluding the ENCODE human blacklisted regions [14] and PCR-duplicated reads identified by GATK-Picard (v 4.1.9.0) were kept for downstream analysis. Aligned reads were shifted + 4 and − 5 bp for positive and negative strands using alignmentSieve of deepTools (v. 3.5.0). The genomic distribution of aligned reads to transcription start sites (TSSs) were analyzed using deepTools. Peaks were identified using MACS2 (v. 2.2.7.1) with the parameters -f BAMPE, -keep-dup all, and -q 0.05. Identified peaks from the same cell line were merged using bedtools (v. 2.29.2) and ATAC peaks associated genes or nearby genes were identified using the ChIPseeker R package. Footprint and transcription factor binding analysis was performed by TOBIAS with transcription factor motifs of JASPAR (8th release). The read count of each ATAC peaks was calculated using bedtools and converted into count per million for further analysis. The NOISeq R package was utilized to identify the differential ATAC peaks.
Human tissue, immunohistochemistry, and Picrosirius red staining
Resection specimens from 17 patients diagnosed as having both conventional and scirrhous part of HCC from 2003 to 2020 in National Taiwan University Hospital were used in this study. These sections were reviewed by a hepatopathologist (Jeng, Y.-M.) to identify the conventional and scirrhous parts. This study was approved under the regulations of the Research Ethics Committee of the National Taiwan University Hospital (approval number: 201911077RINB) and conducted according to the principles of the Declaration of Helsinki.
Immunohistochemical staining for KRT19 and Ki-67 was performed in a Ventana BenchMark XT autostainer (Roche, Basel, Switzerland). After deparaffinization, rehydration, and antigen retrieval using CC1 (pH 8.0) for 20 min at 100 °C, the slides were incubated with the primary antibody for 16 min at 37 °C, followed by visualization using an OptiView DAB IHC Detection Kit (Roche). The antibodies used and their dilutions are listed in Supplementary Table 1. Picrosirius Red staining was conducted using a Picro Sirius Red Stain Kit (Abcam, Cambridge, UK).
Results
HCC cells grown on collagen 1 HAD enhanced cholangiocytic marker expression and reduced hepatocytic marker expression
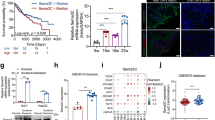
Studies have demonstrated that HCCs with abundant fibrous stroma commonly express cholangiocytic cytokeratin KRT19 [8, 15]. Immunostaining of HCC tumors with both conventional and scirrhous components demonstrated that KRT19 was more likely to be expressed in the scirrhous part. The expression of KRT19 tended to be predominant in the periphery of tumor nests, where tumor cells interact with the extracellular matrix (ECM) (Fig. 1a). The analysis of the Cancer Genome Atlas (TCGA) hepatocellular carcinoma cohort (LIHC, n = 372) showed that KRT19 expression is strongly correlated with the expression of COL1A1, which encodes a major component of type I collagen, in HCC (Fig. 1b).
HCC cell–collagen 1 interaction enhances cholangiocytic marker expression and reduces hepatocytic marker expression. a Immunostaining of HCC tumor sections with both conventional and scirrhous components show that KRT19 is more likely to be expressed in the scirrhous part (S) than in the conventional part (C). Picrosirius Red stain highlights the fibrous stroma. Note that KRT19 expression is predominantly located in the periphery of tumor nests. b Analysis of the Cancer Genome Atlas (TCGA) HCC cohort (LIHC, n = 372) shows that KRT19 expression is strongly correlated with COL1A1 expression. c Liver cancer cells grown on collagen 1- or fibronectin-coated plates have higher levels of KRT19 expression. d–g Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) assays show enhanced expression of the cholangiocytic markers KRT19 (d), KRT7 (e), JAG1 (f), and HES1 (g) in Huh7 and HepG2 cells grown on collagen 1. h–j RT-PCR assays show that the expression of the mature hepatocyte markers albumin (ALB) (h) and transferrin (TF) (i) and the immature hepatocyte marker α-fetoprotein (AFP) (j) is suppressed after interaction with collagen 1. ***p < 0.001, ****p < 0.0001
To study whether interaction with the ECM can induce liver cancer cell differentiation toward the cholangiocytic lineage, we first tested a panel of liver cancer cell lines for the expression of KRT19 and the mesenchymal marker vimentin (Supplementary Fig. 1). We found that HepJ5, HCC36, HA22T, and Mahlava cells expressed vimentin, indicating that these cells had undergone epithelial–mesenchymal transition. HA59T and HCC36 cells demonstrated high KRT19 expression intrinsically. Hep3B and PLC5 cells expressed neither KRT19 nor vimentin. Huh7 and HepG2 cells had low but detectable levels of KRT19, and did not express vimentin. Therefore, these two cell lines were chosen for subsequent studies.
Huh7 and HepG2 cells were grown on collagen 1-coated plates for 2 days. Compared with cells grown on non-coated plates, the cells grown on collagen 1-coated plates had higher KRT19 expression (Fig. 1c). Fibronectin—another major component of ECM—also induced the expression of KRT19 in liver cancer cell lines, indicating that this induction was independent of specific ECM components. Because collagen 1 is the most abundant ECM component in the fibrous tissue and the induction was more prominent in collagen 1-coated plate, we analyzed liver cancer cell–collagen 1 interaction in the subsequent experiments.
To further characterize the effects of collagen 1 on cell differentiation, we performed real-time RT-PCR for analyzing the expression of cholangiocytic and hepatocytic markers and found that Huh7 and HepG2 cells grown on collagen 1 had enhanced expression of cholangiocytic markers KRT19, KRT7, JAG1, and HES1 (Fig. 1d–g). By contrast, the expression of mature hepatocyte markers albumin (ALB) and transferrin (TF) and the immature hepatocyte marker α-fetoprotein (AFP) was suppressed after the interaction with collagen 1 (Fig. 1h–j). These results indicated that interaction with collagen 1 induces a phenotypic change toward the cholangiocytic lineage in liver cancer cells.
To study whether the induction effect is universal, we analyzed the expression levels of these markers in Hep3B, PLC5, HCC36, and HA59T cells and found neither collagen 1 nor fibronectin could induce similar phenotypic switch in these cell lines (Supplementary Fig. 2), indicating the observed phenomenon is cell-specific.
Interaction with collagen 1 enhanced tumor invasion but suppressed cell proliferation
HCCs with KRT19 expression and fibrous stroma had more frequent microvascular invasion and associated with poor prognosis [6, 7, 9, 10]. Therefore, we hypothesized that interaction with collagen 1 induces the invasive phenotype in liver cancer cells. In 3D spheroid culture, both Huh7 and HepG2 cells formed solid spheroid nests with a smooth external contour in Matrigel, which is mainly composed of basement membrane component laminin, entactin, type IV collagen, and heparan sulfate proteoglycan [16]. By contrast, when grown on Matrigel supplemented with collagen 1, these tumor nests became invasive, losing their rounded morphology and becoming stellate and protrusive (Fig. 2a).
Interaction with collagen 1 enhances tumor invasion but suppresses cell proliferation. a After grown in 3D culture for 6–8 days, Huh7 and HepG2 cells form solid spheroid nests with a smooth external contour in Matrigel. The tumor nests lose their spheroid morphology and become stellate and protrusive when grown in Matrigel supplemented with collagen 1. b Huh7 and HepG2 cells are cultured on collagen 1 for 2 days, followed by seeding of these cells on non-coated plates. KRT19 upregulation persisting on non-coated plates for 2 days after the transfer. c, d After preconditioning with collagen 1 for 2 days, Huh7 and HepG2 cells are seeded into the inner chamber of the Boyden chamber, where cells conditioned with collagen 1 have more migration capacity than those grown on non-coated plates. e, f MTT assay shows that Huh 7 cells grown on collagen 1-coated plates have considerably reduced cell proliferation rate but that collagen 1 induces slightly enhanced proliferation rate in HepG2 cells. g, h Immunostaining for the proliferation marker Ki-67 in 17 HCC tumors with both conventional and scirrhous components. The result shows that the scirrhous part (S) tends to have a lower proliferation rate than the conventional part (C). Picrosirius Red stain highlights fibrous stroma. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
A possible explanation for these results is that the contact with collagen 1 provided signals to direct tumor invasion into the ECM. Another possibility is that the interaction with collagen 1 altered the transcriptional program, thus inducing a more invasion phenotype in liver cancer cells. We cultured Huh7 and HepG2 cells on collagen 1 for 2 days and then seeded these cells on non-coated plates. The KRT19 upregulation persisted for 2 days after the cells were transferred and grown on non-coated plates, indicating that the cholangiocytic transdifferentiation can be maintained in collagen 1-free condition for at least 2 days (Fig. 2b). We cultured Huh7 and HepG2 cells on non-coated or collagen 1-coated plates for 2 days and then seeded them into a Boyden chamber. As shown in Fig. 2c, d, cells conditioned with collagen 1 had a larger migration capacity than those grown on non-coated plates.
The effects of collagen 1 on liver cancer cell proliferation were also evaluated. Huh 7 cells grown on collagen 1 had a considerably reduced cell proliferation rate, whereas collagen 1 induced a slight elevation in the proliferation rate of HepG2 cells (Fig. 2e, f). We observed the morphology and found that HepG2 cells formed tight adherent tumor nests on the non-coated plates but the cells spread out as single discohesive cells when they were grown on collagen 1 (Supplementary Fig. 3). The nest growth pattern may be the reason for the limited HepG2 cell proliferation on the non-coated plates. To study the effects of liver cancer cell–stroma interaction on cellular proliferation in human sample, we stained the proliferation marker Ki-67 in 17 samples of HCCs with both conventional and scirrhous components. As shown in Fig. 2g, h, the scirrhous part tended to have a lower proliferation rate than the conventional part.
Interaction with collagen 1 induced expression signature of tumor invasion and suppressed expression of genes associated with hepatocytic differentiation
To identify gene expression signatures associated with interaction with collagen 1, we performed RNA-seq on Huh7 and HepG2 cells grown on non-coated and collagen 1-coated plates. Collagen 1 induced upregulation of 418 genes in Huh7 cells and 331 genes in HepG2 cells (Fig. 3a, Supplementary Table 3 and 4). It also suppressed the expression of 304 genes in Huh7 cells and 199 genes in HepG2 cells (Fig. 3b). The upregulated and downregulated genes highly overlapped in the two cell lines. ORA of the commonly upregulated genes revealed significant enrichment in cell–cell and cell–substrate adhesion, cell motility, angiogenesis, and migration pathways (Fig. 3c). ORA of the commonly suppressed genes revealed a significant enrichment of lipid, nucleoside, ammonium ion, and organic acid metabolism pathways and genes responsive to toxic substance (Fig. 3d). These results supported our hypothesis that interaction with collagen 1 induces a more invasive phenotype and suppressed hepatocytic metabolic function in liver cancer cells.
Liver cancer cell–collagen 1 interaction induces expression signature of tumor invasion and suppresses expression of genes associated with hepatocytic differentiation. a Collagen 1 induces the upregulation of 418 genes in Huh7 cells, 331 genes in HepG2 cells, and 155 genes in both cell lines. b Collagen 1 suppresses the expression of 304 genes in Huh7 cells, 199 genes in HepG2 cells, and 49 genes in both cell lines. c Over-representative analysis (ORA) of the commonly upregulated genes reveals a significant enrichment in cell–cell adhesion, cell–substrate adhesion, cell mortality, angiogenesis, and migration pathways. d ORA of the commonly suppressed genes reveals a significant enrichment of lipid, nucleoside, ammonium ion, and organic acid metabolism pathways and genes responsive to toxic substances. e, f Gene set enrichment analysis of RNA-seq data of 372 HCC and 36 ICC samples in TCGA PanCancer Atlas shows that the genes activated and suppressed by liver cancer cell–collagen 1 interaction are highly enriched in ICC and HCC, respectively
We used the genes upregulated and suppressed by collagen 1 to generate two gene expression signatures. We then used these two signatures to query the publicly available RNA-seq data of 372 HCC and 36 ICC samples in TCGA PanCancer Atlas. Gene set enrichment analysis (GSEA) showed the genes activated and suppressed by liver cancer cell–collagen 1 interaction were highly enriched in genes predominantly expressed in ICC and HCC, respectively (Fig. 3e, f). Taken together, the results suggested that direct contact with collagen 1 induces tumor invasiveness and cholangiocytic differentiation and suppresses hepatocytic differentiation both in the cell culture system and human cancer tissue.
Real-time RT-PCR was used to confirm the RNA-seq data of selected genes in independent samples. We confirmed that the upregulation of 9 genes associated with tumor invasion (AXL, CTHRC1, CYR61, and FOSL1), matrix interaction (ITGA3 and ITGA5), and matrix remodeling (LOXL2, ADAM19, and TIMP2) was induced by collagen 1 in both Huh7 and HepG2 cells (Fig. 4a, b). Cell cycle inhibitors CDKN1A, CDKN2A, and CDKN2B were also upregulated by collagen 1 (Fig. 4c, d). The overexpression of these cell cycle inhibitors may be the reason for the slower proliferation of Huh7 cells grown on collagen 1 and the scirrhous part of HCC. We also confirmed that the interaction with collagen 1 downregulated the expression of hepatocyte differentiation-associated genes MT1H, APOH, and FABP1 (Fig. 4e, f).
Confirmation of the upregulation of invasiveness genes and cell cycle inhibitors and downregulation of hepatocytic differentiation-associated genes identified by RNA-seq data in independent samples. a, b Real-time RT-PCR assay shows nine genes associated with tumor invasion and matrix interaction are induced by collagen 1 in both Huh7 and HepG2 cells. c, d Cell cycle inhibitors CDKN1A, CDKN2A, and CDKN2B are upregulated by collagen 1. e, f Downregulation of hepatocytic differentiation-associated genes MT1H, APOH, and FABP1 by collagen 1. **p < 0.01, ***p < 0.001, ****p < 0.0001
Interaction with collagen 1 induced activation of the Src/Mapk pathway in liver cancer cells to promote cholangiocytic differentiation and invasion
Studies have demonstrated that the activation of the Notch and transforming growth factor-β pathway promotes cholangiocytic differentiation [17,18,19] Therefore, we tested whether these pathways were involved in cholangiocytic differentiation of liver cancer cells by interaction with collagen 1. Treatment with the γ-secretase inhibitor DAPT or the SMAD2/3 inhibitor SB4315542 did not abolish collagen 1-induced KRT19 upregulation (Supplementary Fig. 4). Therefore, the Notch and transforming growth factor-β pathways were not the mechanism underlying collagen 1-induced phenotypic switching.
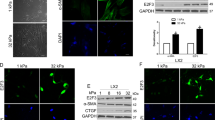
To identify the mechanisms underlying gene regulation by interaction with collagen 1, we used Connectivity Map [20] (https://www.broadinstitute.org/connectivity-map-cmap) to analyze the association of gene expression profile and treatment with bioactive small molecules. The genes commonly upregulated by the liver cancer cell–collagen 1 interaction in Huh7 and HepG2 cells were positively correlated with PKC activator treatment but negatively with MEK and Src inhibitor treatment—indicating that PKC, MAPK, and Src pathways are possible mechanisms underlying the collagen 1-induced gene upregulation (Fig. 5a). The analysis of the transcription factor binding data of chromatin immunoprecipitation databases ENCODE and ChEA demonstrated that FOSL2 had by far the highest score (adjusted p value = 4.5 × 10−7; odds ratio = 8.85, combined score = 164.75; Fig. 5b). FOSL2, a subunit of AP-1 transcription factor, is a major downstream target of the MAPK pathway. FOSL2 promotes tumor progression and invasion in HCC and other cancer types [21, 22]. Notably, FOSL1, a paralog of FOSL2, was one of the most upregulated genes induced by interaction with collagen 1 (Fig. 4a, b). The focal adhesion kinase (FAK)/Src pathway—one of the most important signaling pathways activated by integrin-dependent cell–matrix interaction—is activated in many invasive cancers [23, 24]. FAK/Src-dependent and P21-activated kinase 1(PAK1)-mediated phosphorylation of MEK1 on S298 contribute to the cell–matrix adhesion dependence of growth factor signaling to MAPK [24, 25]. Therefore, we focused on the effects of the Src/MAPK pathway. Seeding cells on collagen 1-coated plates induces phosphorylation of ERK, MEK at Ser 298, and Src (Fig. 5c), indicating the Src and MAPK pathway was activated by liver cancer cell–collagen 1 interaction. The Src inhibitor dasatinib abolished Src, MEK, and ERK phosphorylation induced by liver cancer cell–collagen 1 interaction, indicating that Src mediates the activation of MAPK by collagen 1. The treatment of Huh7 and HepG2 cells with the MEK inhibitor PD98059 abolished the collagen 1-induced invasive phenotype in both 3D spheroid culture and Boyden chamber migration assay (Fig. 5d, e and Supplementary Fig. 5a). Moreover, the collagen 1-induced upregulation of the cholangiocytic marker KRT19 and invasiveness genes (Fig. 5f, g and Supplementary Fig. 5b and 5c) was reduced by PD98059 treatment. By contrast, the hepatocyte marker ALB was restored in PD98059-treated cells (Fig. 5f and Supplementary Fig. 5b). Dasatinib treatment induced similar changes in tumor invasiveness and gene expression—similar to the results of PD98059 treatment (Fig. 5d and Supplementary Fig. 5a and 6).
Activation of the Src/MAPK pathway by collagen 1 in liver cancer cells promotes cholangiocytic differentiation and invasion. a Using the Connectivity Map, genes commonly upregulated by interaction with collagen 1 in Huh7 and HepG2 cells are correlated with PKC activator treatment positively but with MEK and Src inhibitor treatment negatively. b Analysis of the transcription factor binding data from the chromatin immunoprecipitation databases of ENCODE and ChEA identifies the most enriched transcription factor binding sites in the promoters of genes commonly upregulated by the live cancer cell–collagen 1 interaction in Huh7 and HepG2. c Huh7 and HepG2 cells grown on collagen have elevated levels of phosphorylated ERK, MEK at Ser 298, and Src, which can be reduced by treatment with the Src inhibitor dasatinib. d 3D spheroid culture assay shows Huh7 cells have an invasive morphology when grown in Matrigel supplemented with collagen 1. Treatment with the MEK inhibitor PD98059 (50 μM for Huh7 and 20 μM for HepG2) or the Src inhibitor dasatinib (100 nM) reverts the tumor morphology to a noninvasive spheroid. e Boyden chamber migration assay shows Huh7 cells preconditioned with collagen 1 have enhanced migration ability, which is abolished by PD98059 treatment. f, g Huh 7 cells grown on collagen 1 have elevated expression of the cholangiocytic marker KRT19 (f) and invasiveness genes (g) but decreased expression of the hepatocytic marker ALB (F). PD98059 treatment abolishes the changes of gene expression. **p < 0.01, ***p < 0.001
Liver cancer cell–collagen 1 interaction induced chromatin remodeling
To identify the downstream mechanisms for the regulation of gene expression due to liver cancer cell–collagen 1 interaction, we analyzed chromatin accessibility using ATAC-seq. As expected, ATAC peaks were preferentially observed in the proximity of TSSs (Supplementary Fig. 7). Liver cancer cells grown on non-coated and collagen 1-coated plates had similar degrees of chromatin accessibility (Fig. 6a). We assigned each peak to the closest gene based on the annotated TSSs. To investigate whether the gene expression changes caused by liver cancer cell–collagen 1 interaction can be linked to a direct effect on chromatin accessibility, we performed comparison analyses of transcriptomic and ATAC peaks and obtained a scatter plot showing good correspondence (Fig. 6b), which is exemplified by the high accessibility of the KRT19 gene in liver cancer cells grown on collagen 1 (Fig. 6c).
Liver cancer cell–collagen 1 interaction induces chromatin remodeling. a ATAC-seq results show that liver cancer cells grown on non-coated and collagen 1-coated plates have similar degrees of chromatin accessibility. b Comparison of RNA-seq and ATAC-seq data shows a positive correlation between gene expression and chromatin accessibility. c High accessibility of the KRT19 gene is noted in Huh7 and HepG2 cells grown on collagen 1. d, e Motif analysis demonstrates the enrichment of FOSL1/JUN, FOSL1/JUND, BACH1 and NFE2L1 binding sites in Huh7 and HepG2 cells grown on collagen 1-coated plates. KLF15, NRF1, and TCFL5 binding sites are enriched in Huh7 and HepG2 cells grown on non-coated plates
We used motif analysis to study enriched transcription factor binding sites in the loci with differential chromatin accessibility. DNA sequences bound by FOSL1/JUN and FOSL1/JUND—the downstream transcription factor complexes of the MAPK pathway [26]—were among the most commonly enriched motifs in liver cancer cells grown on collagen 1 (Fig. 6d, e). The two other motifs most commonly enriched in liver cancer cells grown on collagen 1 were the binding sites of BACH1 and NFE2L1, which were reported to promote tumor invasion [27, 28]. The three motifs most commonly enriched in liver cancer were binding sites of KLF15, NRF1, and TCFL5. Of these, KLF15 induces the development of mature hepatocyte from hepatoblasts and stimulates liver function gene expression [29]. NRF1 is a liver cancer repressor and regulator of fat metabolism and response to oxidative stress [30].
HCCs with collagen 1-induced signature clustered into a distinct transcriptomic subgroup of tumors
Hierarchical clustering of a pairwise similarity matrix for in the RNA-seq data of the TCGA HCC cohort demonstrated that the vast majority of collagen 1-upregulated and -suppressed genes were positively coexpressed in human HCC tissues, whereas the expression of collagen 1-upregulated genes was negatively associated with collagen 1-suppressed genes (Fig. 7a). The expression of collagen 1-upregulated genes had a strong positive correlation with COL1A1 expression (ρ = 0.82, p < 10−16) but the expression of collagen 1-suppressed genes did not show a correlation with COL1A1 expression—indicating that gene upregulation is the major effector of tumor cell–collagen 1 interaction in HCC samples (Fig. 7b, c). Tumors with high collagen 1-induced gene signature formed a distinct cluster in hierarchical clustering analysis (cluster C1, Fig. 7d). This patient group tended to have worse survival than other patients, but the difference was not statistically significant (Fig. 7e, f).
HCCs with collagen 1-induced signature cluster into a distinct transcriptomic subgroup of tumors in the TCGA HCC cohort. a Hierarchical clustering of the RNA-seq data of the TCGA HCC cohort demonstrates the vast majority of collagen 1-upregulated and -suppressed genes are positively coexpressed in human HCC tissues and that the expression of collagen 1-upregulated genes is negatively associated with that of collagen 1-suppressed genes. b Expression of collagen 1-upregulated genes shows a strong positive correlation with COL1A1 expression. c Expression of collagen 1-suppressed genes has no correlation with COL1A1 expression. d Tumors with high collagen 1-induced gene signature form a distinct cluster in hierarchical clustering analysis (cluster C1). e, f Cluster C1 patients tend to have worse progression-free survival (e) and overall survival (f) than other patients, but the difference is not statistically significant
Discussion
Hepatocytes and cholangiocytes are the two major cell types derived from liver stem cells. Malignant transformation of liver parenchyma cells results in two major cancer types: HCC and ICC. These two types of cancers are considered to be distinct, differing in etiology, genetic change, morphology, prognosis, and response to therapy. However, recent studies have indicated that primary liver carcinoma represents a spectrum of diseases—with conventional HCC composed of cancer cells with hepatocytic differentiation and vascular stroma on one end and large duct-type ICC composed of glandular epithelium in fibrous stroma, similar to hilar and extrahepatic cholangiocarcinoma on the other end [2]. The tumors between the two ends of the spectrum include small duct-type ICC, combined HCC–ICC, and scirrhous HCC [2, 3, 5, 6]. ICC is more aggressive than HCC, with a higher frequency of extrahepatic metastasis. Combined HCC–ICC and scirrhous HCC are also associated with more aggressive behavior than conventional HCC, but the underlying mechanisms warrant elucidation [3, 6].
The tumor mass is composed of not only tumor cells but also a heterogeneous population of inflammatory cells, stromal cells, blood vessels, and the ECM. This complex and dynamic microenvironment influences tumor cell growth, invasion, and metastasis. HCC and ICC have considerably different microenvironments. In general, HCC tumor nests are surrounded by vascular spaces, whereas ICC tumor glands are surrounded by dense collagenous fibrous stroma rich in activated fibroblasts and inflammatory cells. Scirrhous HCC has a microenvironment similar to that of ICC, with similarly aggressive behavior—suggesting that stromal components that are common between the tumor microenvironments of ICC and scirrhous HCC may modulate tumor behavior.
Seok et al. were the first to demonstrate that scirrhous HCC has an ICC-like gene expression profile [7]. The authors also observed that core epithelial–mesenchymal transition-related genes were expressed in scirrhous HCC. Overexpression of transforming growth factor-β was proposed as the mechanism for the observed phenotypic changes, but this hypothesis has not been proven by laboratory experiments. The same group also identified fibroblast-derived HGF promotes KRT19 expression via the MET/ERK1/2/AP1 and SP1 axis [31]. Although their results demonstrated the KRT19 overexpression mechanism in scirrhous HCC, they did not account for the expression of other cholangiocytic markers and aggressive behaviors. Notably, based on their and our observations, both fibroblasts and the ECM use the MAPK pathway to induce KRT19 expression, indicating the importance of this pathway in the regulation of KRT19 expression.
In this study, we observed that direct interaction between liver cancer cells and collagen 1 induced phenotypic changes in liver cancer cells, resulting in reduced expression of genes involved in the metabolic and synthetic functions of hepatocytes but overexpression of genes highly expressed in cholangiocytes. In addition, the liver cancer cell–collagen 1 interaction induced the expression of genes that promote ECM adhesion and invasion. In ICC, the genes induced and inhibited by liver cell–collagen 1 interaction were upregulated and downregulated, respectively. Furthermore, the genes induced by liver cancer cell–collagen 1 interaction were strongly correlated with COL1A1 expression in the TCGA HCC cohort. These observations indicate that the direct interaction between liver cancer cells and the ECM is a major mechanism shaping the behavior of liver cancer cells.
We identified the Src/MAPK pathway as the mechanism underlying liver cancer cell–ECM interaction-induced phenotypic change. Studies have demonstrated that interaction with the ECM induces integrin clustering, which leads to the recruitment and activation of a FAK/Src dual kinase complex [23]. Src phosphorylates Raf-1 on Tyr 340 and Tyr 341 to relieve its autoinhibition and activate the MAPK pathway [32]. FAK and Src signaling also affect PAK1-dependent phosphorylation of MEK1 on Ser 298 induced by cell adhesion to fibronectin [24, 25]. This tumor–ECM interaction–Src/MAPK pathway contributes to tumor progression by enhancing tumor cell proliferation and invasion [23]. Although the role of the MAPK pathway on the differentiation of liver cancer cells toward cholangiocytic differentiation has not been well characterized thus far, clinical and animal studies have provided some suggestions. Mutation of KRAS, the upstream regulator of the MAPK pathway, is observed in cholangiocarcinoma frequently but rarely in HCC [33]. In a mouse model, the induction of mutant KRAS and loss of TP53 in cholangiocytes resulted in the formation of cholangiocarcinoma, but targeting these genes exclusively in adult hepatocytes resulted in rare HCC [34]. Cholestatic injury induced by a 3,5-diethoxycarbonyl-1,4-dihydrocollidine diet accelerated hepatocyte-derived tumorigenesis and promoted phenotypic switching to cholangiocarcinoma—indicating that tumor cells had the potential to transform into cholangiocyte-like cells under appropriate signaling [34]. Mouse hepatoblasts underwent hepatocyte differentiation when transferred from collagen-coated plates to non-coated plates, suggesting that interaction with collagen 1 may be crucial for inhibiting hepatocytic differentiation during embryogenesis [35]. Whether mouse hepatoblasts use the ECM/Src/MAPK pathway to control differentiation requires confirmation.
In addition to invasiveness induction, the liver cancer cell–matrix interaction also inhibits liver cancer cell proliferation. In tumors with both scirrhous and conventional components, the cell proliferation rate is higher in the conventional component. Phenotypic switching between the proliferative and invasive state has also been observed in other cancer types [36, 37]. The current gene expression analysis suggested that the induction of cell cycle inhibitors CDKN1A/B and CKKN2B may be the mechanism for slower proliferation in cells grown on collagen 1. Similar to our discovery, Kamiya et al. found that MEK–ERK activity induced the accumulation of Cdkn2a in mouse fetal hepatoblasts to restrict proliferation [38].
Although the liver cancer cell–ECM interaction induced some ICC features, the tumor cells did not fully recapitulate ICC. For example, we did not observe induction of cholangiocyte-enriched transcription factors SOX9 and HNF-1β in liver cancer cells grown on collagen 1. No enrichment of the progenitor cell markers and epithelial–mesenchymal transition-related genes, which were highly expressed in scirrhous HCC [8, 10], was observed. Other factors, such as cell of origin, genetic alteration, and other microenvironmental factors, may also affect the differentiation status and behavior of liver cancer cells.
The phenotypic switch induced by interaction with ECM was limited to Huh-7 and HepG2 cells. HCC cells with intrinsically undetectable (Hep3B and PLC5 cells) or high (HA59T and HCC36) levels of KRT19 expression didn’t have the induction effects. This is consistent with the clinical observation that not all liver cancer cells in contact with ECM transdifferentiated into cholangiocyte-like cells. In HCCs with KRT19 expression, not all cancer in contact with ECM express KRT19. HepG2 is derived from a hepatoblastoma, which has the potential to differentiate into cholangiocyte-like cells [39]. Cells possessing features of bipotential liver precursors can be derived from Huh7 cell [40]. It is likely that only cancer cells with stem cell features have the capacity for bilineage differentiation.
In conclusion, we found that a direct interaction between liver cancer cells and the ECM results in a phenotypic switching of the cells toward cholangiocytic differentiation, promoting tumor invasion. These observations can explain the association among fibrous stroma, cholangiocytic differentiation, and aggressive behavior in ICC and scirrhous HCC. Therapeutic interventions targeting the tumor cell–ECM interaction may be beneficial to patients with ICC or scirrhous HCC.
Data availability
The RNA-seq data were deposited in NCBI Genome Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under the accession number: GSE176270. Other data are available on request.
Abbreviations
- 3D:
-
Three-dimensional
- AFP:
-
α-Fetoprotein
- ALB:
-
Albumin
- ATAC-seq:
-
Assay of transposase-accessible chromatin using sequencing
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- ECM:
-
Extracellular matrix
- FAK:
-
Focal adhesion kinase
- GESA:
-
Gene set enrichment analysis
- HNF-1β:
-
Hepatocyte nuclear factor 1β
- ICC:
-
Intrahepatic cholangiocarcinoma
- KRT19:
-
Keratin 19
- MAPK:
-
Mitogen-activated protein kinase
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H- tetrazoliumbromide
- ORA:
-
Over-representative analysis
- PAK1:
-
P21-activated kinase 1
- PBS:
-
Phosphate buffered saline
- RNA-seq:
-
RNA sequencing
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- TCGA:
-
The Cancer Genome Atlas
- TF:
-
Transferrin
- TSS:
-
Transcriptional start sites
References
Yu MC, Yuan JM, Govindarajan S, Ross RK. Epidemiology of hepatocellular carcinoma. Can J Gastroenterol. 2000;14:703–709
Liau JY, Tsai JH, Yuan RH, Chang CN, Lee HJ, Jeng YM. Morphological subclassification of intrahepatic cholangiocarcinoma: etiological, clinicopathological, and molecular features. Mod Pathol. 2014;27:1163–1173
Akiba J, Nakashima O, Hattori S, Tanikawa K, Takenaka M, Nakayama M, et al. Clinicopathologic analysis of combined hepatocellular-cholangiocarcinoma according to the latest WHO classification. Am J Surg Pathol. 2013;37:496–505
Mavros MN, Economopoulos KP, Alexiou VG, Pawlik TM. Treatment and prognosis for patients with intrahepatic cholangiocarcinoma: systematic review and meta-analysis. JAMA Surg. 2014;149:565–574
Torbenson MS. Morphologic subtypes of hepatocellular carcinoma. Gastroenterol Clin North Am. 2017;46:365–391
Lee JH, Choi MS, Gwak GY, Lee JH, Koh KC, Paik SW, et al. Clinicopathologic characteristics and long-term prognosis of scirrhous hepatocellular carcinoma. Dig Dis Sci. 2012;57:1698–1707
Huang SC, Liao SH, Su TH, Jeng YM, Kao JH. Clinical manifestations and outcomes of patients with scirrhous hepatocellular carcinoma. Hepatol Int. 2021;15:472–481
Seok JY, Na DC, Woo HG, Roncalli M, Kwon SM, Yoo JE, et al. A fibrous stromal component in hepatocellular carcinoma reveals a cholangiocarcinoma-like gene expression trait and epithelial-mesenchymal transition. Hepatology. 2012;55:1776–1786
Yuan RH, Jeng YM, Hu RH, Lai PL, Lee PH, Cheng CC, et al. Role of p53 and β-catenin mutations in conjunction with CK19 expression on early tumor recurrence and prognosis of hepatocellular carcinoma. J Gastrointest Surg. 2011;15:321–329
Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, et al. Human hepatocellular carcinomas with “stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54:1707–1717
Yuan RH, Lai HS, Hsu HC, Lai PL, Jeng YM. Expression of bile duct transcription factor HNF1β predicts early tumor recurrence and is a stage-independent prognostic factor in hepatocellular carcinoma. J Gastrointest Surg. 2014;18:1784–1794
Lien HC, Jeng YM, Jhuang YL, Yuan RH. Increased trimethylation of histone H3K36 associates with biliary differentiation and predicts poor prognosis in resectable hepatocellular carcinoma. PLoS One. 2018;13: e0206261
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1-21.29.9
Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9:9354
Krings G, Ramachandran R, Jain D, Wu TT, Yeh MM, Torbenson M, et al. Immunohistochemical pitfalls and the importance of glypican 3 and arginase in the diagnosis of scirrhous hepatocellular carcinoma. Mod Pathol. 2013;26:782–891
Emonard H, Grimaud JA, Nusgens B, Lapière CM, Foidart JM. Reconstituted basement-membrane matrix modulates fibroblast activities in vitro. J Cell Physiol. 1987;133:95–102
Jeliazkova P, Jörs S, Lee M, Zimber-Strobl U, Ferrer J, Schmid RM, et al. Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology. 2013;57:2469–2479
Clotman F, Lemaigre FP. Control of hepatic differentiation by activin/TGF-β signaling. Cell Cycle. 2006;5:168–171
Schaub JR, Huppert KA, Kurial SNT, Hsu BY, Cast AE, Donnelly B, et al. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935
Milde-Langosch K, Janke S, Wagner I, Schröder C, Streichert T, Bamberger AM, et al. Role of Fra-2 in breast cancer: influence on tumor cell invasion and motility. Breast Cancer Res Treat. 2008;107:337–347
Li Z, Liu Y, Yan J, Zeng Q, Hu Y, Wang H, et al. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging (Albany NY). 2020;12:2485–2497
Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523
Brown MC, Cary LA, Jamieson JS, Cooper JA, Turner CE. Src and FAK kinases cooperate to phosphorylate paxillin kinase linker, stimulate its focal adhesion localization, and regulate cell spreading and protrusiveness. Mol Biol Cell. 2005;16:4316–4328
Slack-Davis JK, Eblen ST, Zecevic M, Boerner SA, Tarcsafalvi A, Diaz HB, et al. PAK1 phosphorylation of MEK1 regulates fibronectin-stimulated MAPK activation. J Cell Biol. 2003;162:281–291
Jiang X, Xie H, Dou Y, Yuan J, Zeng D, Xiao S. Expression and function of FRA1 protein in tumors. Mol Biol Rep. 2020;47:737–752
Sato M, Matsumoto M, Saiki Y, Alam M, Nishizawa H, Rokugo M, et al. BACH1 promotes pancreatic cancer metastasis by repressing epithelial genes and enhancing epithelial-mesenchymal transition. Cancer Res. 2020;80:1279–1292
Lee YK, Kwon SM, Lee EB, Kim GH, Min S, Hong SM, et al. Mitochondrial respiratory defect enhances hepatoma cell invasiveness via STAT3/NFE2L1/STX12 axis. Cancers (Basel). 2020;12:2632
Anzai K, Tsuruya K, Ida K, Kagawa T, Inagaki Y, Kamiya A. Kruppel-like factor 15 induces the development of mature hepatocyte-like cells from hepatoblasts. Sci Rep. 2021;11:18551
Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci USA. 2005;102:4120–4125
Rhee H, Kim HY, Choi JH, Woo HG, Yoo JE, Nahm JH, et al. Keratin 19 expression in hepatocellular carcinoma is regulated by fibroblast-derived HGF via a MET-ERK1/2-AP1 and SP1 axis. Cancer Res. 2018;78:1619–1631
Tran NH, Frost JA. Phosphorylation of Raf-1 by p21-activated kinase 1 and Src regulates Raf-1 autoinhibition. J Biol Chem. 2003;278:11221–11226
Tada M, Omata M, Ohto M. Analysis of ras gene mutations in human hepatic malignant tumors by polymerase chain reaction and direct sequencing. Cancer Res. 1990;50:1121–1124
Hill MA, Alexander WB, Guo B, Kato Y, Patra K, O’Dell MR, et al. Kras and Tp53 mutations cause cholangiocyte- and hepatocyte-derived cholangiocarcinoma. Cancer Res. 2018;78:4445–4451
Strick-Marchand H, Weiss MC. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology. 2002;36:794–804
Hur H, Lee JY, Yang S, Kim JM, Park AE, Kim MH. HOXC9 induces phenotypic switching between proliferation and invasion in breast cancer cells. J Cancer. 2016;7:768–773
Janssen SM, Moscona R, Elchebly M, Papadakis AI, Redpath M, Wang H, et al. BORIS/CTCFL promotes a switch from a proliferative towards an invasive phenotype in melanoma cells. Cell Death Discov. 2020;6:1
Kamiya A, Ito K, Yanagida A, Chikada H, Iwama A, Nakauchi H. MEK-ERK activity regulates the proliferative activity of fetal hepatoblasts through accumulation of p16/19(cdkn2a). Stem Cells Dev. 2015;24:2525–2535
Zimmermann A. Hepatoblastoma with cholangioblastic features ('cholangioblastic hepatoblastoma’) and other liver tumors with bimodal differentiation in young patients. Med Pediatr Oncol. 2002;39:487–491
Ito M, Sun S, Fukuhara T, Suzuki R, Tamai M, Yamauchi T, et al. Development of hepatoma-derived, bidirectional oval-like cells as a model to study host interactions with hepatitis C virus during differentiation. Oncotarget. 2017;8:53899–53915
Acknowledgements
We thank the 2nd and 3rd Core Laboratory of National Taiwan University Hospital and Translational core facility of Taipei Medical University for technical support.
Funding
The study was supported by grants from Ministry of Science and Technology, Republic of China (grant number: 108-2320-B-002-059-MY3 and 109-2314-B-002-082).
Author information
Authors and Affiliations
Contributions
Study concept and design: JYM and YRH; methodology and technical support: YRH and JYL; analysis and interpretation of data: HCL, LYR and HTH; writing, review and/or revision of the manuscript: JYM, YRH, and HCL.
Corresponding author
Ethics declarations
Conflict of interest
Ray-Hwang Yuan, Chia-Lang Hsu, Yu-Lin Jhuang, Yun-Ru Liu, Tsung-Han Hsieh, and Yung-Ming Jeng have no conflicts of interest relevant to this article.
Ethic approval
This study was approved under the regulations of the Research Ethics Committee of the National Taiwan University Hospital (approval number: 201911077RINB) and conducted according to the principles of the Declaration of Helsinki.
Animal research
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Plant reproducibility
Not applicable.
Clinical trials registration
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. 1.
Supplementary file5 KRT19 and vimentin expression in liver cancer cell lines. HepJ5, HCC36, HA22T, and Mahlava cells express vimentin. HA59T and HCC36 cells have intrinsically high KRT19 expression. Hep3B and PLC5 cells lack KRT19 and vimentin expression. Huh7 and HepG2 have low but detectable KRT19 expression but no vimentin expression. (TIF 244 KB)
Supplementary Fig. 2.
Supplementary file6The effects of interaction with collagen 1 or fibronectin on the expression levels of hepatocellular or cholangiocytic differentiation markers in Hep3B, PLC5, HCC36, and HA59T cells. N.D.: not detected (Ct value >30). (TIF 591 KB)
Supplementary Fig. 3.
Supplementary file7Morphology of Huh7 and HepG2 cells on non-coated and collagen 1–coated plates. Huh7 cells grown on non-coated and collagen 1–coated plates have similar morphology. HepG2 cells forms tightly adherent tumor nests on non-coated plates, but they spread out as single discohesive cells on the collagen 1–coated plate. (TIF 2146 KB)
Supplementary Fig. 4.
Supplementary file8The Notch and transforming growth factor-β pathways are not involved in collagen 1–induced KRT19 expression in Huh7 and HepG2 cells. Treatment with the γ-secretase inhibitor DAPT (a) or SMAD2/3 inhibitor SB4315542 (b) has no effect on collagen 1–induced upregulation of KRT19 expression. (TIF 378 KB)
Supplementary Fig. 5.
Supplementary file9a Three-dimensional spheroid culture assay showing that HepG2 cells have an invasive morphology when grown in Matrigel supplemented with collagen 1. Treatment with MEK inhibitor PD98059 (20 µM) or Src inhibitor dasatinib (100 nM) returns the morphology to a noninvasive spheroid. b, c HepG2 cells grown on collagen 1 have elevated expression of the cholangiocytic marker KRT19 (b) and invasiveness genes (c) but reduced expression of the hepatocytic marker ALB (b). PD98059 treatment abolishes the changes in gene expression. *P < 0.05, **P < 0.01, ***P < 0.001. (TIF 456 KB)
Supplementary Fig. 6.
Supplementary file10Huh7 (a, b) and HepG2 (c, d) cells grown on collagen 1 have elevated expression levels of the cholangiocytic marker KRT19 (a, c) and invasiveness genes (b) but reduced expression levels of the hepatocytic marker ALB (a, c). Dasatinib treatment (100 nM) abolishes the changes in gene expression. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (TIF 383 KB)
Supplementary Fig. 7.
Supplementary file11Fragment size, the location of ATAC peaks related to transcription start site, and the distribution of ATAC peaks. a Huh7 cells grown on a non-coated plate, b Huh7 cells grown on a collagen 1–coated plate, c HepG2 cells grown on a non-coated plate, d HepG2 cells grown on a collagen 1–coated plate. (TIF 426 KB)
Rights and permissions
About this article
Cite this article
Yuan, RH., Hsu, CL., Jhuang, YL. et al. Tumor–matrix interaction induces phenotypic switching in liver cancer cells. Hepatol Int 16, 562–576 (2022). https://doi.org/10.1007/s12072-022-10315-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-022-10315-w