Abstract
This study aimed to identify vernalization responsive genes in the winter wheat cultivar Jing841 by comparing the transcriptome data with that of a spring wheat cultivar Liaochun10. For each cultivar, seedlings before and after the vernalization treatment were sequenced by Solexa/Illumina sequencing. Genes differentially expressed after and before vernalization were identified as differentially expressed genes (DEGs) using false discovery rate (FDR) ≤ 0.001 and |log2 (fold change)| >1 as cutoffs. The Jing841-specific DEGs were screened and subjected to functional annotation using gene ontology (GO) database. Vernalization responsive genes among the specific genes were selected for validation by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and the expression change over the time was investigated for the top 11 genes with the most significant expression differences. A total of 138,062 unigenes were obtained. Overall, 636 DEGs were identified as vernalization responsive genes including some known genes such as VRN-1 and COR14a, and some unknown contigs. The qRT-PCR validated changes in the expression of 18 DEGs that were detected by RNA-seq. Among them, 11 genes displayed four different types of expression patterns over time during the 30-day-long vernalization treatment. Genes or contigs such as VRN-A1, COR14a, IRIP, unigene1806 and Cl18953. Contig2 probably have critical roles in vernalization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Temperature is an important abiotic stimuli, provides diurnal and seasonal cues for plants, thus enabling them to adapt to environmental changes (Winfield et al. 2010). Different wheat cultivars exhibit great genetic variability when exposed to low temperature (Monroy et al. 2007). Requiring a long period of cold treatment to become competent to flower is termed as vernalization (Winfield et al. 2009), which delays the reproductive growth before winter and enhances the capacity of cold acclimation (Limin and Fowler 2002). Winter wheat cultivars need vernalization; however, spring wheat cultivars do not (Winfield et al. 2010). Further, Entz and Fowler (1991) reported that winter wheat outyielded spring wheat by an average 36%.
Vernalization genes, VRN1, VRN2 and VRN3, are recognized to have critical roles in vernalization in wheat (Yan et al. 2004). VRN1 is a positive regulator that converts plants from the vegetative growth phase to the reproductive phase, and maintains this conversion when the plants are shifted to warm condition (Yan et al. 2003), whereas VRN2 is a negative regulator (Yan et al. 2004). MADS box transcription factor TaVRT2 (vegetative to reproductive transition2) is associated with TaVRN2 in the vernalization response and represses the transcription of TaVRN1 (Kane et al. 2007). VRN3 is linked completely to a gene similar to Arabidopsis FT (flowering locus T), and plants with dominant Vrn3 alleles have an early flowering phenotype (Yan et al. 2006). Efforts have been made for fine mapping and epistatic interactions of VRN-D4 in hexaploid wheat (Kippes et al. 2014). Vernalization genes have been considered to regulate the expression of low temperature-induced genes in winter wheat and rye (Fowler et al. 1996). Low-temperature responsive genes have been identified by molecular analysis, such as dehydrins and crystallization genes (Thomashow 1999). Overexpression of TaSnRK2.8 enhanced the tolerance to low temperature in Arabidopsis (Zhang et al. 2010).
Next-generation sequencing can provide substantial genetic information, and transcriptome sequencing is more cost-efficient compared to whole-genome sequencing (Parchman et al. 2010). Spring wheat cultivar Liaochun10 and winter wheat cultivar Jing841 are widely grown with high production yield in North China. In this study, we attempted to identify the vernalization responsive genes by comparing the transcriptome data between a spring wheat cultivar Liaochun10 and a winter wheat cultivar Jing841, each undergoing an artificial vernalization treatment. Further, we also investigated the expression pattern of these vernalization-responsive genes during the vernalization period to unravel their roles in this event.
Material and methods
Plant materials
Liaochun10, a spring wheat cultivar, widely grown in northern and northeast China (Dong 1993; Zhao and Zhao 1998), is a superior bread-making variety, and Jing841, a winter wheat cultivar, mainly grown in northern and northwestern China, is high-yielding and resistant to disease and insects (Dong 1993; Zhao and Zhao 1998). Seeds of Liaochun10 and Jing841 were first placed on moist filter paper in Petri dishes and incubated in the dark at 20°C for 24 h. Next, the germinated seeds of each cultivar were incubated in moist sterilized vermiculite in the dark at 4°C for 30 days for vernalization treatment. For each wheat cultivar, seedlings before vernalization and after 30-day-long cold treatment were sampled for constructing cDNA libraries. Meanwhile, during the 30-day-long cold vernalization treatment, the seedlings were sampled every 6 days (at day 6, 12, 18, 24 and 30), followed by immediate storage in liquid nitrogen at −80°C for later qRT-PCR.
Solexa/Illumina sequencing, data assembly and annotation
For each wheat cultivar, total RNA was extracted from leaves using the RNAiso Plus Kit (Takara, Tokyo, Japan) according to the manufacturer’s instruction. After ethanol precipitation, total RNA was dissolved in DEPC-treated water and stored at −80°C. RNA quality was checked on 1.0% agarose gel; RNA integrity was determined using Agilent 2000 Bioanalyser CE system. For each wheat cultivar, cDNA libraries were constructed using seedlings before vernalization and the vernalized seedlings, respectively. In total, four cDNA libraries were constructed. Then, each cDNA library was sequenced using Illumina HiSeq 2000.
Paired-end reads of 90 bp in length were generated. The raw reads were first filtered by removing adaptor reads, reads contaning N of >5%, as well as low-quality reads that contained more than 20% bases with Q ≤ 10. Then, de novo assembly of the clean reads was carried out using Trinity, a software package for short-read RNA-seq assembly (Grabherr et al. 2011). The assembled unigenes were aligned to protein databases nr, Swiss-Prot, Kyoto encyclopaedia of genes and genomes (KEGG), and the database of clusters of orthologous groups of proteins (COG) by BLASTx (E-value threshold of 1e-4); the optimal alignment result was used to decide the sequence direction of a unigene. A priority order of nr, Swiss-Prot, KEGG and COG was followed to decide the sequence direction when the results of different databases were not consistent. Software ESTScan was used to decide the sequence direction of a unigene that was not aligned to any of the four databases (Grabherr et al. 2011).
Differentially expressed genes (DEGs) identification
Gene expression level was estimated by the value of fragments per kb million fragments (FPKM) using the Cufflinks software (Mortazavi et al. 2008). For each wheat cultivar, genes differentially expressed after and before vernalization were identified as DEGs using false discovery rate (FDR) ≤ 0.001 and |log2 (fold change) | >1 as cutoffs (Audic and Jean-Michel 1997). FDR value was obtained by adjusting the raw P value using the Benjamin–Hochberg (BH) method (Benjamini and Hochberg 1995).
Screening of Jing841-specific DEGs and functional annotation
DEGs identified between the two wheat cultivars were further compared to screen the common ones between them, as well as those specifically expressed in the winter wheat cultivar Jing841.
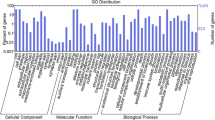
Gene ontology (GO) functional classification of genes specifically expressed in Jing841 was performed using bioinformatics platform Blast2GO (https://www.blast2go.com/), supporting multiple informatics analyses such as GO annotation and KEGG pathway enrichment analysis (Conesa et al. 2005).
qRT-PCR validation
Based on the result of functional annotation, DEGs that were specifically identified in Jing84 were grouped into different functional categories. Vernalization-related genes in each category were chosen to validate the changes in their expression level through qRT-PCR according to the following criteria: transcription factors were randomly selected; methylation genes with the highest relative expression levels were selected; for other genes, only those with larger absolute value of log2 ratio were selected. Finally, a total of 18 genes were further subjected to qRT-PCR, including six methylation-related genes, five transcription factors, two flower development-related genes, one vernalization-related gene, one cold-responsive gene, one dehydrin-encoding gene, one ice recrystallization inhibition protein and one ABA-related gene (table 1). MRNAs were extracted from the seedlings collected before vernalization and at day 30, and then transcribed into cDNA using reverse transcriptase (Takara). The qRT-PCR was performed using the diluted cDNA samples in a 20 μL mixture, containing 10 μL 2 × SYBR\(^{{\circledR }}\) Premix Ex Taq TM II (Takara), 0.4 μM of both forward and reverse primers and 100 ng cDNA template. q-RT-PCR was performed in triplicate using Bio-Rad iQ5 instrument (Biorad, Hercules, USA) following the protocol: initial denaturation for 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 20 s at 60°C. The 2 −ΔΔCt method was used to calculate the fold change in gene expression (Livak and Schmittgen 2001) and β-actin was used as an internal control. The primer pairs were shown in table 1.
Expression pattern analysis of 11 Jing84-specific vernalization-related genes
Finally, 11 genes with the most significant expression differences were selected for further analysis of their expression pattern during vernalization (the top 11 listed in table 1). For each gene, its expression level was detected through qRT-PCR at day 6, 12, 18, 24 and 30 during the 30-day-long vernalization treatment. qRT-PCR was performed as described above, with primers listed in table 1 (the top 11).
Results
Transcriptome sequencing, assembly and annotation
Totally, 69,751,192 raw reads and 5,787,106,380 (5.78 Gb) nucleotides were obtained, with Q20 percentage (namely, with an error rate of 1%) and GC percentage of 97.42% and 51.78%, respectively; 213,862 contigs were assembled from the raw short reads, with an average length of 256 nt. Finally, 138,062 unigenes were obtained with an average length of 509 nt (table 2). The average reads per kilobases per million reeds (RPKM) value of all unigenes was 14.28 with the minimal and maximal values of 0 and 5,856.67, respectively.
Identification of DEG and Jing841-specific DEGs
In total, 54,699 significantly changed tags were identified between the vernalized (J-V) and unvernalized (J-NV) Jing841 samples, which were mapped to 29,888 unigenes, including 14,647 upregulated unigenes and 15,241 downregulated unigenes (figure 1). Meanwhile, 15,908 significantly changed tags were obtained after comparing vernalized (LC-V) samples and the unvernalized (LC-NV) Liaochun10 samples, which were mapped to 2818 unigenes, including 740 upregulated unigenes (figure 2, B1) and 2078 downregulated unigenes (figure 1). Later, we further compared the DEGs between the two wheat cultivars, and found 97 common upregulated genes (figure 2, C1) and 293 common downregulated genes between the two cultivars (figure 2, C2).
Venn diagrams showing the number of DEGs (P< 0.05) in plants exposed to long period of cold treatment. (a) number of upregulated genes in A1 (J-NV versus J-V comparison) and B1 (LC-NV versus LC-V comparison); (b) number of downregulated genes in A2 (J-NV versus J-V) and B2 (LC-NV versus LC-V). C1, the shared genes of A1 and B1; C2, the shared genes of A2 and B2. J-NV, Jing841 nonvernalization; J-V, Jing841 vernalization; LC-NV, Liaochun10 nonvernalization; LC-V, Liaochun10 vernalization.
Then, a total of 636 DEGs of eight categories were further identified to be specifically expressed in Jing841, including 14 genes related to flower development, two cold-responsive genes, two vernalization-related genes, 10 genes encoding ice recrystallization inhibition protein, nine dehydrin-encoding genes, 304 transcription factors, 286 methylation-related genes and nine ABA (abscisic acid)-related genes (table 3).
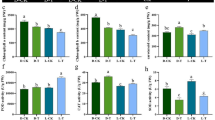
Relative expression of 18 selected DEGs in Jing841 on day 0 and 30 after cold treatment by qRT-PCR. A, methylation-related genes; B, transcription factors; C, flower development-related genes; D, vernalization-related gene; E, cold-responsive genes; F, dehydrin; G, ice recrystallization inhibition proteins; H, ABA-related gene; V0d, vernalization treatment for 0 day; V30d, vernalization treatment for 30 days.
qRT-PCR validation
Among the 636 DEGs specifically detected in Jing841, which were grouped into eight categories, 18 genes were selected for qRT-PCR. Among them, four methylation-related genes, five transcription factors and one flower development-related gene were downregulated after vernalization, while the rest were upregulated, all agreeing with their differential expression profiles based on sequencing data (figure 3).
Next, 11 genes (unigene1806, CL18953.contig2, CL19373. contig1, unigene12389, unigene8329, unigene41092, CL19305. contig2, unigene3230, CL3094.contig3, CL17784.contig1 and unigene5440) with the most significant expression differences were subjected to further analyses of their expression patterns during the 30-day-long vernalization. The results showed that their expression patterns can be divided into four categories (figure 4). Genes of the first category, unigene41092 displayed fluctuating decline in expression pattern with one peak (figure 4a); genes of the second category, including CL19305.contig2 (VRN1) exhibited a constant increase in expression over time during the 30-day-long vernalization (figure 4b); genes of the third category including unigene8329, unigene3230 (cold-responsive protein COR14a), CL3094.contig3 (dehydrin-/LEA group 2-like protein), CL17784.contig1 (ice recrystallization inhibition protein 1 precursor), unigene5440 (stress-related bZIP transcription factor) and CL19373.contig1 (obtusifoliol 14-alpha demethylase-like), showed fluctuating and rising trend with one or two peaks (figure 4, a, c–g); genes of the fourth category showed a constant decline in expression, including one methylation-related gene unigene12389 (obtusifoliol 14-alpha demethylase-like) (figure 4g), two transcription factors unigene1806 (ribulose bisphosphate carboxylase/oxygenase activase B, chloroplastic) and CL18953.contig2 (ribulose bisphosphate carboxylase activase B) (figure 4h).
Expression of 11 selected DEGs in the Jing841 from 0 to 30 days of cold treatment by qRT-PCR. (a) flower development-related gene, unigene8329 and unigene41092; (b) vernalization-related gene, CL19305.contig2 (VRN-A1); (c) cold-responsive gene, unigene3230 (COR14a); (d) CL3094.contig3 (dehydrin-/LEA group 2-like protein); (e) CL17784.contig1 (ice recrystallization inhibition protein 1 precursor); (f) ABA-related gene, unigene5440 (bZIP); (g) methylation-related gene, CL19373.contig1 and unigene12389; (h) transcription factors unigene1806 (ribulose bisphosphate carboxylase/oxygenase activase B, chloroplastic) and CL18953.contig2 (ribulose bisphosphate carboxylase activase B).
Discussion
Genes involved in the acclimation to low temperatures might be closely related to vernalization response (Brule-Babel and Fowler 1988). In this study, we attempted to identify the vernalization responsive genes by finding out the DEGs specifically detected in the winter wheat cultivar Jing841, namely, first identifying the genes differentially expressed before and after vernalization (DEGs) in each wheat cultivar and then subtracting the common DEGs between the two cultivars from the total DEGs identified in the winter wheat cultivar Jing841. Based on the sequence similarity with known proteins, some unigenes or contigs assembled from sequences from transcriptome sequencing were annotated, while some others were not.
Here, it was found that DEGs involved in the vernalization response in the spring wheat cultivar Liaochun10 were relatively fewer than those in the Jing841, which were consistent with a previous finding that the number of cold responsive DEGs in the leaves of spring wheat (paragon) was smaller than that of winter wheat (Harnesk and Solstice) (Winfield et al. 2010). This may be attributed to the speculation that spring wheat does not require a cold period to flower as winter wheat, hence, the vernalization response is not so strong as that in the winter wheat, accordingly activating fewer vernalization responsive genes.
The vernalization-responsive genes that were found specifically expressed in the winter wheat cultivar Jing841 included vernalization-related genes, cold-responsive genes, dehyrins (group II late embryogenesis abundant (LEA) proteins)—encoding genes, ice recrystallization inhibition protein encoding gene, ABA-related genes and methylation-related genes in addition to some unannotated unigenes or contigs. The altered expression of these unigenes or contigs revealed that plants are acclimated to low temperature through regulating the expression of genes involved in reproductive growth-related metabolisms, such as VRN-A1, COR14a, IRIP, unigene1806 and Cl18953.contig2.
VRN-A1 (CL19305.contig2) showed a constant response to vernalization, which is consistent with that of Norstar and Manitou at 6°C (Laudencia-Chingcuanco et al. 2011). Thus, VRN-A1 may play an important role in the vernalization response in the wheat. COR14 showed a rapid response to vernalization response, but decreased later, which has been reported in spring Norstar (Laudencia-Chingcuanco et al. 2011). Unigene8329, COR14a (unigene3230), CL3094.contig3 (dehydrin-/LEA group 2-like protein), CL17784.contig1 (ice recrystallization inhibition protein 1 precursor), unigene5440 (stress-related bZIP transcription factor) and CL19373. contig1 (obtusifoliol 14-alpha demethylase-like) showed a fluctuating rise in expression with one or two peaks during the 30-day-long cold treatment. Among them, COR14a expression has been reported to be accumulated in the frost-tolerant wheat and barley, leading to the acquisition of frost resistance (Crosatti et al. 1996; Vágújfalvi et al. 2000), and its expression was regulated by two loci on wheat chromosome 5A in both frost tolerant and sensitive genotypes (Vágújfalvi et al. 2000). Expression of COR14a and CL3094.contig3 remained at high level on the last day of long-term vernalization treatment compared with that on day 0 (figure 4, c&d), whereas the expression of unigene41092 was dropping compared to that on day 0. Many dehydrins have been identified in response to cold stress in plants (Borovskii et al. 2002). During a long-term cold acclimation, dehydrin accumulation is greatly influenced by VRN1/Fr1 locus and the expression of the major vernalization gene VRN1, respectively (Kosová et al. 2008; Kosová et al. 2011). Therefore, the constant expression of dehydrin may be attributed to VRN1. The binding of IRIP to ice could restrict ice crystal growth in the apoplast, allowing plants to survive under freezing conditions (Kuiper et al. 2001). So far, there is limited evidence on the functions of IRIP in vernalization. Here, we found the expression of IRIP (CL17784.contig1) remained at a high level on day 30, indicating the important role of IRIP in vernalization. Therefore, IRIP may act as a protection protein in the vernalization response in the wheat through enhancing the freezing tolerance. Previous reports have shown that endogenous ABA is involved in low-temperature response (Ling et al. 1994). Many bZIP transcriptional factors are involved in ABA signalling (Nakamura et al. 2001). The elevated expression level of stress-related bZIP transcriptional factors during vernalization in our transcriptome analysis indicates that ABA signalling plays an important role in the vernalization response. Several methylation-related genes, including unigene6399 (DNA(cytosine-5)-methyltransferase 1B-like)), unigene2999 (DNA(cytosine-5)-methyltransferase 1B-like)), and CL19373.contig1 (predicted obtusifoliol 14-alphademethylase-like) were found to be related with vernalization in our results. In Arabidopsis, epigenetic silencing of flowering locus C (FLC) by histone methylation is required for vernalization (Bastow et al. 2004), while in winter wheat, site-specific DNA hypermethylation at the VRN-A1 locus was induced by the vernalization treatment (Sherman and Talbert 2002; Khan et al. 2013). The above methylation genes might be responsible for the VRN-A1 DNA hypermethylation. Detailed molecular studies are needed to illustrate the underlying mechanism.
Unigene1806 and CL18953.contig2 encode ribulose bisphosphate carboxylase/oxygenase activase B, chloroplastic, respectively, both displaying a significant decrease in expression level during the 30-day-long vernalization treatment. Rubisco activase regulates the activity of Rubisco in the light. The activation state of Rubisco in leaves reflects a balance between sequestration of Rubisco active sites in a closed, inactive conformation and the reactivation of these sites by conformational changes induced by Rubisco activase (Portis 2003). A decrease in the expression of Rubisco activase indicates that low temperature impairs the expression of Rubisco activase, but this may be a vernalization-induced acclimation to low temperatures in winter wheat cultivar.
Conclusions
Here, through transcriptome sequencing, we detected some genes or contigs that probably have critical roles in vernalization, such as VRN-A1, COR14a, IRIP, unigene1806, Cl18953.contig2, which may advance our insight into this significant event.
References
Audic S. and Jean-Michel C. 1997 The significance of digital gene expression profiles. Genome Res. 7, 986–995.
Bastow R., Mylne J. S., Lister C., Lippman Z., Martienssen R. A. and Dean C. 2004 Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167.
Benjamini Y. and Hochberg Y. 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57, 289–300.
Borovskii G. B., Stupnikova I. V., Antipina A. I., Vladimirova S. V. and Voinikov V. K. 2002 Accumulation of dehydrin-like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BMC Plant Biol. 2, 5.
Brule-Babel A. and Fowler D. 1988 Genetic control of cold hardiness and vernalization requirement in winter wheat. Crop Sci. 28, 879–884.
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M. and Robles M. 2005 Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676.
Crosatti C., Nevo E., Stanca A. and Cattivelli L. 1996 Genetic analysis of the accumulation of COR14 proteins in wild (Hordeum spontaneum) and cultivated (Hordeum vulgare) barley. Theor. Appl. Genet. 93, 975–981.
Dong H. -H. 1993 Spring wheat liaochun 10 with high quality and yield. New Agric. 12.
Entz M. and Fowler D. 1991 Agronomic performance of winter versus spring wheat. Agron. J. 83, 527–532.
Fowler D., Chauvin L., Limin A. and Sarhan F. 1996 The regulatory role of vernalization in the expression of low-temperature-induced genes in wheat and rye. Theor. Appl. Genet. 93, 554–559.
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I. et al. 2011 Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652.
Kane N. A., Agharbaoui Z., Diallo A. O., Adam H., Tominaga Y., Ouellet F. et al. 2007 TaVRT2 represses transcription of the wheat vernalization gene TaVRN1. Plant J. 51, 670–680.
Khan A. R., Enjalbert J., Marsollier A. -C., Rousselet A., Goldringer I. and Vitte C. 2013 Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol. 13, 209.
Kippes N., Zhu J., Chen A., Vanzetti L., Lukaszewski A., Nishida H. et al. 2014 Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Mol. Genet. Genomics 289, 47–62.
Kosová K., Prášil I. and Vitámvás P. 2008 The relationship between vernalization- and photoperiodically-regulated genes and the development of frost tolerance in wheat and barley. Biol. Plant. 52, 601–615.
Kosová K., Vítámvás P. and Prášil I. T. 2011 Expression of dehydrins in wheat and barley under different temperatures. Plant Sci. 180, 46–52.
Kuiper M. J., Davies P. L. and Walker V. K. 2001 A theoretical model of a plant antifreeze protein from Lolium perenne. Biophys. J. 81, 3560–3565.
Laudencia-Chingcuanco D., Ganeshan S., You F., Fowler B., Chibbar R. and Anderson O. 2011 Genome-wide gene expression analysis supports a developmental model of low temperature tolerance gene regulation in wheat (Triticum aestivum L.). BMC Genomics 12, 299.
Limin A. and Fowler D. 2002 Developmental traits affecting low-temperature tolerance response in near-isogenic lines for the vernalization locus Vrn-A1 in wheat (Triticum aestivum L. em Thell). Ann. Bot. 89, 579–585.
Ling V., Mantyla E. and Welin B. 1994 Alterations in water status, endogenous abscisic acid content, and expression of rab78 gene during the development of freezing tolerance in Arabidopsis thalia. Plant Physiol. 104, 1341–1349.
Livak K. J. and Schmittgen T. D. 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408.
Monroy A. F., Dryanova A., Malette B., Oren D. H., Farajalla M. R., Liu W. et al. 2007 Regulatory gene candidates and gene expression analysis of cold acclimation in winter and spring wheat. Plant Mol. Biol. 64, 409–423.
Mortazavi A., Williams B. A. and Mccue K. 2008 Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628.
Nakamura S., Lynch T. J. and Finkelstein R. R. 2001 Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635.
Parchman T. L., Geist K. S., Grahnen J. A., Benkman C. W. and Buerkle C. A. 2010 Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics 11, 1.
Portis Jr A. R. 2003 Rubisco activase–Rubisco’s catalytic chaperone. Photosynth. Res. 75, 11–27.
Sherman J. D. and Talbert L. E. 2002 Vernalization-induced changes of the DNA methylation pattern in winter wheat. Genome 45, 253–260.
Thomashow M. F. 1999 Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599.
Vágújfalvi A., Galiba G., Dubcovsky J. and Cattivelli L. 2000 Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost-tolerant and frost-sensitive genotypes. Mol. Gen. Genet. 263, 194–200.
Winfield M. O., Lu C., Wilson I. D., Coghill J. A. and Edwards K. J. 2009 Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol. 9, 55.
Winfield M. O., Lu C. and Wilson I. D. 2010 Plant responses to cold: transcriptome analysis of wheat. Plant Biotech. J. 8, 749–771.
Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T. and Dubcovsky J. 2003 Positional cloning of the wheat vernalization gene VRN1. Proc. Nat. Acad. Sci. USA 100, 6263–6268.
Yan L., Loukoianov A. and Blechl A. 2004 The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644.
Yan L., Blechl A., Tranquilli G. and Dubcovsky J. 2006 The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Nat. Acad. Sci. USA 103, 19581–19586.
Zhang H., Mao X., Wang C. and Jing R. 2010 Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One 5, e16041.
Zhao H. and Zhao H. 1998 Wheat varieties and questions and answers of related technologies. China Agriculture Press, Beijing, China.
Acknowledgement
This work was financially supported by the Twelfth Five-Year National Science and Technology Pillar Programme (2011BAD16B07).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Arun Joshi
[Feng Y., Zhao Y., Wang K., Li Y. C., Wang X. and Yin J. 2016 Identification of vernalization responsive genes in the winter wheat cultivar Jing841 by transcriptome sequencing. J. Genet. 95, xx–xx]
Rights and permissions
About this article
Cite this article
FENG, Y., ZHAO, Y., WANG, K. et al. Identification of vernalization responsive genes in the winter wheat cultivar Jing841 by transcriptome sequencing. J Genet 95, 957–964 (2016). https://doi.org/10.1007/s12041-016-0724-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0724-0