Abstract
The chloroplast genome sequence of Nitraria tangutorum, a desert shrub, was sequenced using high-throughput sequencing technology and analysed phylogenetically in the present study. The chloroplast genome is 159,414 bp in length, including a large single copy region of 87,924 bp and small single copy region of 18,318 bp, and a pair of inverted repeat regions of 26,586 bp. The chloroplast genome contains 110 unique genes, including 77 protein-coding genes, four ribosomal RNA genes, and 29 tRNA genes. Most of these genes are present as a single copy and in two or more copies 19 genes occurred. Seventeen genes have one intron, and clpP and ycf3 genes contain two introns. A total of 81 simple sequence repeats (SSRs) were identified, most of them were found to be mononucleotide repeats composed of A/T. In addition to SSRs, 66 repeats were identified, including 41 tandem repeats, 10 palindromic repeats, and 15 forward repeats. The phylogenetic analysis based on 54 protein-coding genes demonstrated a close relationship between N. tangutorum and other plant species in Sapindales. The complete chloroplast genome sequence of N. tangutorum will provide important data for further study of taxonomy and systematics of the genus Nitraria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitraria tangutorum, a member of genus Nitraria, is a wild shrub distributed in the desert and semi-desert areas of northwest China. It exhibits high tolerance to high salinity and drought stresses, and plays a key role in maintaining the fragile ecosystems in the desert areas of central Asia (Yang et al. 2010). In addition, N. tangutorum is of great economic value for the local people (Liu et al. 2014), e.g., fruits and seeds are used to make medicines and drinks (Zhao et al. 2017), and the dry branches are often used as firewood by locals.
As a shrub with ecological and economic importance in harsh environment, N. tangutorum has attracted the attention of many researchers in recent years. Studies have been conducted to investigate the ecological adaption and stress tolerance mechanism by using molecular biological technology and biochemistry methods (Yang et al. 2013; Zheng et al. 2014; Yan et al. 2018). However, the phylogenetic relationship of the genus Nitraria remains an open question. N. tangutorum was classified into different family by Liu and Zhou (2003), and Xu and Huang (1998). (http://frps.iplant.cn/frps/Nitraria). Nitraria is one of the six genus in Zygophyllaceae of Geraniales (Xu and Huang 1998), but by Liu and Zhou (2003, http://foc.iplant.cn/), genus Nitraria was classified into Nitraria of Sapindales. Thus, more molecular evidences are needed to clarify the evolutionary position of genus Nitraria.
Recent studies have shown that the chloroplast genome sequences are essential data for plant phylogenetic and genetic population analyses (Parks et al. 2009). Thus, the phylogenetic analysis using the complete chloroplast genome of N. tangutorum should be an appropriate way to get a better understanding of the evolution of this plant species. Here, we present a complete chloroplast genome of N. tangutorum based on the next-generation sequencing data.
Materials and methods
Plant materials
Leaf sample of a wild individual of N. tangutorum were collected by Fei Gao from Mengxi Town, Erdos City, Inner Mongolia Autonomous Region (\(106^{\circ }\,79'\hbox {E}\), \(39^{\circ }83'\hbox {N}\)). The sample (PM20181001-Nta-1) was deposited in College of Life and Environmental Sciences, Minzu University of China, Beijing.
Genome sequencing and annotation
The genomic DNA was extracted from the leaves using the modified CTAB method (Doyle 1987). DNA sequencing was performed using an Illumina Hiseq2500 (Illumina, San Diego, USA) at Shenzhen Huitong biotechnology (Shenzhen, China). After adapter trimming and filtering of the low quality reads (read has >5% unidentified nucleotides and >50% of its bases with a quality value of <20.), the resulting clean reads were assembled into contigs using the assembler SPAdes v3.9.0 (Bankevich et al. 2012) using the default parameters. The contigs were aligned to chloroplast genome sequences of Arabidopsis thaliana and Nicotiana tabacum using BLAST program (E value \(<1\hbox {e}^{-10})\) to find the fragments of chloroplast genome of N. tangutorum and the contigs supported by higher sequencing depth were used for assembling chloroplast genome of N. tangutorum.
The genes in chloroplast genome of N. tangutorum were annotated using the DOGMA tool with default parameters (Wyman et al. 2004). Online program OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html; Lohse et al. 2013) was used to draw the gene map of the N. tangutorum chloroplast genome. The finally annotated chloroplast genome of N. tangutorum was deposited in GenBank with the accession number MK341053.
Repeat and SSR analysis
Simple sequence repeats (SSR) are some DNA repeats formed by one or several tandemly arranged nucleotides, which spread widely in eukaryotic genomes. Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/misa.html) was used to detect microsatellites with minimal repeat numbers of 10, 5 and 4 for mononucleotide, dinucleotide and trinucleotide repeats, respectively.
Phylogenetic analysis
To get more knowledge about phylogenetic analysis of N. tangutorum, we chose 18 plant species (table 1) from five orders (Sapindales, Geraniales, Malpighiales, Zygophyllales and Oxalidales) which belong to the same evolutionary branch (rosids) as N. tangutorum, and the phylogenetic relationships of these species and N. tangutorum were analysed. To analyse the phylogenetic tree of N. tangutorum, we downloaded their whole chloroplast genome sequences from the NCBI Organelle Genome and Nucleotide Resources. The software MAFFT 7.380 (Katoh and Standley 2013) was used to align the genome sequence, and RAxML 8.2.4 (Stamatakis 2014) was used to analyse the evolution of these species. The significance level for the phylogenetic tree was assessed by bootstrap testing with 1000 replications.
Results and discussion
Organization and features of the N. tangutorum chloroplast genome
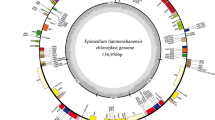
Gene map of the N. tangutorum chloroplast genome. The genes shown inside of the circle indicates transcriptional direction is clockwise, while those shown outside are counterclockwise. Genes belonging to different functional groups are labelled with different colours. The GC content of the genome shown with grey histogram in the inner circle, and the grey line depicts the 50% threshold line.
The chloroplast genome of N. tangutorum was assembled using \(\sim \)3.98 G sequencing reads. The length of chloroplast genome of N. tangutorum was 159,414 bp, and the average sequencing depth was 2494.8X. The large single copy (LSC) region, small single copy (SSC) region and the two inverted repeat regions (IRs), IRa and IRb, were 87,924, 18,318, and 26,586 bp in length, respectively (figure 1). A total of 110 unique genes were annotated from the chloroplast genome of N. tangutorum, including 77 protein-coding genes, four ribosomal RNA genes, and 29 tRNA genes. Most of these genes were present as single copy and in two or more copies 19 genes occurred. Of the 110 unique genes, 58 were involved in self-replication of chloroplast genome, 12 genes encode ribosomal small subunit proteins, nine genes encode ribosomal large subunit proteins, and four genes encode RNA polymerase subunits. Forty-three genes in N. tangutorum chloroplast genome encode proteins associated with photosynthesis, including six ATP synthase subunits, 11 subunits of NADH dehydrogenase complex, six components of cytochrome b/f complex, five subunits of photosystem I, 14 subunits of photosystem II, and one large chain of rubisco (table 2). A total of 17 genes with introns were found in the chloroplast genome of N. tangutorum. Among these genes, atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC had one intron, and clpP and ycf3 gene contained two introns. The overall GC content of the N. tangutorum chloroplast genome was 37.3%.
Repeat and SSR analysis
SSR markers in the N. tangutorum chloroplast genome were predicted using MISA, and compared with the chloroplast genomes of E. carvifolium, Pelagronium x hortorum, L. usitatissimum and A. carambola. In total, 81 SSRs were identified in the N. tangutorum chloroplast genome, including 78 mononucleotide repeats and three dinucleotide repeats, and no other type of SSR markers was found. The total numbers of the SSR repeats were 31, 74, 35 and 65 in E. carvifolium, P. x hortorum, L. usitatissimum and A. carambola, respectively (figure 2a). Similar to N. tangutorum, only mononucleotide repeats and dinucleotide repeats were found in the chloroplast genomes of E. carvifolium, P. x hortorum, L. usitatissimum and A. carambola. The total number of SSRs predicted from the N. tangutorum chloroplast genome were comparable to the chloroplast genomes of P. x hortorum and A. carambola, and were higher than those of E. carvifolium and L. usitatissimum.
A total of 66 repeats were identified from N. tangutorum chloroplast genome, including 41 tandem repeats, 10 palindromic repeats, and 15 forward repeats. The distribution of the repetitive sequences of different species in the chloroplast genomes of N. tangutorum, E. carvifolium, L. usitatissimum and A. carambola were similar: the tandem repeats is also the most abundant repeat category, followed by forward repeats and palindromic repeats. However, in the chloroplast genome of P. x hortorum, the numbers of the three categories of repeats (tandem repeat, forward repeat and palindromic repeat) are very similar. In addition, no reverse repeat was found in all these chloroplast genomes (figure 2b).
Phylogenetic analysis of N. tangutorum based on conserved protein sequences
A phylogenetic analysis was performed based on 19 complete chloroplast genomes of plant species in Sapindales, Geraniales, Malpighiales, Zygophyllales, and Oxalidales. The phylogenetic tree was constructed from the 54 protein-coding genes presented in all the 13 species using maximum likelihood (ML). RAxML was used to construct the ML tree with 1000-bootstrap replicates (Stamatakis 2014). The results indicated that N. tangutorum was clustered into a monophyletic group with the other eight plant species in Sapindales, and the five species in Geraniales were clustered in another clade (figure 3). Our results supported the taxonomic status of N. tangutorum defined in Liu and Zhou (2003, http://foc.iplant.cn/) and Angiosperm phylogeny website (Stevens 2001). In brief, the present study characterized the complete chloroplast genome structure of N. tangutorum, and clarified phylogenetic relationships of N. tangutorum and relative taxa in rosids, which may be useful for further study of taxonomy and systematics of genus Nitraria.
References
Bankevich A., Nurk S., Antipov D, Gurevich A. A., Dvorkin M., Kulikov A. S. et al. 2012 SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477.
Doyle J. J. 1987 A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Katoh K. and Standley D. M. 2013 MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780.
Liu Y. and Zhou L. 2003 Nitrariaceae. In Flora of China (ed. Z. Wu and P. Raven), vol. 11, pp. 41–42. Science Press, Beijing.
Liu Z., Mei L., Wang Q., Shao Y. and Tao Y. 2014 Optimization of subcritical fluid extraction of seed oil from Nitraria tangutorum using response surface methodology. LWT- Food Sci. Technol. 56, 168–174.
Lohse M., Drechsel O., Kahlau S. and Bock R. 2013 Organellar genome DRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, 575–581.
Parks M., Cronn R. and Liston A. 2009 Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7, 84.
Stamatakis A. 2014 RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Stevens P. F. 2001 Angiosperm phylogeny website, version 14, July 2017. http://www.mobot.org/MOBOT/research/APweb/ (accessed 30 June 2019).
Wyman S. K., Jansen R. K. and Boore J. L. 2004 Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255.
Xu L. and Huang C. (ed.) 1998 Zygophyllaceae. In Flora reipublicae popularis sinicae, vol. 43, pp. 122–123. Science Press, Beijing.
Yan Y. Q., Pan C. H., Du Y. L., Li D. Y. and Liu W. 2018 Exogenous salicylic acid regulates reactive oxygen species metabolism and ascorbate–glutathione cycle in Nitraria tangutorum Bobr. under salinity stress. Physiol. Mol. Biol. Plants. 24, 577–589.
Yang Y. L., Yang F., Li X. N., Shi R. X. and Lu J. 2013 Signal regulation of proline metabolism in callus of the halophyte Nitraria tangutorum Bobr. grown under salinity stress. Plant Cell Tiss. Org. Cult. 112, 33–42.
Yang Y., Shi R., Wei X., Fan Q. and An L. 2010 Effect of salinity on antioxidant enzymes in calli of the halophyte Nitraria tangutorum Bobr. Plant Cell Tiss. Org. Cult. 102, 387–395.
Zhao J. Q., Wang Y. M., Yang Y. L., Zeng Y., Wang Q. L., Shao Y. et al. 2017 Isolation and identification of antioxidant and \(\upalpha \)-glucosidase inhibitory compounds from fruit juice of Nitraria tangutorum. Food Chem. 227, 93–101.
Zheng L. L., Dang Z. H., Li H. Y., Zhang H. R., Wu S. B. and Wang Y. C. 2014 Isolation and characterization of a \(\Delta \)1-pyrroline-5-carboxylate synthetase (NtP5CS) from Nitraria tangutorum Bobr. and functional comparison with its Arabidopsis homologue. Mol. Biol. Rep. 41, 563–572.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31670335, 31570407, and 31770363) and the Ministry of Education of China through 111 and ‘Double First-Class’projects, grant number B08044 and Yldxxk201819.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Rights and permissions
About this article
Cite this article
Abla, M., Zha, X., Wang, Y. et al. Characterization of the complete chloroplast genome of Nitraria tangutorum, a desert shrub. J Genet 98, 91 (2019). https://doi.org/10.1007/s12041-019-1135-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1135-9