Abstract
Genome editing (GE) technology has emerged as a multifaceted strategy that instantaneously popularised the mechanism to modify the genetic constitution of an organism. The clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) protein-based genome editing (CRISPR/Cas) approach has huge potential for efficacious editing of genomes of numerous organisms. This framework has demonstrated to be more economical in contrast to mega-nucleases, zinc-finger nucleases (ZFNs), and transcription activator-like effector nucleases (TALENs) for its flexibility, versatility, and potency. The advent of sequence-specific nucleases (SSNs) allowed the precise induction of double-strand breaks (DSBs) into the genome, ensuring desired alterations through non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathways. Researchers have utilized CRISPR/Cas-mediated genome alterations across crop varieties to generate desirable characteristics for yield enhancement, enriched nutritional quality, and stress-resistance. Here, we highlighted the recent progress in the area of nutritional improvement of crops via the CRISPR/Cas-based tools for fundamental plant research and crop genetic advancements. Application of this genome editing aids in unraveling the basic biology facts in plants supplemented by the incorporation of genome-wide association studies, artificial intelligence, and various bioinformatic frameworks, thereby providing futuristic model studies and their affirmations. Strategies for reducing the ‘off-target’ effects and the societal approval of genome-modified crops developed via this modern biotechnological approach have been reviewed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 CRISPR/Cas system: a prospective strategy for crop improvement
Genome editing (GE) is an efficient toolkit for altering a target genome, which turns out to be explicitly effective, and targeted change from an individual nucleotide to a broad-range sequence alteration. This task is usually performed with numerous endonucleases that cleave a specific gene at a distinct locus. Clustered regularly interspaced short palindromic repeats (CRISPR) or CRISPR associated (Cas) proteins perform as a molecular cleaving tool-kit in an agreement with the cell’s endogenous restoration system, intended to cut and modify DNA at a particular point within the genome. CRISPR/Cas mechanism is comprised of a CRISPR arsenal harbouring many Cas nucleases (Cas9, Cpf1, Cas13a, Cas14a) (table 1). This technique utilizes a single effector Cas endonuclease with two RNAs, i.e., CRISPR RNA (crRNA) that contains 20-nucleotide target-specific sequence and partial alternative trans-activating crRNA (tracrRNA) to fulfill its function. The assembled CRISPR/Cas complex fastens to the target recognition position upstream of the protospacer adjacent motif (PAM) and introduces a double-strand break (DSB) 3 bp prior to the PAM (Jinek et al. 2014) (figure 1). Recently, a different hypercompact CRISPR/Cas system identified in numerous bacteriophages harboured the CasΦ enzyme that has magnified the genome editing toolbox in both human and plant systems. It invariably exhibited higher target recognition capabilities in contrast to other CRISPR/Cas proteins, i.e., Cas9, and Cas12a and possessed half their molecular weight. CasΦ enzyme utilized an identical active site for processing mature CRISPR RNA (crRNA), that is responsible for cleavage of foreign nucleic acids (Pausch et al. 2020). According to phylogenetic classification, CRISPR/Cas system may be categorized into 2 classes, namely Class 1 that included types I, III, and IV, whereas, Class 2 comprised of types II, V, and VI that employs a particular protein to target overwhelming genetic materials (Makarova and Koonin 2015; Kaul et al. 2020). Among all the type II proteins, Cas9 has been universally accepted as an innovative tool-kit for genome altering functions. It revealed that Cas9 endonuclease consisted of a conserved core, bilobed structure juxtaposed to an active site with two major nucleic acid binding grooves (Jinek et al. 2014). The bilobed structure has a big globular recognition lobe known as the REC lobe and is connected with the small nuclease lobe (NUC). The determinate functional domain of Cas9 of REC-lobe has bi-partite domains, i.e., REC1 and REC2; and bridge helix cd domain (arginine-rich alpha-helical domain). NUC lobe acts as a seat for two nuclease domains; RUV C & HNH and, PI domain (PAM interacting domain) (Jinek et al. 2014; Hsu et al. 2014). Among two nucleic acid binding grooves, the primary large groove is positioned within the REC lobe and the exiguous major groove is positioned within the NUC lobe (Jinek et al. 2014; Nishimasu et al. 2014). Without the ligand DNA molecule, Cas9 behaves in an auto-inhibited confirmatory state and converts into the active state when loaded on the ligand DNA molecule via the guide RNA. This leads to the induction of conformational change in the NUC and REC lobes of Cas9 and creates a proper channel for the generation of a DNA-RNA heteroduplex complex (Jinek et al. 2014). The primary interaction of guide RNA with the REC lobe of Cas9 leads to the synthesis of the bicameral sgRNA-Cas9 complex. This binary complex examines the PAM site on the sense strand of the DNA double helix (Jinek et al. 2014; Sternberg et al. 2014). Various studies revealed that the PI domain of the NUC lobe of Cas9 plays an essential role in the recognition of the PAM site via its unsaturated tryptophan-rich flexible loop (Jinek et al. 2014). Due to the energy-independent helicase activity of Cas9, the PAM recognition triggers the destabilization of DNA sequences that are juxtaposed to the PAM site resulting in the formation of R-loop (Nishimasu et al. 2014; Sternberg et al. 2014). Several reports also concluded that the two arginine residues within the major groove of the DNA binding domain of Cas9 (R13.33 and R13.35) recognize the guanine dinucleotide of the PAM site on the non-complementary strand of a ligand DNA molecule. Besides, the two amino acid residues within the minor groove of DNA binding domain, lysine (K1107) and arginine (S1109) interact with the PAM sequences’ phosphate group (position +1) in the complementary strand of ligand DNA molecule in the form of a loop known as phosphate lock loop, which in turn plays an important role in the correct orientation for the hybridization between complementary DNA strand and guide RNA, may lead to the unwinding of DNA double helix (Sternberg et al. 2014). It was revealed through experimentation that the base pairing between the ligand DNA strand and the seed region of guide RNA (up to 8–12 bp) triggers the simultaneous stepwise destabilization of DNA duplex and the development of RNA-DNA heteroduplex. The RNA-DNA heteroduplex was occupied by NUC and REC lobe of Cas9, which resulted in a four-way node that ascended the arginine-rich alpha-helical bridge helix (Anders et al. 2014). As a result of the two domains of the NUC lobe, first is mobile HNH domain, always ready to cleave the complementary ligand DNA strand of tertiary complex, and the second is Ruv C domain which nicks the non-complementary strand, hence produces the double-strand breaks (Nishimasu et al. 2014). However, it is still obscure by what means recycling of Cas9 and its dissociation from sg-RNA occurs (Sternberg et al. 2014). The CRISPR/Cas system led to DNA double-strand breaks (DSBs) at defined positions that in turn introduced numerous genomic alterations employing DNA repair pathways i.e., HDR and NHEJ (Symington and Gautier 2011). Numerous customized engineered nucleases have been employed for targeted genome modifications, for instances, disruptions, additions and/or substitutions within genes, and over-expression of the gene (Xie and Yang 2013; Kim and Kim 2014; Sedeek et al. 2019; Chen et al. 2019; Hua et al. 2019a; Kaul et al. 2020). Various Cas9 and gRNA variants are accessible, which might be used for neoteric purposes, especially in the enhancement of traits within crops, such as, the increment of stress resistance level, crop turnout, and particularly nutritional up-gradation of crops.
Overview of CRISPR/Cas9-sgRNA targeted genome editing. sgRNA complexes with Cas9 nuclease on the targeted genomic site containing an adjacent PAM sequence. Nucleotide hybridization of sgRNA-Cas9 complex to targeted loci generated a conformational change that activates Cas9 nuclease activity, resulting in DNA double-strand breaks.
2 Recent progress of genome editing tools in crop improvement
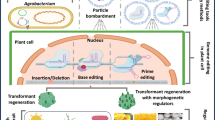
GE tools developed rapidly during the previous decade are being reliably utilized by researchers for crop improvement, globally. They offer extensive applications with a massive impact on life sciences and agriculture. The CRISPR/Cas system has already created a prominent position in several disciplines of basic and applied biology, genetic engineering and biotechnology because of its ease and robustness in modifying genomes, in order to comprehend the underlying regulatory processes of gene expression, transcriptional activation and repression, promoter activity and, so on. Here, we have compiled the numerous tools available for GE (figure 2), highlighting their robust implications on advancements in the area of crop improvement and the regulatory concerns related to their viability.
Potential genome editing outcome of CRISPR/Cas9-based technique based on NHEJ and HDR-mediated repair pathways. In NHEJ, random nucleotide insertions and/or deletions occur as the cell ligates the DSB within DNA, resulting in gene disruption. In HDR, the DSB is repaired by using an externally supplied homologous donor repair template resulting in directed precision repairing.
2.1 NHEJ-mediated knockout of genes
Creation of double-strand breaks (DSBs) led to the generation of gene knockout/s (KO) by means of fragment excision or deletions/insertions of few base pairs of a gene from a crop genome and ensuing repair by NHEJ, in this manner incorporating a mutation within the gene that modifies the expression of the targeted gene. CRISPR/Cas-based gene KO strategy has been utilized to interrogate the functional significance of a particular gene/s of interest. As of now, these tools have been efficiently set up in numerous crop species, for instance, S-gene (disease susceptible genes) KO strategy was successfully employed in hexaploid bread wheat that generated fungal-resistant wheat (Wang et al. 2014; Wang et al. 2018a). DuPont Pioneer (2016) revealed that CRISPR/Cas-based KO of corn Wx1 gene (waxy gene), produced amylopectin rich (waxy) corn grains by Corteva Agriscience. Wx1 gene that encoded the granule-bound starch synthase (GBSS), protein stimulated the development of amylopectin enriched corn. The development of two best glutinous sticky japonica rice varieties was achieved by the Waxy (OsWx) gene KO (Yunyan et al. 2019). Reports revealed targeted modification of starch branching enzyme SBEIIb in rice induced high amylose content in grains (Sun et al. 2017). Moreover, blast-resistant rice lines were generated by the OsERF922 gene KO (Wang et al. 2016). CRISPR/Cas-based KO of genes associated with floral development, i.e., AP1, SVP, and TFL evoked the advancement of floral attributes in Arabidopsis (Liu et al. 2019). According to Gaoneng et al. (2017), rice betaine aldehyde dehydrogenase (BADH2) gene acts as a negative regulator of the aroma production in rice. Targeted KO of the BADH2 gene generated fragrance enriched rice. Similarly, the use of CRISPR/Cas-based frameworks for the introduction of explicit hereditary changes in plant genomes prompted the rectification of complex metabolic mechanistics that further offers tremendous scope for the generation of newfangled crop germplasm with desired agronomic attributes.
2.2 HDR-mediated gene replacement
The robust CRISPR/Cas9, gene editing system is eminent for its stringent editing efficiency and user-friendliness. A new array of genome editing techniques have been developed for accurately and efficiently inserting desired mutations into a gene. Knocking-in of desired nucleotide sequences has been more challenging than knocking them out. Knocking out involved insertion of CRISPR/Cas9-sgRNA into a cell that targets the gene of interest, followed by fixing the cut (DSBs) using a cell’s regular DNA repair mechanism or NHEJ repair pathway. However, in case of knocking a gene in, the cuts must be fixed accurately, with no additional insertions/deletions, which needs harnessing of a second alternative DNA repair approach known as Homology-Directed Repair (HDR). Many agronomic traits may be conferred by gene expression changes via single-nucleotide insertions/substitutions or the expansion of novel gene activities. Meticulous gene alteration via knock-in and replacement may boost plant breeding by the introduction of new alleles without producing allelic variants, which do not occur normally (Chen et al. 2019). Unfortunately, HDR-mediated editing frequency is quite low and its utilization in trait advancement has hitherto been constrained. Shi et al. (2017) employed the CRISPR/Cas-based genome altering technique via swapping of the GOS2 promoter by the native ARGOS8 promoter through HDR-mediated genome editing for enhanced drought tolerance in maize. Yu et al. (2017) also generated edited tomato lines with extended shelf life via T317A substitution in the ALC gene. Geminivirus-mediated-DNA replicon has been utilized for enhanced gene-targeting efficiency in various crops, for instance, tomato (Cermak et al. 2015; Dahan-Meir et al. 2018), potato (Butler et al. 2016), wheat (Gil-Humanes et al. 2017), rice (Wang et al. 2017), cassava (Hummel et al. 2017) via increased copy number of repair templates. The replacement of principle amino acids via HDR-mediated pathway in the endogenous genes, for instance, acetolactate synthase (ALS) and 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) bestowed resistance to non-selective, systemic herbicides, i.e., sulfonylurea and glyphosate, respectively (Li et al. 2015; Sun et al. 2016). Correspondingly, double amino acid T102I/P106S (TIPS) and T102I/P106A replacements were incorporated into the EPSPS gene the flax plant (Linum usitatissimum) (Sauer et al. 2016) and cassava plant (Hummel et al. 2017). Another new RNA-mediated genome editing technique CRISPR/Cpf1 effectuated efficient and targeted gene insertions, in contrast to the HR-guided recombination. Therefore, the CRISPR/Cpf1 in conjunction with the CRISPR/Cas9 system may indomitably activate the focused gene knock-in mechanism for crop advancement.
2.3 Gene targeting
CRISPR/Cas technique is an efficacious strategy for the transmutation of crop genomes. In the light of homologous recombination (HR), a gene fragment substitution may be obtained via gene targeting (Capecchi 2005). To beat the low rate of HR frequency in crops, DSBs in DNA has been achieved at the defined loci (Steinert et al. 2016). Besides, co-delivery of the SSN altering system and an HDR template into the plant cell facilitated gene targeting in a specific way (Li et al. 2013). The rearranging of intrinsic TAL effector-binding sites in the promoters of R genes bestowed resistance transversely to various Xanthomonas sp. strains (Romer et al. 2009). To upgrade the negligible HR frequencies of somatic cells, Cas9 expression riveted to the young embryo and egg cells conferred a higher gene targeting potency in Arabidopsis (Miki et al. 2018; Wolter et al. 2018). As reported by Li et al. (2019), in vivo fabrication of RNA transcripts existing within the nucleus, assisted as repair templates for transcript-template HDR (TT-HDR)-based explicit gene-altering manifested an optimistic mutation frequency in rice. Although genome editing propounds differentially useful aspects, nonetheless complicated alleles demanded high accuracy. CRISPR/Cas system enforces HDR-based gene targeting to overcome such limitations. Nevertheless, HDR repair pathway potency in a plant cell is still inadequate and that may be ascribed to the inadequacy of efficacious delivery techniques for DNA repair templates (Steinert et al. 2016; Yin et al. 2017; Sedeek et al. 2019; Chen et al. 2019). In line with this, numerous advancements had been developed, for instance, increasing the quantity of donor DNA (Baltes et al. 2014); also a repair protein, KU70/80, and LIG4 may have suppressed NHEJ repair pathway (Endo et al. 2016). Therefore, the CRISPR/Cas system represents a better frequency of HDR-mediated gene targeting in plants.
2.4 Base editing with programmable nucleases
Precise genome editing is troublesome to attain with NHEJ- and HDR-mediated correction because of its lower editing frequency. Recently, an alternative tool, base editors or programmable deaminases, were developed to beat those limitations. Base editing is a novel genome modifying technique that empowers the irretrievable change of one DNA base to another at target position without DSBs as in the case of HDR (Komor et al. 2016; Nishida et al. 2016; Eid et al. 2018; Li et al. 2018a; Moon et al. 2019). DNA base editors were divided into two categories: adenine base editors (ABEs) and cytosine base editors (CBEs), wherein, ABEs sustain the switching of A•T pairs into G•C base pairs and CBEs interchanging the C•G into T•A base pairs. Both base editors (BEs) utilize nickase Cas9 (nCas9) and catalytically inactivate Cas9 (dCas9), and then fused to deaminases, thus hydrolyzes the amine group of A (Gaudelli et al. 2017) and C (Komor et al. 2016). In this methodology, nucleoside deaminases (cytidine, adenine) may be superintended by the sgRNA-Cas9 loaded complex to a target position, that culminated in the reversal of adenine or cytosine into inosine or uracil by hydrolysis. Eventually, following rectification or DNA replication, the edited strand had thymine or guanine instead of cytosine or adenine (i.e., single nucleotide bubble generated by Cas9n) (Li et al. 2017a; Hua et al. 2018; Ren et al. 2018; Kang et al. 2018). In cytosine base editors, a uracil glycosylase inhibitor is also employed in combination with Cas9n for expanded accuracy and potency of base-editing technique (Komor et al. 2016; Ren et al. 2018; Wu et al. 2020). dCas9 conjointly with cytidine deaminase enzyme may prompt the immediate change of cytidine to uridine. So, by following replication of the duplex a C -to- G was changed into T- to -A. It was reported that for catalyzing the first ‘A (adenine) to I (inosine)’ E. coli tRNA adenosine deaminase enzyme (TadA) was deployed for ABEs. The created I is recognized as G by DNA polymerase. Both CBEs and ABEs were employed earlier in numerous plant species, which required no double-stranded DNA breaks or HDR templates. Gao et al. 2017 demonstrated the base editing potency using BE3 in rice, wheat, and maize were up to 43% with an indel frequency of 0.01–0.22%. Each base editing tool offered various editing choices depending upon the deaminase, type of nuclease, and gRNA (Rees and Liu 2018). Various rice genes, for instance, OsSLR1, OsPDS, OsSBEIIb, and OsNRT1.1B modified with the aid of the BE3 technique effectuated an altering potency up to 20% (Komor et al. 2016; Nishida et al. 2016; Ren et al. 2018; Zhu et al. 2019). The earliest narrated CBEs that encouraged the C-to-T changes were generated via a combination of Cas9n with activation-induced cytidine deaminase orthologs PmCDA1 or rat cytidine deaminase (rAPOVEC1) (Li et al. 2017a; Lu and Zhu 2017; Ren et al. 2018; Shimatani et al. 2017; Zong et al. 2017; Zhong et al. 2018; Zuo et al. 2019; Endo et al. 2019; Kaul et al. 2020). Wu et al. (2019) further extended their applications by widening the editing frame of numerous base pairs from the canonical PAM (NGG) sequence. They combined cytidine deaminase1 from Petromyzon marinus, i.e., PmCDA1 with Cas9 D10A nickase of numerous SpCas9 variants, likewise as uracil DNA glycosylase inhibitor (UGI), and thereby generated two novel potent PmCDA1-based cytosine base editors (pBEs). Using this methodology, they broadened the base modifying efficiencies (4–90%) in T0 lines that commensurated to expanded base editing efficiencies from 1.3 to 7.6 fold (Wu et al. 2019; Wang et al. 2019a). Various engineered Cas9 variants that recognized non-canonical PAM sequences have been utilized with rAPOVEC1 or activation-induced cytidine deaminase to generate novel CBEs in rice (Negishi et al. 2019; Jin et al. 2019). Additionally, Zhou et al. (2019) tentatively assessed RNA single nucleotide variations (SNVs) that were acquired via either CBEs or ABEs and highlighted that both CBEs and ABEs produced numerous off-target RNA SNVs. Umpteen off-target effects in the DNA altering system may be erased through such robust engineering of deaminases. Gene alterations and write over employing mitochondrial DNA (mtDNA) have emerged as an encouraging option to orthodox therapy for mitochondrial dysfunction that may culminate in a broad range of ailments and inadequacy of organ and tissue functions. Recently, CRISPR-free DddA-derived cytosine base editors (DdCBEs) have been employed to target the alterations in human mtDNA, which proved significantly crucial for the therapeutic research related to mitochondrial diseases (Mok et al. 2020).
2.5 ‘Search-and-replace’ genome editing or prime editing
CRISPR-mediated genome editing may alter any base within the entire genome with utmost precision. However, despite the breakneck advancements in the genome editing machinery, most of the known human genetic variants (>75,000) related to diseases remains problematic to address or introduce in the majority of therapeutically vital cell types. One amongst the several reasons that may be caused by imperfect genome editing systems is the off-target effect. Even the most effective available CRISPR-based editing system that employed HDR may introduce unwanted mutations. Whereas escalating the potency and preciseness of DSBs-mediated editing is still the fundamental target, these difficulties may motivate the investigation of other precise genome editing techniques. A new genome altering tool referred to as prime editing may render the CRISPR system more precise than existing genome editing approaches in both animal and plant systems (Lin et al. 2020). Prime editors are often considered as word processors that are competent at searching precise DNA sequences and replacing them (Cohen 2019). It’s a fusion between Cas9n and reverse transcriptase (RT), which has similar effectiveness to CRISPR/Cas9 but showed less off-target effects. It used the same Cas9 nuclease but combined the enzyme with two new reagents: a longer-than-usual guide RNA referred as prime editing guide RNA (pegRNA) and a fusion protein consisting of Cas9 H840A nickase combined to a specially designed RT enzyme that initiated the addition of a new sequence or base into the genome. Primer binding site (PBS) sequence complementary to the nicked genomic DNA hybridized to the target site and acted as a template for reverse transcription. Host primer binding the RT stimulated the incorporation of a novel sequence or base into the genome, the new genetic material integrated into the cleaved strand of DNA, and the prime editor nicked the unedited strand, directing the cell to reconstruct by matching with the edited strand. Liu and his team (Anzalone et al. 2019) used prime editing to target genes underlying Tay-Sachs disease and RBC anemia. Prime editors (PEs), in the first instance represented by PE1, where an RT combined with a nickase Cas9 or RNA-guided nickase (Cas9n) and a pegRNA directly duplicated heritable data from the appended region on the pegRNA into the target genomic site. Whereas, PE2 exploited an assembled RT to regulate altering potencies, while PE3 nicked the non-edited strand, induced its substitution and augmented modifying potency, usually from 20–50% and 1–10% indel development in human HEK293T cells (Anzalone et al. 2019). Prime editing generated fewer by-products and effectuated similar potency in comparison to Cas9-initiated HDR as well as corresponding virtues and flaws in comparison to base editors. Prime editing is less repressed by the location of the PAM sequence and has immense potential towards the advancement of genome editing technology via rectification of most pathogenic alleles.
2.6 Single-cell genome engineering
Generating the whole plant from a single cell remains challenging because of the genotype-dependent low regeneration frequency and lack of elite germplasm for use in agriculture (Altpeter et al. 2016). Therefore, an efficient single-cell genome engineering evoked significant accomplishment in plant biotechnology by embarking research on numerous forefronts. Recent, endeavours employing protoplast transformations in lettuce and tobacco highlighted the transfer of CRISPR/Cas9 constructs in the form of RNP (Ribonucleoprotein)-complexes (Woo et al. 2015; Kim et al. 2017; Lee et al. 2018a). In many plant species, the regeneration frequency was quite low (<2%) from protoplasts after transformation, thus different morphogenic parameters have been utilized to boost the regeneration efficiencies (Altpeter et al. 2016; Lowe et al. 2016). Temporal expression of shoot-related transcription elements employing protoplast single-cell transformation, recognizing efficient procedures to pump up regeneration competency of the CRISPR/Cas modified recalcitrant cells and/or equating cells of germplasm with robust regeneration potency to those that are recalcitrant for screening regeneration amplifiers are approaches that may fire-up the capability of regenerating plants from single cells. In this manner, the single-cell genome engineering approach may be significantly effective in boosting up the CRISPR/Cas genome editing system.
2.7 CRISPR/Cas9-based germline engineering
Plant genome editing endeavours are generally regulated via conventional tissue culture, as well as transgenesis techniques. However, all these classical strategies confined the repurposing of the CRISPR/Cas machinery in numerous crop plants, in particular, those which were recalcitrant to regeneration or Agrobacterium-mediated transformation (Yadava et al. 2017; Agarwal et al. 2018; Sandhya et al. 2020). So, an alternative technique needs to be developed, which is independent of the classical tissue culture system. Germline cells are the precise target cell kinds for this technique, where transport of DNA or protein required for CRISPR/Cas9 methodology can permanently modify the genotype. Foreign DNA free RNP-mediated genome modifying of germline cells would be an ideal approach for germline engineering. Several approaches may be used including, polyethylene glycol (PEG)-mediated transfection, biolistic gene gun, microinjection, magnetofection, or electroporation that are suitable to few germline cells corresponding to the developmental stage of plant and plant species (Mao et al. 2016; Mohanty et al. 2016). Improving delivery methods may ensure the amplification of the programmes in plant genome engineering. Some viral systems may also be employed to supply sgRNAs to germ cells (Sedeek et al. 2019).
2.8 Genome editing for a single trait
CRISPR/Cas9-based genome modification has crucially empowered crop breeding via targeted genome editing of numerous agronomically significant traits (Chen et al. 2019; Schindele et al. 2020). Agronomically significant trait regulated by only one gene is termed as a single-gene trait, for instance, several qualitative traits, i.e., oil quality, rice grain aroma. Mutations of this single gene generally do not affect other important agronomic traits but the targeted one. For instance, CRISPR/Cas-based KO of the rice metal transporter gene OsNRAMP5 culminated into low Cd accretion in grains without influencing other significant agronomic characters (Tang et al. 2017a). Editing a single gene ZmLG1 led to erect architecture in corn and these plants showed enhanced density in the field (Li et al. 2017b; Tian et al. 2019). Additionally, umpteen single gene controlling traits, for instance, OsWAXY gene that controlled the rice amylose content (Sun et al. 2017); OsBADH2 gene that codes for rice grain aroma (Shan et al. 2015); FT2 gene, i.e., GmFT2a and GmFT5a that regulated the photoperiod in soybean (Cai et al. 2018, 2020) and phytate in corn (Liang et al. 2014) have been controlled by directed mutagenesis employing CRISPR/Cas9 editing.
2.9 Multiplexing of genes for trait stacking
Installation of some agronomic traits may be dependent upon the precise engineering of complex metabolic pathways that require a conjunct expression of multiple genes which in turn often needed multiple loci editing. Hence, a molecular tool-kit with an aptitude to alter numerous genes concurrently, pose immense potential to interrogate gene functions, and introduce quantitative traits. CRISPR/Cas-based genome modification offers robust multiplexing abilities (Adiego-Pérez et al. 2019). Multiple sgRNAs were assembled into a single sgRNA-Cas9 expression vector utilizing the Golden Gate Cloning/Gibson Assembly method (Xie et al. 2015; Silva and Patron 2017). The endogenic tRNA-processing technique was also engineered by scientists as a platform for increased targeting via the multiplexing capacity of the CRISPR/Cas9/Cpf1 system (Ding et al. 2018). Wang et al. (2017) reported the feasibility of multiplexing gene modification. Further, several gRNAs might be tailored to target a unique gene for increased crop editing rates with meagre trans-modification efficiencies by the customization of numerous sgRNAs, for instance in rice, engineering CRISPR/Cpf1 with a crisp DR-guide assembly (Hua et al. 2019b; Wang et al. 2019c). Lately, plant MGE has shown promising results via enabling modified homeoalleles/numerous alleles or concurrent incorporation of around 107 genes of a single gene family, to create umpteen target sites within a gene, to target numerous genes, and to generate discrete gene variants (Kannan et al. 2018). Despite, genome editing capabilities and benefits of ZFNs and TALENs, the CRISPR/Cas9 framework provided far more adaptability for the modification of dissimilar targets due to its simplicity, precision, and relative cost-effectiveness to deliver multiple sgRNAs (Xing et al. 2014; Ma et al. 2015; Kaul et al. 2020). Recently, it was revealed that two gRNAs were concurrently employed to change two targets lying in the conserved domain of endogenous acetolactate synthase gene (ALS1) in rice that resulted in amino acid replacements, i.e., W548L and S627I (Sun et al. 2016). The MGE machinery has proved efficacious in numerous practical applications with different forms. MGE randomly stimulated sizable fragment deletions and/or insertions in the non-coding regions that generated a vast range of genetic variants with differential expressions (Najera et al. 2019). Targeting various positions of a 2 kb fragment upstream of the CLAVATA3 gene in tomato via MGE mechanism generated heterozygous loss-of-function in terms of variations in fruit dimensions and architecture of the inflorescence (Rodriguez-Leal et al. 2017). Lately, the optimization of the MGE framework in Arabidopsis thaliana generated concurrent mutations in the genes via excision of 450 bp controlling regions inside the second intron of AGAMOUS gene that regulated flowering (Yan et al. 2016). In addition, alterations in genes of different hormone biosynthesis pathways within umpteen number of crops were achieved employing MGE. For instance, mutants with excessive accumulation of gibberellic acid were obtained by simultaneous KO of four paralogs of rapeseed RGA family utilizing two gRNAs (Yang et al. 2017). Similarly, simultaneous mutation of two paralogs of the barley HvPM19 gene hindered the gibberellic acid biosynthesis pathway (Lawrenson et al. 2015). In addition, the re-modeling of plant developmental features was attempted in numerous crop species. This may be enumerated as follows: (a) various seed storage proteins were effectively altered utilizing the MGE of paralogous genes; (b) In sorghum, the simultaneous modification of numerous genes from the KIC family employed a unique gRNA that introduced multiple mutations (Li et al. 2018b). (c) MAP kinase signaling pathway was targeted to determine the role of partially inessential genes, i.e., MPK1, MPK2, MPK5, and MPK6 that produced double (45%) and quadruple (86%) mutants in rice (Minkenberg et al. 2017). The CRISPR/Cas-based method has been comprehensively used for the promotion of enormous technological breakthroughs in other crops, which was not accomplished, formerly.
2.10 Molecular farming in plants
Molecular farming is a biotechnological programme that features the genetic alteration of agricultural merchandise, manufacture chemicals and proteins for pharmaceutical, more so for commercial purposes by enhancing crop quality. In line with this, field plants may pose as beneficial model systems over alternative mammalian systems (Buyel 2019). The CRISPR/Cas system provided a far better way to generate biopharmaceuticals in plants by altering the salient features of a recombinant macromolecule. The alteration of plant metabolism sans the synthesis of glycans is a notable example of molecular farming (Ma et al. 2003). Generation of human therapeutic proteins in plants many a time import a plant-like glycan instead of human-like that may be viewed as unwanted as they may influence numerous biological functions, immunogenicity, and protein stability. KO of the desired gene, which encoded the catalysts, i.e., as b(1,2) xylosyltransferase (XylT) and a(1,3)-fucosyltransferase (FucT) generated preferred recombinant protein instead of plant glycan synthesis. These two catalysts were responsible for the generation of principle glycan impressions in crop proteins that are distinct than those generated in mammals. Reports revealed the KO of two genes that encoded for 2 XylT and 4 FucT enzymes led to the production of a recombinant antibody that lacked synthesis of plant glycans in N. benthamiana mutant (Jansing et al. 2019). On similar lines, CRISPR/Cas-based KO of two genes encoding enzyme XylT and FucT in tobacco, cv Bright Yellow 2 (BY2) cell suspensions generated plant glycan-deficient recombinant proteins. Subsequently, the MGE method provided a brilliant forum for the development of important biopharmaceutical products (Mercx et al. 2017; Hanania et al. 2017). This may offer an enormous probability of enhanced credible target for genome engineering.
2.11 CRISPR and plant domestication
Modern crops have been specifically bred for several centuries for the incorporation of significant attributes in order to develop nutrient-rich, better-quality foodstuffs. Thus, this method has culminated into reduced diversity amongst crops that may have an effect on human fitness (Østerberg et al. 2019). Currently, tools for editing genomes may be used to re-design and reconstitute genetic variations in distinct genes offering an efficient solution for current demands for food. Combined with classical breeding, CRISPR/Cas-based editing of genomes may accelerate the fusion of favourable attributes, thereby minimizing the expenses efficiently. Subsequently, the genetic components incorporated may be removed from the genome via breeding or developing segregating lines that resulted in null segregates (Mao et al. 2019). The effectiveness of targeting traits that are controlled by a single gene employing CRISPR/Cas-based editing of genomes has been validated in several crops, for instance, enriched amylose content in rice and oil quality of soybean (Haun et al. 2014; Zhang et al. 2017a). However, reports showed that quantitative traits were controlled by numerous QTLs, many of which have been screened, mapped, and cloned (Zuo and Li 2014; Xing and Zhang 2010) and editing of such QTLs may lead to negative effects. CRISPR/Cas breeding has emerged as an innovative and efficacious approach for plant domestication, thereby ensuring increased crop diversity and agricultural sustainability. To exemplify, CRISPR/Cas-empowered aping of domestication events in wild or semi-domesticated plants resulted in the development of crops harbouring neoteric traits and generated enormous sources of germplasm for breeding, thereby creating a new sort of evolution. Improvement of potato has faced enormous difficulties using the conventional breeding process. Recently, a self-compatible diploid potato has been re-domesticated via disruption of the self-incompatibility S-RNase gene employing the CRISPR/Cas9 technique (Ye et al. 2018). Wild tomato plants, those that are naturally biotic/abiotic stress-resistant may emerge as a starting point for de novo domestication. Based on this wild tomato was domesticated without losing the stress resistance capability of the first wild germplasm (Li et al. 2018c; Zsögön et al. 2018). One group of scientists revealed that a wild relative of tomato referred to as husk tomato (Physalis pruinosa) was domesticated with higher yield and larger fruit (Lemmon et al. 2018). Novel domesticated crops amoured with increased tolerance to a variety of harsh environments would augur agricultural diversity and resolve several issues related to agricultural sustainability in the future.
2.12 Metabolic engineering via CRISPR
Metabolic engineering through the CRISPR/Cas system is an evolutionary approach for modulating metabolism, wherever plant cells are focused to generate desired metabolites (Lau et al. 2014). A regulatory gene, O-methyltransferase (OMT2) facilitated the biosynthesis of different metabolites (noscapine, papaverine, codeine, and morphine) through benzylisoquinoline alkaloids (BIAs) pathway. Alagoz et al. (2016) altered the biosynthesis of BIAs in Papaver somniferum by KO of 30 OMT2 genes employing the CRISPR/Cas9 mechanism. Moreover, GABA shunt metabolic pathway regulating genes, i.e., SSADH, CAT9, GABA-TP1, -TP2, and -TP3 were modified employing six-gRNAs that led to increased GABA levels by 19-fold as compared to wild type (Li et al. 2018d). Li et al. (2017c), targeted the SmCPS1 (diterpene synthase) gene from Salvia miltiorrhiza involved in tanshinone (well-known for antiarrhythmic and vasorelaxation effects) biosynthesis. With the aid of the CRISPR/Cas9 technique, eight chimeric and three homozygous mutants were generated from twenty-six distinct transgenic hairy root lines of Salvia. The chimeric mutants exhibited reduced accumulation of tanshinone, whereas the homozygous mutants showed no accumulation. Therefore, such a genome editing technique may be employed for transforming plants to biofactories for the generation of desired metabolites, just by altering gene sequences in a targeted manner. Thus, the new advanced tools in the CRISPR/Cas genome editing tool-kit have expanded the avenues to design different strategies for the development of preferable agronomically significant attributes in crops of interest.
3 Novel approaches in genome editing for precision crop improvement
CRISPR/Cas genome editing platform has emerged as a multifunctional tool-kit for genetic improvement of important traits, thereby conferring a revolutionary effect on plant research and crop breeding. Mainly three novel genome modification methods may be recruited for defined desirable mutational events in crop genomes i.e., (i) Loss of function mutants (ii) Gain/change of function mutants and, (iii) Epigenemone modified mutants: fine-tuning gene regulation
3.1 Loss of function mutants
Over the past decade, loss-of-function mutations were studied extensively with the development of RNAi technology for knock-down of the desired gene. Due to incomplete loss-of-function and a high percentage of off-target effects, the RNAi technique is less effective than the CRISPR/Cas system (Xu et al. 2006; Barrangou et al. 2015). The loss-of-function mutation by the CRISPR-based system is the simplest mechanism for a much better understanding of gene function. Desired loss of function may be accomplished by DSB to the targeted loci, which then would be repaired by the NHEJ-mediated repair pathway. Alterations within the genome may be attained via frame-shift mutations. In frameshift mutations, indels are produced by ZFNs/TALENs/Cas9 rendering loss of function alleles. Due to its efficacious and versatile nature, indels developed by this approach have evolved as a distinctive technique to analyze the functions of uncharacterized genes in plants (Chen et al. 2018). Numerous economically important agronomic traits have been developed via site-directed mutagenesis, for instance, 41.2% LbCpf1- derived mutations were recognized via KO of the bentazon sensitive lethal (OsBEL) gene (Xu et al. 2017). Shan et al. (2013) revealed that disruption of the OsPDS gene using CRISPR/Cas9-assisted mutagenesis resulted in expected phenotypes in rice. Similar results had been highlighted in Arabidopsis and tobacco (Li et al. 2013). The CRISPR/Cas-mediated KO of ZmIPK in Zea mays protoplasts revealed 13.1% mutation efficiency (Liang et al. 2014) along with the reduction in phytic acid and simultaneous increment in inorganic phosphate levels in maize seeds (Shi et al. 2003; Liang et al. 2014). sgRNA-Cas9-effectuated loss of function mutation in the promoters of two genes (OsSWEET14 and OsSWEET11) resulted in bacterial blight resistance in rice (Jiang et al. 2013; Xu et al. 2019). Soyk et al. (2016) reported that mutations in the self-pruning 5G (SP5G) gene enabled the development of early-yielding varieties of tomato.
3.2 Gain/change of function mutants
Besides loss-of-function analysis, the CRISPR/Cas9 machinery may also be utilized for gain or change of function mutations by knock-in of desired sequences via HDR-mediated repair mechanism. HDR or homology donor repair involved an exogenously supplied DNA donor template with ends homologous to the targeted DNA, accurately modified the genes thereby creating a new phenotype or function. HDR-mediated knock-in has been achieved in different plants, for instance, tobacco and rice by using protoplast transformation (Shan et al. 2013; Li et al. 2013) and in Arabidopsis using Agrobacterium-mediated transformation (Schiml et al. 2014). Zhao et al. (2016) effectively standardized a gene deletion/replacement method to develop stably transformed Arabidopsis plants that may be employed to incorporate desired traits for prospective precision-targeted crop enhancement. They developed a vector carrying the dual-sgRNA-Cas9 constructs that efficaciously deleted miRNA gene regions from MIR169a, MIR827a and highlighted deletion efficiencies for MIR169a (20%) and MIR827a (24%) loci, respectively. Subsequently, they created another construct that comprised of sites homologous to Arabidopsis TERMINAL FLOWER 1 (TFL1) to enable HDR with sequences identical to the two sgRNAs. The co-transformation of the two constructs into Arabidopsis plants led to both precision deletion and donor guided repair for desired gene substitution. Results exhibited that 0.8% of the transformed Arabidopsis plants harboured the swapped templates. They also incorporated an eGFP expression cassette flanked by left and right homologous arms of the AtTFL1 (A. thaliana terminal flower 1) gene into a targeted site within this gene via CRISPR/Cas9-mediated gene replacement. Further, it has been noted that amino acid substitutions via the base editing approach may generate partial gain or change of functional alleles. Active sites within distinctly-characterized proteins might be targeted by base editors (ABE/CBE), to evoke specific amino acid alterations for the creation of loss or gain-of-function alleles (Nishida et al. 2016; Shimatani et al. 2017; Zhong et al. 2018; Li et al 2018a). Different herbicide-resistant soybean (Li et al. 2015), maize (Svitashev et al. 2016), and rice (Endo et al. 2016; Sun et al. 2016; Li et al. 2016; Butt et al. 2017) plants were developed via HDR-guided base transversions.
3.3 Epigenemone modified mutants: fine-tuning gene regulation
Beyond gene disruption via knock-out/knock-in, genome modification tools may control the expression of genes. Gene regulation may be attained by a fusion of transcriptional repressors or activators to the DNA-binding domains thereby directing gene activation or repression (Qi et al. 2016; Kaul et al. 2020). Expression of genes may be influenced at different levels, such as transcription, mRNA maturation, and translation. Portions of promoter regions of desired genes may serve as potential target sites for CRISPR/Cas-based genome modification. Non-specific disruption in the cis-elements modulated the expression of the desired gene, in a dose-dependent way (Birchler 2017). Besides, the non-specific mutations, the targeted disjunction of the cis-elements may regulate the gene transcription. Insertion/s of a transposon in promoters or upstream of the promoter sequences might modify the genes’ epigenetic status. dCas9-equipped epigenome modifying tools may regulate gene expressions via epigenomic modulations. The idea of controlling the epigenetic status of the promoter for the modulation of gene transcription has been utilized in plant systems for the advancement in crop breeding (Hilton 2015; Liu et al. 2016; Dominguez et al. 2016; Hua et al. 2019a). In addition to targeting the promoter regions, gene expression may be regulated via spatio-temporal targeting of the gene. In this strategy, the dCas9 can be fused to any activator such as VP16 and VP64 (Gilbert et al. 2013; Moradpour and Abdulah 2020) or repressor such as Kruppel-associated box (KRAB) (Lawhorn et al. 2014; Li et al. 2017c) and thereby, activating (CRISPRa) or inhibiting (CRISPRi) the gene expression, respectively. Gene expression may be regulated even at the post-transcriptional level. By far microRNAs (miRNAs) regulated the gene expression at the post-transcriptional level by a translational blockage of the coding mRNA. Gene expression may be regulated by interfering with miRNA-mRNA binding through the introduction of a point mutation in the miRNA-binding site of the target gene that may in turn be achieved without disruption in the amino acid sequence of the protein. The position and frequency of the mismatches in the miRNA-mRNA pair, guide the efficacy of miRNA directed downgradation of the mRNA, wherein, both CBEs and ABEs may be used; thus effectuating the post-transcriptional expression of the gene in plants (Hua et al. 2019a). The regulation of gene expression through CRISPR/Cas tools played a potent role at the gene translational level also. In plants, numerous transcripts harbour upstream open reading frames (uORFs) in conjuction with the standard open reading frame. CRISPR/Cas9-based hindrance of the uORFs’ translation due to the inception of mutation/s in the start codon of the uORF consequently accelerated protein translation downstream of the standard open reading frame (Zhang et al. 2018). Conjointly, the incorporation of an NHEJ or HDR guided translational enhancer in the 5’ UTR region increased the expression of the target gene (Hua et al. 2019a), although this technique has not been employed in crop plants yet. Neoteric approaches employing CRISPR/Cas-mediated genome editing tools have ushered in an era of crop trait improvements, such as enhanced yield, upgraded nutritional traits, resistance/tolerance to biotic/abiotic factors, and so on.
4 Strategies alleviating off-target effects
Despite many advantages of CRISPR/Cas technology, it still faces various challenges. One of the potent features that scientists require to appropriately analyze is its off-target impacts in the CRISPR/Cas modified genomes. Generation of unbidden mutations that may arise due to off-targets at random sites in the target gene sequence or at any other site within the genome may hamper precise genome modifications. Nevertheless, accuracy and efficacy of the CRISPR/Cas machineries may be influenced by numerous parameters, for instance, target site recognition and designing of sgRNAs, the frequency of HDR-mediated repair, different delivery techniques, and Cas9 inactivity induced by anti-CRISPR proteins, etc. These themes have been discussed in a nutshell in the following sections.
4.1 Stringency in sgRNA designing
Various approaches have been proposed to alleviate off-target mutation/s. Firstly, the effect may be reduced by the use of highly specific Cas nuclease or stringent sgRNA design, which is different from the other genomic sites by three mismatches in combination with one mismatch present in PAM proximal region. The success of any CRISPR experiment relies enormously on the specificity of the guide RNA (sgRNA). The sgRNAs with more than 50% GC content were sufficient to promote on-target mutagenesis due to their enhanced target-site binding capability (Kim et al. 2015). Efficient designing of sgRNAs enabled the targeted mutagenesis, regardless of whether several homologous loci existed in the genome that are being interrogated (Ren et al. 2014; Baysal et al. 2016; Mao et al. 2016).
Currently, different computer algorithms are freely available, for instance, Cas-OFFinder (http://www.rgenome.net/casoffinder) that identifies unique target sequences and possible off-target sites in the genomes of targeted species and organisms (Cong et al. 2013; Hsu et al. 2013; Gerashchenkov et al. 2020). Among several bioinformatic tools, CRISPR-P and CRISPR-PLANT are explicitly structured for Cas9-based genome modifications. CRISPR-P facilitated gRNA designing of practically all plant species whose genome sequences are accessible, and furthermore provided both off-target- and restriction-site analysis (Lei et al. 2014). Consequently, CRISPR-PLANT has provided a genome-wide analysis of extremely specific RNAs in eight or more plant species and also facilitated restriction enzyme analyses of target sites (Xie et al. 2015). Moreover, many improved bioinformatic software tools are also available for designing gRNAs, for instance, CRISPR Design (http://crispr.mit.edu), which provided a scoring procedure for selecting sgRNAs with least off-target sites. sgRNA designing tools, for instance, Chop-Chop (http://chopchop.re.fas.harvard.edu) and sgDESIGNER, a freely accessible application (http://crispr.wustl.edu) provided an easy-to-use sgRNA designing interface (Hiranniramol et al. 2020). The four distinct forms of sgRNA may generate maximum on-target and minimum off-target effects. Modified sgRNAs may decrease off-target mutations with increased efficacy without renouncing mutation efficiencies at an on-target site. Lin et al. (2014) described various guidelines to design sgRNAs for reducing the off-target effects in numerous crop species. Results revealed that sgRNA bulges to a maximum of 4 bp can be sustained by the CRISPR/Cas9 machinery. The correspondence between cleavage function and position of DNA or sgRNA bulges in correlation to the PAM seems to be loci- and sequence-dependent. Firstly, (i) target sequences should be selected strictly with GC level (~40%–60%), (Hiranniramol et al. 2020) (ii) target DNA sequences should be distally placed from either NGG- or NAG-PAMs with ≤3 mismatches at 5′- and 3′- ends of DNA bulges or around 7–10 bp upstream of PAM, (iii) moreover, efficient sgRNA bulges should persist within 12 bp from PAM (Lin et al. 2014). The sgRNAs with minimum mismatches beyond seed sequences may prompt off-target changes (Shan et al. 2013). Few mismatches may be tolerated by the sgRNA-Cas9 complex, with mismatches being furthermore endured within regions downstream of the seed sequence (Cong et al. 2013; Lino et al. 2018). To avoid off-target effects, important components of CRISPR/Cas9, the PAM, and seed sequences require designing with utmost precision. For enhanced targeted cleavage efficiency, the designed sgRNAs should be complemented with the proper selection of promoters and terminators. Undesired cleavage of DNA target sites may arise due to promiscuous PAM regions. To avoid these events, bioinformatic tools, for instance, E-CRISPR (Heigwer et al. 2014) and CasOT (Xiao et al. 2014) may be employed to ensure sgRNAs designing based on whole-genome sequence information. Therefore, the specific designing of the sgRNAs fostered targeted mutation efficacies and alleviated unintended off-target mutation/s, which has been accounted for in numerous plants, for instance, A. thaliana (Peterson et al. 2016), Oryza sativa (Feng et al. 2014) and many more.
4.2 Factors affecting RNA efficacy
Length, mismatches, and GC content of gRNAs are important factors that regulate off-target effects. The distinctive target sequence choice that may vary from any other regions within the genome by a minimum of two or three nucleotides in a 20-nt sequence is a significant factor for reducing off-target effects (Cho et al. 2014). Cas9 off-target efficiency may raise significant concerns as it can re-write a target DNA harbouring five mismatches to its sgRNAs (Cradick et al. 2013; Fu et al. 2013) and alters the fidelity of Cas9-gRNAs. Generally, Cas9-gRNA was unable to recognize a DNA site that harboured greater than three mismatches close to a PAM (within10–12 bp). Therefore, the length, mismatches, and GC content of sgRNAs may influence the rate of targeted mutations. Sugano et al. (2014), described the impact of sgRNA nucleotide lengths (~16, 17, 18, and 20 bp) on genome editing efficacy and off-target mutation frequency. It was revealed that sgRNA sequences with 18 bp nucleotides reduced the off-target mutation efficiency. Doench et al. (2016) re-inforced that CRISPR-gRNA sequences directly influenced the on-target DNA cleavage efficiency, thereby avoiding unintended off-target binding and cleavage (Doench et al. 2016). Li et al. (2017a), assessed the effect of off-target mutations utilizing four sgRNAs in rice. Tang et al. (2018) also revealed that greater than two mismatches in sgRNAs on the target position may avoid unexpected mutations in rice and many other crops.
4.3 Concentration of sgRNA-Cas9 and Promoter selection
Another crucial parameter that modulates off-target effects is the concentration of sgRNA-Cas9, which may be controlled by titrating the amount of sgRNAs and Cas9. Lower concentrations of the sgRNA-Cas9 complex led to reduced off-target mutations. This strategy effectuated high-efficiency rates of on-target mutagenesis in targeted gene/s within plant species (Jia and Wang 2014). The choice of the promoter is a crucial step towards the regulation of Cas9 expression. Wang et al. (2015a) revealed that employing the egg cell-specific promoter in place of a constitutive CaMV35S promoter led to stronger expression of the Cas9 gene that in turn resulted in higher genome editing efficiency in A. thaliana. Contrastingly, on the utilization of high amounts of sgRNAs, lower editing efficiencies of the CRISPR/Cas9 system were reported in A. thaliana and tomato plants. Zhang et al. (2017b) demonstrated that the use of YAO like embryo-specific promoters played a crucial role in the Citrus sinensis genome via enhancement of the sgRNA-Cas9 expression. In few monocots, the expression of Cas9 controlled by their endogenous promoters resulted in higher on-target mutations than the CaMV35S promoter (Shan et al. 2015). Moreover, it has been investigated by Sun et al. (2015) that the U6-10 promoter-based on-target mutation frequency increased by 2–4 fold when compared to the AtUbi promoter in case of Glycine max. Nevertheless, the AtUbi and 2×CaMV35S promoters’ mediated expression of Cas9 generated similar on-target mutation efficiencies in tomato plants (Pan et al. 2016). Numerous studies conducted showed enhanced on-target mutations, for instance, Xu et al. (2015) showed that segregation of T1 rice lines harbouring sgRNA-Cas9 exhibited lower off-target mutations. An appropriately balanced ratio of sgRNA:Cas9 led to an increase or decrease of on- and off-target variation rates, respectively. Li et al. (2013) investigated the effect of sgRNA:Cas9 ratios (1:1, 19:1, and 20:1) on the mutation rate in A. thaliana and revealed that sgRNA:Cas9 (1:1) resulted in targeted mutations in the desired genes AtPDS3 (5.6%) and AtFLS2 (1.1%). Further, they compared two methods, firstly, the cgRNA-Cas9 ratio (chimeric guide RNAs) at 1:1 and secondly, the cgRNA-Cas9 co-delivery into cells for exploring mutagenesis efficiency in Nicotiana benthamiana. They demonstrated that the gene modification frequency of the co-delivered Cas9 and sgRNA was higher (12.7%) than in the case of the mixed ratio of (1:1) cgRNA:Cas9 (1.8%). This suggested that the presence of Cas9 and cgRNA in a single construct and their efficient co-delivery led to higher on-target mutation efficiency than employing a ratio of cgRNA:Cas9 at 1:1. It has been evaluated by Malnoy et al. (2016) that the mutation efficiency varied with the targeted gene locus and the ratio of sgRNA and Cas9. The best ratio of sgRNA-Cas9 as well as the use of RNP (ribonucleoprotein) complexes contributed to maximum mutation efficiencies in apple and grape. A few reports suggested that high level of sgRNAs led to low editing potentials, therefore, it was concluded that selection of suitable CRISPR/Cas9 edited plants wherein the CRISPR components were segregated, conferred low or no off-target mutations. Likewise, different ratios of sgRNA-Cas9 may increase or decrease on- and off-target mutations frequencies in various crops.
4.4 Novel players in genome editing: Cas9 orthologs and Cpf1 variants
Interestingly, Cas9 proteins reported from different organisms, including Neisseria meningitides (NmCas9) (Hou et al. 2013); Staphylococcus aureus (SaCas9) (Ran et al. 2013) and S. thermophiles (StCas9) (Kleinstiver et al. 2015) were employed for gene editing. These Cas9 variants identified distinct PAM sequences and displayed differential potencies. However, the choice of an optimal and specific ortholog of Cas9 may ensure enhanced gene editing efficacy for a target sequence. Recently, two important SpCas9 variants, for instance, engineered high accuracy xCas9 and Cas9-NG (targets non-canonical PAM) were identified. Amongst them, xCas9 variant revealed nearly indistinguishable editing efficiency as compared to wild-type Cas9 at almost every canonical PAM (NGG) site, while it revealed reduced activity at a non-canonical PAM site NGH (H = A, C, T). In the case of CBE systems, xCas9 at canonical PAM (NGG) sites resulted in higher editing efficiency. Contradictorily, Cas9-NG variants revealed increased editing efficiencies exclusively at non-canonical AT-rich PAM sites (GAT, CAA, and GAA) as compared to xCas9 variants. However, Cas9-NG variants showed significantly diminished activity at the sites harbouring canonical PAM (Hua et al. 2019b; Ren et al. 2019; Zhong et al. 2019). Furthermore, the efficiency of certain variants to act upon non-canonical NGCG, NG, and NGA PAMs has broadened the editable range of ABEs (Jeong et al. 2019). Besides, both the variants xCas9 and Cas9-NG, engineered SpCas9 (XNG-Cas9) also efficiently mutagenized constitutive target sites with GAA, NG, GAG, and GAT like PAMs in the genomes of Arabidopsis and tomato (Nishimasu et al. 2019). Further, other identified nucleases such as AsCpf1 (BV3L6) and LbCpf1 (ND2006) displayed restricted accessibility of relevant target loci, which reduced their functional utility. Cpf1 commands a definite prerequisite of a PAM (TTTV) to be present in their DNA substrates, where V might be A, C, or G. Two variants of AsCpf1 protein engineered to harbour mutations of S542R/K548V/N552R and, S542R/K607R accepted TATV and TYCV as PAMs, respectively, which enhanced their target sequence recognition specificities via the reduction in off-target effects. In addition, the introduction of a subsidiary non-PAM-interacting mutation further increased their efficacies. Similarly, the variant of LbCpf further showed increased target specificity and recognized altered PAM sites (Gao et al. 2017). Recently, FnCpf1-RR and FnCpf1-RVR, the two variants of FnCpf1 recognized canonical PAMs and exhibited increased target specificity in rice protoplasts than their wild type (FnCpf1) (Zhong et al. 2018).
4.5 Anti-CRISPRs
Anti-CRISPR (Acr) proteins referred to as natural ‘off switches’ de-activate the Cas9 protein by directly disarming its activity. The protein sequences and the functional mechanisms of these anti-CRISPRs vary widely and potentially interrupted several stages of the CRISPR/Cas machinery that include, crRNA maturation including processing and assembly, spacer acquisition, Cas protein expression, target DNA recognition and binding, and cleavage of target DNA (Marino et al. 2018). Initially identified Acrs were able to silence type I-E and I-F systems in phages that effectively infected Pseudomonas aeruginosa in combination with active type I CRISPR/Cas machinery and complementary CRISPR spacer (Bondy-Denomy et al. 2013). Eventually, numerous other Acr proteins were discovered and some of them actively inhibited types I-D, I-C, II-C, II-A, and V CRISPR/Cas machineries (Rauch et al. 2017; Marino et al. 2018; Lee et al. 2018b; Watters et al. 2018). Presently, more than 50 Acr proteins have been characterized that serve as inactivators of CRISPR’s molecular scissors by inactivating the cut-and-paste features of CRISPR machineries (Dolgin 2019). AcrIIA4Lmo (type II-A) actively blocked the nuclease activity of the most frequently used SpCas9 by counteracting Cas9-DNA interaction by sterically occupying the PAM-interacting domain (PID) and rendering the RuvC nuclease domain inaccessible, in part via DNA mimicry (Dong et al. 2017; Shin et al. 2017; Yang and Patel 2017). It has been revealed that type II and V Acrs drastically reduced the activity of Cas9 and Cas12a-mediated applications. Furthermore, sixteen families of the type I Acr proteins were routinely encoded next to the putative transcriptional regulatory genes but did not inherit nucleotide sequences or common structural resemblances to that of anti-CRISPR-associated (Aca) genes (Marino et al. 2018; Pawluk et al. 2018). Till now, the important functions of Aca proteins remain ambiguous. Recently, it was deciphered that Aca2 acted as an auto regulator, which suppressed the anti-CRISPR–aca2 operon. Helix-turn-helix domain of Aca2 protein generated a homodimer, which through interaction with dual inverted repeats in the promoter region of the anti-CRISPR gene inhibited DNA binding. Thus, it was highlighted that auto regulator Aca2-mediated the anti-CRISPR suppression (Pawluk et al. 2016; Birkholz et al. 2019). Recently, scientists incorporated Acr proteins into numerous tools, for instance, biosensors where Acrs may track the activity of a therapeutic gene editor inside the cells, and their optogenetic control strategies. It was revealed that if Cas9 remains active for long spans, it may lead to enhanced peril of unwarranted edits in the genome. Incidentally, Acr proteins may overcome this problem by blocking the activity of the CRISPR system (Dolgin 2019). Therefore, Acr proteins may limit the incidence of the undesired effects especially the off-targets within genomes.
4.6 Genome-wide mutant library screening
Mutant libraries employing whole-genomes pose as a significant functional genomic analysis approach for genetic improvement of crop traits. The CRISPR/Cas9 machineries are efficacious tools for genome-wide screening, that offered an innovative ‘gene discovery platform’ due to the simplicity of designing and precision mutagenesis enabled via sgRNA-Cas9 complex for modifying target gene/s expression (Sharma and Petsalaki 2018). To identify desirable traits, for instance, screening of biotic and/or abiotic stress tolerance, nutritional improvement, and other attributes may be performed in the expressed progeny. In the past genetic improvement of crop traits was time-intensive and tedious, which required consistent mapping endeavours. Large scale CRISPR/Cas9-mediated KO mutant libraries were constructed by two major groups, that covered the majority of the rice genes. Lu et al. (2017), have identified 34,234 genes and developed over 90,000 transgenic plants. Similarly, Meng et al. (2017) screened 12,802 genes and developed greater than 14,000 transgenic lines. For characterizing the unknown proteins, key functional residues of those proteins may be characterized by functional screening using pooled libraries of tiling array of sgRNAs by BEs or Cas9. In case of proteins that are uncharacterized, the crucial functional residues may be scanned via functional evaluation and screening employing methods for the transformation of pooled libraries of a tiling microarray of sgRNAs (utilizing either Cas9, CBE or ABE). The tiling microarray of sgRNAs was devised to comprise several hundreds of sgRNAs that spanned the complete exon of the targeted gene to be edited. In order to screen beneficial alleles, the tiling microarray of sgRNAs may be merged for the development of vector and plant transformation (Hua et al. 2019a; Wang et al. 2018b). With the emergence of the CRISPR framework, the breeder’s toolkit would be updated towards yield improvement and other trait improvements by the introduction of desired mutations in crops.
4.7 Enhancing HDR-mediated knock-in efficiency
Amongst two DNA repair pathways, the frequency of HDR-mediated precision DNA repair of DSB is typically lower than the error-prone NHEJ repair pathway. However, several approaches have been attempted for the enhancement of HDR efficiency by minimizing the frequency of NHEJ pathway, including synchronization of the cell cycle (Lin et al. 2014), utilization of minute molecular inhibitors of NHEJ repair pathway (Vartak and Raghavan 2015; Yu et al. 2015), silencing of the targeted gene (Chu et al. 2015) and, employment of NHEJ deficient cell lines (Weinstock and Jasin 2006). The CRISPR/Cas-based DSB repair efficacy employing the HDR pathway enhanced 19-fold by inhibition of DNA ligase IV enzyme of the NHEJ pathway by Scr7 inhibitor (Srivastava et al. 2012; Chu et al. 2015; Vartak and Raghavan 2015). A dominant-negative mutant of 53BP1, DN1S, when combined to Cas9 nucleases led to DN1S-Cas9 fusion protein that efficiently enhanced HDR frequency (86%) in K562 cells through masking off the NHEJ episodes precisely at Cas9 cleavage loci (Jayavaradhan et al. 2019).
4.8 CRISPR/Cas delivery systems
Diverse gene-modifying reagents have been transferred into plant cells, employing varied methods, i.e., transformations via protoplast, Agrobacterium, and particle bombardment (Zhang et al. 2016; Liang et al. 2018). Regardless of the delivery techniques, genome-modified cells must be regenerated into whole plants, which employs time-consuming and tedious tissue culture protocols. Majority of crops, lack a distinct and standardized tissue culture-based transformation systems, and developing one may be tedious, time-intensive, and infeasible (Altpeter et al. 2016). Even for crops with standardized transformation protocols, several new varieties remain recalcitrant to transformation owing to meagre regeneration capacity, for instance, several cereals (Altpeter et al. 2016). A recent report revealed scientific breakthrough employing ectopic production of morphogenic regulators, i.e., Baby Boom and WUSCHEL through Agrobacterium-based transformation, which significantly increased the regeneration frequency of various explants, for instance, mature seeds and leaf segments (maize) (Agarwal et al. 2018); immature embryos (sorghum, sugarcane, indica rice) (Lowe et al. 2016).
Plant viruses lead to enormous losses in yield, globally. The CRISPR/Cas machinery may be repurposed to function as defense mechanism effectuating cleavage of both DNA and RNA viruses, in order to generate virus-resistant plants. Numerous geminiviral replicons have been recommended as transport vehicles, wherein REP (replication initiation protein) gene is co-delivered with sgRNA-Cas9 constructs in to plants (Baltes et al. 2014). Yin et al. (2015) reported that a geminivirus, i.e., Cabbage Leaf Curl virus (CaLCuV) is capable to transfer gRNA and can be employed to accurately target genomic sites to induce systemic gene mutations in plants. Tripathi et al. (2019) generated streak virus-resistant banana. Similarly, targeting the tomato yellow leaf curl virus (TYLCV) encoded coat protein sequences or replicase ensured robust virus interference, exhibited by a significant reduction of TYLCV disease in transgenic tomato plants (Tashkandi et al. 2018). As opposed to the majority of Cas proteins, single-stranded (SS) RNAs were cleaved by Cas13a (C2c2) that interceded with turnip mosaic virus (TuMV) replication in plants. Employing viral promoters for driving Cas9 expression minimized off-target effects to an unassuming level (Chen et al. 2018).
The choice of a transformation method posed as a crucial factor for increasing on-target efficiency. Generally, gRNAs and Cas9 were incorporated into plant cells either via Agrobacterium-based transformation of T-DNA regions or physical delivery systems such as PEG-mediated transformation (protoplasts), biolistic approach (callus). Agrobacterium-mediated transformation has emerged as a popular strategy harnessed for transmission of various Cas nucleases (Ali et al. 2015). Nonetheless, geminiviral DNA replicons enhanced gene targeting efficiencies by one to two-fold, in contrast to traditional Agrobacterium-mediated T-DNA transformation. Recently highlighted TRV or Tobacco Rattle Virus-a RNA virus that replicated within the cell’s cytoplasm efficiently transported sgRNAs into Cas9 expressing tobacco lines, in order to accomplish systemic editing of genome. However, on similar lines DNA-virus-based vector delivery system is elusive for CRISPR/Cas genome modification. TRV-assisted CRISPR/Cas system is a speedy, affordable, and effective approach that may infect desired plant species in a comprehensive manner. The development of a virus-mediated genome editing system led to the stable transmission of targeted changes to the next generation.
RNP strategy has arisen as another innovative method to reduce the off-target effects, wherein, the sgRNAs and RNP complexes were delivered either via biolistic or electroporation methods into the protoplast of plants that revealed minimum off-target mutation/s. This system was successfully reported in various plant species, for instance, rice, maize, tomato, and many others (Woo et al. 2015; Lee et al. 2018a). However, Agrobacterium- and biolistic-based delivery methods may be used for stable transformation of sgRNA-Cas9-RNP complexes. Newly developed gesicle technology is a potent tool-kit for reducing off-target impacts with the delivery of active Cas9 protein complexed with sgRNAs by using nanocarriers. Guide-it CRISPR/Cas9 gesicle system is a novel technique for the delivery of active sgRNA-Cas9-RNP complexes to target cells. Cong et al. (2013) revealed that the delivery of active RNP complexes in this manner arrested both the genomic integration and overexpression. Integration of these active RNP complexes via gesicles resulted in reduced off-target effects with increased editing efficiency at targeted loci (Hsu et al. 2013). Nanoparticle-mediated RNP delivery system has been successfully adopted in plants by decreasing the unwanted changes. Numerous CRISPR/Cas delivery systems in different crop species are mentioned in table 2. The recommended CRISPR/Cas delivery and transformation systems are affordable, cost-effective, and species-independent and led to the generation of efficient genome edited crops harbouring desired trait/s.
5 CRISPR achievements: Improving the nutritional content in crop plants
Providing adequate nutritious food emerges as a major challenge for agriculture. Plants are the primary source of the required nutrients for humans and livestock. During the last few decades, improvement of crop quality was achieved via conventional breeding and transgenic approaches, however, recently gene editing tools, viz., mega-nucleases, TALENs and ZFNs have been used. To hasten this improvement technique, an unconventional, efficient, and precise strategy was required (Turcotte et al. 2017). More recently, the trend of using modern genome editing strategies like the CRISPR/Cas technique progressively focussed on genes for the improvement of crop quality (Wada et al. 2020). The CRISPR/Cas9 framework creates permanent heritable modifications without influencing the agronomic features of the prevailing valuable traits (Feng et al. 2014; Pan et al. 2016). CRISPR/Cas9 offered a potent mechanism with comparatively higher editing potential in contrast to ZFNs and TALENs (Johnson et al. 2015; Gaj et al. 2013). Here, we have highlighted the progress and perspectives of nutritionally enhanced food crops developed via CRISPR/Cas-mediated genome engineering (figure 3).
Gene editing in primary crops such as rice and wheat that constitute over 80% source of world’s nutritive index. This illustration shows different approaches that can be incorporated by plant breeders that target yield under normal, drought conditions and result in seeds with high nutritive quotient.
This potent editing innovation has already been enforced in several plant species for its explicit genome modifying capacities. The implementation of the CRISPR/Cas9 machinery in major food crops, for instance, rice and wheat were initially reported by Shan et al. (2013). Wang et al. (2014) reported the sgRNA-Cas9-mediated precise genome altering in sweet orange. Florida’s Institute of Food and Agricultural Sciences (UF/IFAS) generated sweeter strawberries employing the CRISPR/Cas technique (Seonghee et al. 2018). Researchers in the USA had engineered anti-browning mushrooms utilizing the CRISPR/Cas9 technique, wherein minimization of browning in white truffles led to extension of their life span, thereby offering a business advantage. A minor deletion in the polyphenol oxidase gene via genome editing encouraged the oxidation of polyphenol when exposed to air, bringing about an appetizing and attractive appearance in the mushrooms (Waltz et al. 2016). Halterman et al. (2015) reported the classic work of the development of acrylamide free potatoes. Starch present in potato tubers degrades into glucose and fructose, during cold storage time. Cold storage potatoes revealed a reduced amount of sugars transforming into dark-brown pigments that led to generation of a strong carcinogen ‘acrylamide’. During the processing of potato chips or french fries at high temperatures, this acrylamide created a major problem. On the contrary, at high-temperatures, the genome edited potatoes during cold storage revealed hardly any brown pigments and negligible production of acrylamides as compared to wild-type. Similarly, non-browning apples have been developed via mutating the gene that encoded the polyphenol oxidase (PPO) enzyme (Nishitani et al. 2016). Liang et al. (2014) developed edited maize lines with low phytic acid content. Maize/corn grains contain significant levels of phosphorous. However, this phosphorous was stored in the form of phytate, which was ineffectively digested by humans. Employing the CRISPR/Cas9 approach, the reduction of phytic acid/phytate (an anti-nutritional compound) in maize grains was performed by targeting two key enzyme coding genes of its biosynthetic pathway (Liang et al. 2014). Rice being the principle food crop has been the primary target for quality enhancement. Amylose is regarded as an important nutrient quality parameter in rice grains. Targeting the starch branching enzymes, resulted in lower amylose content of rice grains (Sun et al. 2017). Another report revealed, amylose content was increased via the editing of OsWaxy gene, whereas the targeted genome editing in the OsBADH2 enhanced the aroma in rice grains, without affecting the yield (Shan et al. 2015; Sun et al. 2017; Zhang et al. 2018). The Cd content in rice grains was diminished, via the CRISPR/Cas12a-based KO of a Cd transporter gene OsNRAM5 (Tang et al. 2017a). Recently, two key sweet potato genes (IbGBSSI and IbSBEII) of its starch biosynthesis pathway were KO independently via the CRISPR/Cas9 approach, which revealed mutants with an increased and decreased amounts of amylose content, respectively (Wang et al. 2019b). Miyamoto et al. (2019) generated lignin enriched rice lines by CRISPR/Cas9-mediated site-directed mutagenesis of the transcriptional repressor OsMYB108 that enhanced the levels of p-coumaroylated and tricin lignin units in culm cell walls of rice. Incontestibly, high concentrations of α- linolenic and linoleic acids were the vital reasons for the oxidative instability of soybean oil and the foodstuffs thereby prepared. Soybean omega-6 desaturase (GmFAD2) genes perform a significant role in the conversion of oleic to linoleic acids. On similar lines, targeted modification of the GmFAD2 gene via the CRISPR/Cas9 method resulted in the increased accumulation of oleic acids via a reduction in the linoleic and α- linolenic acids (Amin et al. 2019). Isoflavonoids enacted an important function in plant-environment interactions, being widely present in pulses. Of late, the gene modification technique has uncovered considerable accomplishments in the soybean isoflavone pathway via concurrent targeting of three genes. Implementation of this GE framework led to augmentation of isoflavones in soybean seeds and leaves, consequently elevating the protection against soybean mosaic virus (Zhang et al. 2019). A similar CRISPR/Cas-based approach targeting two iron sensing genes to increase the contents of requisite micronutrients, i.e., Fe and Zn in the endosperms and one Cd transporter gene to decrease the accumulation of Cd in white-rice grain has been achieved in our nutritional improvement of crops (NIC) laboratory at ICGEB. Crops such as, grain amaranths of Amaranthus sp. and Vigna umbellata (rice-bean) have emerged as superfoods possessing remarkably high health benefits with tremendous nutritional value, including a vast amount of essential amino acids and minerals in contrast to umpteen number of cultivated crops. Moreover, they hold enormous potential for the reduction of micronutrient malnutrition in populations, globally. The NIC group at ICGEB has spearheaded the whole genome sequencing (NCBI SRA-SRP-132447) as well as transcriptome analyses to expedite the trait advancements (palatability, flowering, and habit) of the underutilized rice-bean crop (Kaul et al. 2019a).
6 CRISPR: a fast forward way for global food security
Conventional plant breeding techniques use naturally accessible resources harbouring desirable traits for crop improvement; however, this is a less effective approach due to its repercussions, i.e., loss of genetic diversity and stress susceptibility. Across the years, several biotic and abiotic cues adversely affected crop plantations thereby threatening global food security. In contrast to TALENS for targeted DNA cleavage, CRISPR/Cas9 emerged as a highly economical strategy that facilitated efficacious genome modifications, in order to upgrade food and feed crops, thereby sustaining food security (table 3). Preliminary studies demonstrated the potential genome editing method in the first generation and robust transformations with targeted site analysis in subsequent plant generations. Twelve different target regions in seven genes were examined in plants for biallelic, chimeric, heterozygous, or homozygous mutations in three generations, respectively. Nowadays, molecular biologists have exploited the CRISPR/Cas technique from bacteria and repurposed this system for functional studies in animals and plants. Biotechnologists have highlighted its crucial practical implications while transfecting into the model plants like Nicotiana tabacum (Gao et al. 2014), Nicotiana benthamiana (Li et al. 2013; Nekrasov et al. 2013; Jiang et al. 2013; Belhaj et al. 2013), Arabidopsis (Li et al. 2013; Jiang et al. 2013; Feng et al. 2013; Mao et al. 2013) and numerous crops, for instance, wheat (Upadhyay et al. 2013; Shan et al. 2013); rice (Feng et al. 2013; Mao et al 2013; Shan et al. 2013); sorghum (Jiang et al. 2013); sweet orange (Jia and Wang 2014); and maize (Liang et al. 2014). Two genes NtPDS and NtPDR6 were mutated in N. tabacum via the CRISPR/Cas system employing transient editing assays in tobacco protoplast via the transfection of sgRNA-Cas9 that led to a mutation frequency rate of 16.2–20.3% (Gao et al. 2014). The gRNA-Cas9-mediated mutations of NtPDS and NtPDR6 obtained in the transgenic tobacco plants were gRNA specific and exhibited a mutation ratio of 81.8% for NtPDS gRNA4 and 87.5% for NtPDR6 gRNA2. A clear phenotypic effect was found in both the mutants, i.e., etiolated leaves in the PDS mutant, and higher number of branches in the PDR6 mutant. Results indicated a biallelic mutation in both transgenic lines (Gao et al. 2014). In addition to CRISPR/Cas-based protoplast transient assays, agro-mediated transformations of sgRNA-Cas9 and non-functional green flurescence protein (GFP) into modern plants like Arabidopsis and tobacco offered an effective alternative approach.
Cleavage of the target site present at the 5′ coding region of the non functional GFP via sgRNA-Cas9 was followed by NHEJ repair that resulted in functional mutations within GFP genes (Jiang et al. 2013). The chimeric guide RNA (cgRNA) with Cas9 endonuclease enact a crucial function in generating the deletion mutation by targeting single or multiple sites in the target gene (Upadhyay et al. 2013). Ron et al. (2014) revealed the conserved nature of SHORT-ROOT and SCARECROW gene functions in Arabidopsis and tomato via the transfection of the CRISPR/Cas9 machinery. Liang et al. (2014) reported the CRISPR/Cas-mediated KO of ZmIPK gene in maize protoplasts with an efficiency rate of 13.1% that led to the reduction of phytic acid content, and accumulation of inorganic phosphate in the seeds, which was similar to the results found by Shi et al. (2003) in case of maize IPK mutants developed using mutator insertion KO technology. The table 4 represented the editing efficiencies of target genes in different crops via the CRISPR/Cas system. However, numerous agriculturally significant traits (yield, crop quality, resistance to abiotic and biotic cues and male sterility) of wheat were altered by means of gene editing (Borisjuk et al. 2019). The CRISPR/Cas9 approach was successfully employed in wheat to alter of a powdery mildew-resistance gene TaMLO (Shan et al. 2013). In another effort, the CRISPR/Cas9 machinery has been successfully implemented in order to target a homolog of TaCe (ECERIFERUM) to induce drought tolerance trait in wheat (Liang et al. 2018). In addition, numerous genes, for instance, TaGASR, TaGW, and TaDEP have been modified via the CRISPR/Cas9 approach for the enhancement of yield characteristics (Zhang et al. 2016). Recently, Lyzenga et al. (2019) targeted the homologs of CRUCIFERIN C (CsCRUC) genes of Camelina sativa, which led to an altered seed storage protein profile. The knockout profile of seeds revealed modified fatty acid content and proportionate profusion of all saturated fatty acids. In recent years, modification of TALE responsive promoter elements of two susceptible genes, i.e., OsSWEET11 (PthXo1) and OsSWEET14 (PthXo3/AvrXa7), in rice cv. Kitaake, a recessive stress tolerant allele of Xa25/OsSWEET13 enhanced resistance to bacterial blight disease (Xu et al. 2019; Oliva et al. 2019). Enhanced repair ratio and non-transformational approaches for plants with precise alterations may be generated via the use of geminiviruses for the delivery of templates and constructs to all parts of the plant (Ali et al. 2015). Exploration of direct delivery of viruses initiated in 2015 demonstrated the acheivements of virus-mediated sgRNA-Cas9 delivery (Yin et al. 2015). In another report, the foxtail mosaic virus (FoMV) derived viral vectors have been used in monocots for virus-enabled gene editing (VEdGE), virus-based over-expression (VOX) and virus-induced gene silencing (VIGS). Furthermore, these vectors were employed for temporal expression of genes, employing single sgRNA-Cas9-based gene modifications in N. benthamiana, Setaria viridis, and Zea mays (Mei et al. 2019). CRISPR interference (CRISPRi) exhibited RNA-guided, systematic, stable regulation of transcription by fusion of inactivated dCas9 to effector domains. Combining of dCas9 C-terminus to the EDLL and SRDX domains during alteration of the native tobacco PDS gene effectuated activation and repression of PDS gene, respectively (Piatek et al. 2015). Genome-scale CRISPRi and CRISPRa libraries may be used to comprehend the intricate stress signaling pathways for interrogation of gene functions (Liu et al. 2015). Bacterial blight resistant indica rice (IR24) was generated via utilization of the CRISPR/Cas9-based tool-kit to target OsSWEET13 promoter regions (Zhou et al. 2015a). Furthermore, a targeted mutation in the ethylene-responsive factor, OsERF922 led to generation of rice varieties that were tolerant to blast disease (Liu et al. 2012). KO of Annexin gene (OsAnn3) employing CRISPR/Cas9 machinery conferred cold stress-resistance in rice (Shen et al. 2017). Rice yield may be enhanced by single or complex genome modifications of numerous QTLs. Shen et al. (2018) revealed the function of numerous QTLs that were crucial for grain size (GS3) and number (Gn1a) in rice crops via the CRISPR-mediated QTL altering strategy. CRISPR/Cas-mediated engineering of herbicide-tolerant crop varieties facilitated the utilization of non-selective herbicides to target diversified weed populations without hampering the crop. The NIC Group at ICGEB has established the effectual genome editing of the PEP-binding active site in EPSPS genes of maize, rice, and pigeonpea via CRISPR-mediated knock-out of a conserved region and homology donor repair via knock-in of a desired mutated fragment to achieve tolerance to glyphosate-a non-selective herbicide. EPSPS gene edited lines of maize and rice led to prominently amplified aromatic amino acid (Phe, Tyr, and Trp) content in contrast to the wild progeny. This approach may be extrapolated to other crops, for example, wheat, soybean, onion, and those crops that face weed constraints.
Along with the CRISPR/Cas9 approach, CRISPR-Cpf1 has emerged as a significantly flexible editing system for genome alterations in crops (Zetsche et al. 2017). Recruiting CRISPR-Cpf1 employing the Pol III driven ribozyme-fused with crRNA showed 100% mutation (biallelic) frequency in rice (Tang et al. 2017b). Enzymatic activity of the CRISPR/Cas13a empowered efficacious RNAi activity in the crops, in the face of diseases due to RNA viruses and also transformed lengthy pre-crRNA copies into active crRNAs. CRISPR/Cas13a approaches led to RNAi via Turnip Mosaic Virus (TuMV) in tobacco (Aman et al. 2018). Transient assays employing CRISPR/Cas13a created interference in GFP-expressing TuMV that in turn led to stable overexpression in tobacco. crRNAs targeted to HC-Pro and GFP sequences revealed improved interference than those targeted to regions for instance; coat protein (CP) sequence. CRISPR/Cas13a approach magnified research intercessions in crops and other eukaryotic organisms using viral RNAi. The CRISPR/Cas14a approach expanded genome modifications of both plant ssDNA and dsDNA viral replicates (Ali et al. 2015; Watters et al. 2018). Besides crop trait advancements, fermentation activities of microorganisms belonging to the Lactobacillus family also profited from gene editing. Genome alterations of microorganisms empowered by CRISPR-based approaches pose huge potential, predominantly as probiotics and psychobiotics in the food sector (Misra and Mohanty 2017) and remarkably enhanced their tolerance to stress (Ismail et al. 2019). Kaul et al. (2019b) and Raman et al. (2019) demonstrated a comprehensive assorted encapsulation of multifarious perks employing neoteric CRISPR associated proteins for instance; Cas9/12/13, outlined for wheat, maize, rice and other crucial crops. Therefore, the recent quantum leap in the genome modifying strategies, if integrated would transform the field of precision breeding by elevating its stature to precision CRISPR/Cas breeding. Recently, compendious and robustious testaments have endorsed the applications of the CRISPR/Cas9 approaches in crops for the generation of agronomically improved varieties.
7 Machine learning and CRISPR/Cas framework
The CRISPR/Cas9 genome editing has emerged as a pivotal mechanism in the governance of the genes, by employing a complementary sequence as the single guide RNA. However, it is impractical to have an exact homology sequence at the target site, so there is a possibility of cleaving fortuitous off-targets. Machine learning (ML) and Artificial Intelligence (AI) propound neoteric approaches to utilize the CRISPR/Cas9 technology for the assessment of altered plant lines empowered with superior attributes for example, abiotic and/or biotic stress tolerance, altered flower and root architectures, higher nutritional values, palatability, etc. Alteration of both genomic regions and target genes remarkably depends upon pre-determination of sgRNA target positions, nevertheless, the efficacy of genome editing may be not elucidated as sgRNAs may lead to extensive off-target effects. Currently, umpteen procedures available may enable designing of accurate sgRNAs, recruiting elementary rules. Here, the two novel algorithms known as CRISPR target assessment (CRISTA) and Elevation were introduced within the framework of machine learning, which performed a significant task in the determination of a particular genomic site to be precisely cleaved via given sgRNAs. The CRISTA and Elevation assisted predictions were found to be more accurate than the conventionally predictable thresholds (Abadi et al. 2017; Listgarten et al. 2018). Furthermore, to the forecast off-target sites, these inferred novel algorithms hold significance for instance, prediction of gRNA secondary structure, genome location, and prediction of GC-rich sites, which were hypersensitive to DNase I enzyme. Target site prediction played a significant role in the differentiation between the wobble pairing assisted mismatches and the mismatches caused through DNA/RNA bulges that in turn had structural impacts (Abadi et al. 2017; Listgarten et al. 2018). Moreover, different experimental designs had a distinctive prediction model that differed in the rules, which governed the activity of CRISPR/Cas9. It has been found that the Bowtie and Burrows-wheeler Alignment (BWA) were the two important bioinformatic tools that were used to achieve the arrangement of short target sequences in comparison to the BLAST-like traditional tools. However, the identification of feasible off-target positions required the perquisition of short sequence motifs up to 20 bp, in addition to the PAM with frequent mismatches. However, only three to five mismatches were allowed by the Bowtie and BWA alignment, respectively, which led to omission of highly mismatched off-targets (Tsai et al. 2015; Doench et al. 2016). To overcome this problem, the new bidirectional alignment like methods were implemented, which played a crucial role in the accurate identification of all potential off-targets (Canzar and Salzberg 2017). In most cases, the aligners first performed the matching of seed sequence and extended the seed sequence in a direction and then tested the match. However, in bidirectional alignment, the aligners lengthened the seed sequence in both the directions. Among all, the bidirectional alignment tool was the most powerful one for the identification of all potential off-targets. The MIT broad score and the cutting frequency determination (CFD) score (Hsu et al. 2013; Doench et al. 2016) were the two most prominent scoring methods based on the synthetic datasets. In these two algorithm-based scoring methods, the mutation in the gRNAs target sequence took place via a specific dataset represented as one, two or three mismatch combinations. The capability of gRNA-assisted target site cleavage frequencies was quantified and the outcome was utilized to design a linear regression algorithm to point out the off-target sites. Athough, both scoring algorithms were based on similar hypotheses however, they differed at the level of definite model development. The MIT broad algorithm assessed up to 20 bp target sequences excluding PAMs, while the CFD score algorithm assessed target sequences including PAM sequences but passively scored the non-canonical PAM harbouring target sequences. In a bid to accurately predict off-target sites through distinct experimental datasets, a comparison between these two scoring algorithms proved that the CFD score algorithm executed the best results as compared to MIT broad score algorithm (Haeussler et al. 2016). In addition, common off-target prediction tools have been mentioned in table 5. Thus, ML and AI analyzed the probable regression point, which may either converge or diverge the plot of both off-target and on-target specificities.
8 Regulatory concerns related to genome edited (GE) crops
Engineering of genomes employing CRISPR/Cas machinery enabled a synchronized approach for the integration of elite traits in crops via base substitutions, additions, deletions, and gene integrations or re-instatements in an arbitrary manner, resulting in characteristics that were identical to those generated employing induced mutations, natural genetic variations and breeding techniques. Regulation of genetically modified organisms (GMOs) is in line with the Cartagena Protocol, which is under the control of international trade of living modified organisms (LMOs). In line with this protocol, LMOs encompassed an altered genetic material modified via modern molecular biology techniques. However, the regulation of this protocol has been a controversial topic over the years. The societal approval to accommodate genome edited crop varieties as non-GMOs requires a re-categorization of the regulatory framework. A fundamental feature of existing regulation is either based on the end product or the process involved (Araki and Ishii 2015). Globally, CRISPR/Cas framework debatably extended from the present GMO laws, however, new specialized difficulties and government methodologies are yet the greatest issues in the method of social endorsement of genome modified crops (Araki and Ishii 2015; Jones 2015; Kanchiswamy et al. 2015). One of the most extraordinary features of genome-edited organisms remains that it is without an exogenous gene fragment/s, in contrast to GMOs. Genome modified crops may be socially more admissible than plants that have transgene insertions in their genomes. On these lines, the USDA and FDA have acknowledged genome edited plants (mushrooms and corn), which have transgene-free DNA and are considered as equivalent to products generated from conventional plant breeding methods. These altered food products do not fall into the category of GMOs and would follow deregulation from conventional GMOs policy and regulation (Waltz 2016). Conversely, the European Court of Justice (EU) has cognominated to consider GE crops as equivalent to GMOs, and the commercialization of GE crops has been restricted in the European countries (Callaway 2018). This might represent a significant issue for intercontinental business between such nations, including India. The upsides of GE innovation for the world’s food security might be retained as a result of public opinion and the current success of plant breeding (Wolt et al. 2016). To surmount these difficulties, straightforwardness in decision-making is quintessential for GE employment.
9 Perspectives on nation-wide policies regulating GE crops
GE crops generated by gene-editing, gene replacement, or Site-Directed Nuclease (SDN) technology showcased hereditary variations by the introduction of significant alterations to their genomes. The prominent features of the SDN system comprises the integration of unique and minute modifications at the DSB loci by the utilization of the host’s inherent repair machinery, which has evolved as a potent tool-kit for acquiring desired traits via targeted modifications. Targeted genome modifications enabled nucleotide additions and/or deletions that procreated neoteric desirable characteristics, for instance, enhanced nutrient content, or reduced generation of allergens. SDN approaches have been typecasted into three classes; firstly, Class-I/SDN-1 created via a DSB within the host cell of a genome without any transgene insertions, and spontaneously reconstructed by a inherent repair machinery, which resulted in nucleotide deletion/s and/or insertion/s (indels), mutation/s, gene repression, removal of a gene. Secondly, Class-II/SDN-2 system that is identified and reorganized by the cell’s inherent repair machinery via a short DNA fragment possessing the beneficial mutations that exhibited sequence complementarity to the targeted area in the genome. Finally, Class-III/SDN-3 approach involved creation of a DSB in the DNA followed by the re-instatements of a fragment harbouring a gene or any other genomic sequences. Interestingly, the opinions concerning organisms that resulted from SDN-1 and SDN-2 approaches were overlapping for both the National Technical Working Group (NTWG) and the German Central Commission of Biological Safety (ZKBS). The subsequent organisms carried mutations, which originated from the inherent DNA repair mechanisms either by NHEJ and/or HDR. According to Jones (2015) SDN-1 may be contemplated as a mutant derived from mutagenic agents like chemicals or radiation utilized in conventional mutational breeding. New plant varieties generated by mutations induced employing SDN-1 and SDN-2 approaches were exempted from the GMO regulations as no external DNA remained in the genome. However, SDN-3 approaches possess a stable insertion of foreign DNA and the developed crop varieties contain ≥ 20 base pair fragments of DNA integration and would be regulated as the conventional GMOs (Lusser et al. 2011; ZKBS 2012).
Normally, two categories of regulatory frameworks are present; few nations regulate the process, while others follow the product. Whereas, guidelines under the systematized framework is often named as product-based, the legislative framework of genome edited crops showed an existing process-based spark in umpteen number of illustrative cases (Wolt 2017). Small and Medium Enterprises (SMEs) that formed Europe’s unprecedented plant breeding area would especially be profited from SDN (site-directed nuclease) framework to co-ordinate market requests and create eco-accommodating and highly sustainable advanced varieties exhibiting enhanced productivity. Although, this unpredictability may be addressed only if the EU clears its vision towards existing SDN innovations. We comprehend that every existing regulation has both pros and cons. In addition, no existing approach has fully comprehended how GE technology works that involved clean deletions, insertions, or base-pair swapping. Henceforth, the regulating legislations should be re-designed for the aforementioned products akin to the products arising from conventional breeding methods. Few countries like South and North America have welcomed the commercialization of GE crops by removing regulatory constraints. Moreover, Japan and Australia, have recently revisited and revised their normative approval procedures for GE organisms/products that involved SDN-1 type modification. A phenomenal degree of uncertainity related to regulatory stature of GE crops, yet remains in most Asian and European nations (Wolt 2017).
10 Conclusions
Genome editing via CRISPR/Cas based system offers applications involving alteration and regulation of genomes for crop advancement through gene KO, knock-in, point mutations, nucleotide substitution, and alterations at any gene locus. The latest developments in CRISPR/Cas technologies for instance prime editing and so on, enabled expansion of genome editing capabilities to encompass base substitutions, precision gene targeting, and manipulation of gene expression. Incidentally, such advancements have elaborated an arrangement of crop enhancement tools accessible to agronomists. Nevertheless, the successful implementation of any genetic method for crop betterment is reliant on its complete genome sequence and functional genomics data including the genetic networks governing crucial agronomic traits. In numerous crop species, such information is still elusive. The emergence of innovative and comprehensive DNA sequencing know-how and the inception of umpteen ‘omics’ databases will be instrumental in the screening of significantly novel targets for genome editing in plants, for instance, negative regulators of positive traits. Genome editing innovation, especially the CRISPR/Cas frameworks have changed the plant science in a gigantic way because of their approach capacity, simplicity, and capacity to concurrently modify numerous attributes in various organisms, including plants. Employing CRISPR/Cas innovation in plants to augment high yields, disease resistance, and nutrition content improvement has proved to be propitious in contrast to genome engineering technologies. The agricultural sector has bloomed in a rapid manner owing to the CRISPR/Cas system that has fostered the functional genomics research to provide nutritionally enhanced, disease-resistant, high yielding crop lines, in order to produce more food with good nutritional value, thereby ensuring global food and nutritional security. Additionally, it has been effectively utilized for the development of comprehensive mutant libraries and anti-viral breeding strategy. Eventhough, striking progress has been achieved in streamlining genome editing, the efficacy of novel genome editing systems would depend on pre-requisites, for instance, genotype-independent delivery techniques, functional genomics, and minimal off-target effects. Prospects of genome editing techniques, especially the CRISPR/Cas systems, have shown multi-fold progression over the years, but these are still not acceptable to the public yet. Presently, genome editing being at its inception, scientists should practice self-restraint and forbearance while implementing gene drives, until legislative policies and public acceptance for GE approaches are in place. Synergistic application of functional genomics in consonance with robustious GE methodologies and NGS, alongside synthetic- and systems-biology approaches may be utilized for the production of crops exhibiting improved qualitative and quantitative traits. Re-addressing the issue of synchronization of crops that are generated via genome editing techniques, in general. Thus far, tracing the footprints of techno-savvy nations for instance, Japan, Australia, and Vietnam, India is on the verge of GE crops’ approval with the representative quintessential regulatory machinery being fine-tuned by the Ministry of Science and Technology and its sister concern, the Department of Biotechnology.
References
Abadi S, Yan WX, Amar D and Mayrose I 2017 A machine learning approach for predicting CRISPR/Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comp. Biol. 13 e1005807
Adiego-Pérez B, Randazzo P, Daran JM, Verwaal R, Roubos JA, Lapujade PD and Oost J 2019 Multiplex genome editing of microorganisms using CRISPR/Cas. FEMS Microbiol. Lett. 366 fnz086
Agarwal A, Yadava P, Kumar K, Singh I, Kaul T, Pattanayak A and Agrawal PK 2018 Insights into maize genome editing via CRISPR/Cas9. Physiol. Mol. Biol. Plants 24 175–183
Alagoz Y, Gurkok T, Zhang B and Unver T 2016 Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR/Cas9 genome editing technology. Sci. Rep. 6 30910
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M and Mahfouz MM 2015 CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16 238
Altpeter F, Springer NM, Bartley LE, et al. 2016 Advancing crop transformation in the era of genome editing. Plant Cell 28 1510–1520. https://doi.org/10.1105/tpc.16.00196
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S and Mahfouz M 2018 RNA virus interference via CRISPR/ Cas13a system in plants. Genome Biol. 19 1
Amin N, Ahmad N, Wu N, Pu X, Ma T, Du Y, Bo X, Wang N, Sharif R and Wang P 2019 CRISPR/Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max L.). BMC Biotechnol. 19 9
Anders C, Niewoehner O, Duerst and Jinek M 2014 Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513 569–573
Anderson M, Turesson H, Nicolia A, Fält A, Samuelsson M and Hofvander P 2017 Efficient targeted multi allelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR/Cas9 expression in protoplasts. Plant Cell Rep. 36 117–128
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan, LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A and Liu DR 2019 Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. https://doi.org/10.1038/s41586-019-1711-4
Araki M and Ishii T 2015 Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 20 145–149
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA and Voytas DF 2014 DNA replicons for plant genome engineering. Plant Cell 26 151–163
Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung and Smith AV 2015 Advances in CRISPR/Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 2015 gkv226
Baysal C, Bortesi L, Zhu C, Farré G, Schillberg S and Christou P 2016 CRISPR/Cas9 activity in the rice OsBEIIb. Mol. Breed. 36 1–11
Belhaj K, Chaparro-Garcia A, Kamoun S and Nekrasov V 2013 Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9 39
Birchler JA 2017 Editing the phenotype: a revolution for quantitative genetics. Cell 171 269–270
Birkholz N, Fagerlund RD, Smith LM, Jackson SA and Fineran PC 2019 The autoregulator Aca2 mediates anti-CRISPR repression. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz721
Bondy-Denomy J, Pawluk A, Maxwell KL and Davidson AR 2013 Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493 429–432. https://doi.org/10.1038/nature11723
Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, Kurishbayev A and Shavrukov Y 2019 Genetic modification for wheat improvement: from transgenesis to genome editing. BioMed Res. Int. 18. https://doi.org/10.1155/2019/6216304
Brooks C, Nekrasov V, Lippman ZB and Van Eck J 2014 Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR associated 9 system. Plant Physiol. 166 1292–1297
Butler NM, Atkins PA, Voytas DF and Douches DS 2015 Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PloS One 10 e0144591
Butler NM, Baltes NJ, Voytas DF and Douches DS 2016 Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 71045
Butt H, Eid A, Ali Z, Atia MAM, Mokhtar, MM, Hassan N, Lee CM, Bao G and Mahfouz MM 2017 Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front. Plant Sci. 8 1441
Buyel JF 2019 Plant molecular farming-integration and exploitation of side streams to achieve sustainable biomanufacturing. Front. Plant Sci. https://doi.org/10.3389/fpls.2018.01893
Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T and Hou W 2018 CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Nature 16 176–185
Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, Han T and Hou W 2020 Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18 298–309. https://doi.org/10.1111/pbi.13199
Callaway E 2018 CRISPR plants now subject to tough GM laws in European Union. Nature 560 16
Canzar S and Salzberg SL 2017 Short read mapping: an algorithmic tour. Proc. IEEE Inst. Electr. Electron. Eng. 105 436–458
Capecchi MR 2005 Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6 507–512
Cermak T, Baltes NJ, Cegan R, Zhang Y and Voytas DF 2015 High-frequency, precise modification of the tomato genome. Genome Biol. 16 232
Chen D, Tang J, Li B, Hou Li, Wang H and Kang L 2018 CRISPR/Cas9-mediated genome editing induces exon skipping by complete or stochastic altering splicing in the migratory locust. BMC Biotechnol. 18 60
Chen K, Wang Y, Zhang R, Zhang H and Gao C 2019 CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70 667–697
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S and Kim JS 2014 Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24 132–41
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K and Kühn R 2015 Increasing the efficiency of homology-directed repair for CRISPR/Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33 543–8
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N and Zhang F 2013 Multiplex genome engineering using CRISPR/Cas systems. Science 339 819–823
Cohen J 2019 Prime editing promises to be a cut above CRISPR. Science 366 406
Cradick TJ, Fine EJ, Antico CJ and Bao G 2013 CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucl. Acids Res. 41 9584–9592
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Bocobza S, Czosnek H, Aharoni A and Levy AA 2018 Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95 5–16
Ding D, Chen K, Chen Y, Li H and Xie K 2018 Engineering introns to express RNA guides for Cas9- and Cpf1-mediated multiplex genome editing. Mol. Plant 11 542–52
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C and Orchard R 2016 Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR/Cas9. Nat. Biotechnol. 34 184–191
Dominguez AA, Lim WA and Qi LS 2016 Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17 5–15
Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z and Huang Z 2017 Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546 436–439
Dolgin E 2019 Finding the CRISPR off-switch. Nature 577 309
DuPont Pioneer 2016 DuPont Announces Intentions to Commercialize First CRISPR/Cas Product. Press Release. https://www.pioneer.com/home/site/about/news-media/newreleases/template.
Eid A, Alshareef S and Mahfouz MM 2018 CRISPR base editors: genome editing without double-stranded breaks. Biochem. J. 475 1955–1964
Endo A, Masafumi M, Kaya H and Toki S 2016 Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisellanovicida. Sci. Rep. 6 38169
Endo M, Mikami M, Endo A, Kaya H, Itoh T, Nishimasu H, Nureki O and Toki S 2019 Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat. Plants 5 14–17
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y and Luo K 2015 Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5 12217
Fauser F, Schiml S and Puchta H 2014 Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79 348–359
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL and Zeng L 2014 Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Nat. Acad. Sci. USA 111 4632–4637
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P and Zhu JK 2013 Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23 1229
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK and Sander JD 2013 High-frequency off-target mutagenesis induced by CRISPR/Cas nucleases in human cells. Nat. Biotechnol. 31 822–826
Gaj T, Gersbach CA and Barbas CF 2013 ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31 397–405
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P and Xia Q 2014 CRISPR/Cas9 mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. http://dx.doi.org/10.1007/s11103014-0263-0
Gao L, Cox DBT, Yan WX, Manteiga, JC, Schneider, MW, Yamano T, Nishimasu H, Nureki O, Crosetto N and Zhang F 2017 Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35 789–792
Gaoneng S, Lihong GJ, Xiangjin W, Zhonghua S, Shaoqing T and Peisong HU 2017 CRISPR/CAS9-mediated Editing of the Fragrant Gene Badh2 in Rice. Chinese J. Rice Sci. 31 216–222
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI and Liu DR 2017 Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551 464–471
Gerashchenkov GA, Rozhnova NA, Kuluev BR, Kiryanova OY, Gumerova GR, Knyazev AV, Vershinina ZR, Mikhailova EV, Chemeris DA, Matniyazov RT Baimiev AK, Gubaidullin IM, Baimiev AK and Chemeris AV 2020 Design of guide RNA for CRISPR/Cas plant genome editing. Mol. Biol. 54 24–42
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres S and Eand Lim WA 2013 CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 442–451
Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, Baltes NJ, Starker C, Barro F, Gao C and Voytas DF 2017 High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89 1251–62
Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB and Joly JS 2016 Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17 148
Halterman D, Guenthner J, Collinge S, Butler N and Douches D 2015 Biotech potatoes in the 21st century: 20 years since the first biotech potato. Am. J. Potato 93 1–20
Hanania U, Arie T, Tekoah Y, Fux L, Sheva M, Gubbay Y, Weiss M, Oz D, Azulay Y, Turbovski A, Forster Y and Shaaltiel Y 2017 Establishment of a tobacco BY2 cell line devoid of plant specific xylose and fucose as a platform for the production of biotherapeutic proteins. Plant Biotechnol. J. 15 1120–1129
Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF and Zhang F 2014 Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12 934–940
Heigwer F, Kerr G and Boutros M 2014 E-CRISP: fast CRISPR target site identification. Nat. Methods 11 122–123
Hiranniramol K, Chen Y, Liu W and Wang X 2020 Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency. Bioinformatics 36 2684–2689
Hilton IB 2015 Epigenome editing by a CRISPR/Cas9-based acetyl transferase activates genes from promoters and enhancers. Nat. Biotechnol. 33 510–517
Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer, EJ and Thomson JA 2013 Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad Sci. USA 110 15644–9
Hsu PD, Lander ES and Zhang F 2014 Development and applications of CRISPR/Cas9 for genome engineering. Cell 157 1262–1278
Hsu PD, Scott DA, Weinstein, JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G and Zhang F 2013 DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 827–832
Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF and Taylor NJ 2017 Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16 1275–1282
Hua K, Tao X, Han P, Wang R and Zhu JK 2019b Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 12 1003–1014
Hua K, Tao X, Yuan F, Wang D and Zhu JK 2018 Precise A-T to G-C base editing in the rice genome. Mol. Plant 11 627–630
Hua K, Zhang J, Botella JR, Ma C, Kong F, Liu B and Zhu JK 2019a Perspectives on the application of genome editing technologies in crop breeding. Mol. Plant 12 1047–1059
Ismail E, Gavahian M, Marti-Quijal FJ, Lorenzo JM, Khaneghah AM, Tsatsanis C, Kampranis SC and Barba FJ 2019 The application of the CRISPR/Cas9 genome editing machinery in food and agricultural science: current status, future perspectives, and associated challenges. Biotechnol. Adv. 37 410–421
Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M and Toki S 2015 CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467 76–82
Jacobs TB, LaFayette PR, Schmitz RJ and Parrott WA 2015 Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15 16
Jansing J, Sack M, Augustine S, Fischer R and Bortesi L 2019 CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking b-1,2-xylose and core a-1,3-fucose. Plant Biotechnol. J. 17 350–361
Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR and Malik P 2019 CRISPR/Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 10 2866
Jeong YK, Yu, Y and Bae S 2019 Construction of non-canonical PAM-targeting adenosine base editors by restriction enzyme-free DNA cloning using CRISPR/Cas9. Sci. Rep. 9 4939
Jia H and Wang N 2014 Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9 e93806. https://doi.org/10.1371/journal.pone.0093806
Jiang W, Zhou H, Bi H, Fromm M, Yang B and Weeks DP 2013 Demonstration of CRISPR/Cas9-sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucl. Acid Res. 41 188
Jin S, zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qui J, Zhang FGC 2019 Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364 292–295
Jinek M, Jiang F, Taylor DW, Sternberg, SH, Kaya E, Ma E and Kaplan M 2014 Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343 1247997
Johnson RA, Gurevich V, Filler S, Samach A and Levy AA 2015 Comparative assessments of CRISPR/Cas nucleases’ cleavage efficiency in planta. Plant Mol. Biol. 87 143–156
Jones HD 2015 Regulatory uncertainty over genome editing. Nat. Plants 1 10–1038
Kanchiswamy CN, Sargent DJ, Velasco R, Maffei ME and Malnoy M 2015 Looking forward to genetically edited fruit crops. Trends Biotechnol. 33 62–64
Kang B, Yun J, Kim ST, Shin Y, Ryu J, Choi M, Woo JW and Kim JS 2018 Precision genome engineering through adenine base editing in plants. Nat. Plants 4 427–431
Kannan B, Jung JH, Moxley GW, Lee SM and Altpeter F 2018 TALEN mediated targeted mutagenesis of more than 100 COMTcopies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16 56–866
Kaul T, Eswaran M, Thangaraj A, Meyyazhagan A, Nehra M, Raman N, Bharti J, Gayacharan BC and Balamurali B 2019a Rice Bean (Vigna umbellata) draft genome sequence: unravelling the late flowering and unpalatability related genomic resources for efficient domestication of this underutilized crop. bioRxiv. http://dx.doi.org/10.1101/816595
Kaul T, Raman NM, Eswaran M, Thangaraj A, Verma R, Sony SK, Sathelly KM, Kaul R, Yadava P and Agrawal PK 2019b Data mining by pluralistic approach on CRISPR gene editing in plants. Front. Plant Sci. 10 801
Kaul T, Sony SK, Raman NM, Eswaran M, Verma R, Thangaraj A, Bharti J, Motelb KFA and Kaul R 2020 How crisp is CRISPR? CRISPR/Cas mediated crop improvement with special focus on nutritional traits; in Advancement in Crop Improvement Techniques (eds) N Tuteja, R Tuteja, N Passricha, S Saifi (New Delhi: Woodhead Printing) 20 159–197
Kaur N, Alok A, Shivani KN, Pandey P, Awasthi P and Tiwari S 2018 CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 18 89–99
Kim D, Bae S, Park J, Kim E, Kim S, Yu HR and Kim JS 2015 Digenome-seq: genome-wide profiling of CRISPR/Cas9 off-target effects in human cells. Nat. Methods 12 237
Kim H and Kim JS 2014 A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15 321
Kim H, Kim ST, Ryu J, Kang BC, Kim JS and Kim SG 2017 CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8 14406
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, Aryee, MJ and Joung JK 2015 Engineered CRISPR/Cas9 nucleases with altered PAM specificities. Nature 523 481–485
Komor AC, Kim YB, Packer MS, Zuris JA and Liu DR 2016 Programmable editing of a target base in genomic DNA without double stranded DNA cleavage. Nature 533 420–424
Lau W, Fischbach MA, Osbourn A and Sattely ES 2014 Key applications of plant metabolic engineering. PLoS Biol. 12 e1001879
Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N and Uauy C 2015 Harwood W: Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16 258
Lawhorn IE, Ferreira JP and Wang CL 2014 Evaluation of sgRNA target sites for CRISPR-mediated repression of TP53. PLoS One 9 e113232
Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim D, Kim S and Kim JS 2018a Directed evolution of CRISPR/Cas9 to increase its specificity. Nat. Commun. 9 3048
Lee O, Mir A, Edraki A, Garcia B, Amrani N, Lou HE, Gainetdinov I, Pawluk A, Ibraheim R, Gao XD, Liu P, Davidson AR, Maxwell KL and Sontheime EJ 2018b Potent Cas9 inhibition in bacterial and human cells by AcrIIC4 and AcrIIC5 anti-CRISPR proteins. mBio. 9 02321–18
Lei Y, Lu L, Liu HY, Li S, Xing F and Chen LL 2014 CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7 1494–1496
Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Van Eck, J and Lippman ZB 2018 Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4 766–770
Li Z, Liu Z, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC and Cigan, AM 2015 Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 169 960–970
Li A, Jia S, Yobi A, Ge Z, Sato SJ, Zhang C, Angelovici R, Clemente THRDR 2018b Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177 1425–1438
Li C, Liu C, Qi X, Wu Y, Fei X, Mao L, Cheng B, Li X and Xie C 2017b RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 15 1566–1576
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R and Gao C 2018a Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19 59
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church G M and Sheen J 2013 Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 688–691
Li, J, Sun, Y, Du, J, Zhao, Y and Xia L 2017a Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 10 526–529
Li M, Li X, Zhou Z, Wu P, Fang M, Pan X, Lin Q, Luo W, Wu G and Li H 2016 Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front Plant Sci. 7 377
Li R, Li R, Li X, Fu D, Zhu B, Tian H, Luo Y and Zhu H 2018d Multiplexed CRISPR/Cas9-mediated metabolic engineering of g-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16 415–427
Li S, Li J, He Y, Xu M, Zhang J, Du W, Zhao Y and Xia L 2019 Precise gene replacement in rice by RNA transcript templated homologous recombination. Nat. Biotechnol. 37 445–450
Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C 2018c Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36 1160–1163
Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J and Li JF 2017c A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 3 930–936
Liang Z, Zhang K, Chen K and Gao C 2014 Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. Genet. Genomics 41 63–68
Liang J, Chen K, Yan Y, Zhang Y and Gao C 2018 Genotyping genome edited mutations in plants using CRISPR ribonucleoprotein complexes’. Plant Biotechnol. J. 16 2053–2062
Lin S, Staahl BT, Alla RK and Doudna JA 2014 Enhanced homology directed human genome engineering by controlled timing of CRISPR/ Cas9 delivery. eLife 3 04766
Lino CA, Harper JC, Carney JP and Timlin JA 2018 Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25 1234–1257
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone A V, Raguram A, Doman JL, Liu DR and Gao C 2020 Prime genome editing in rice and wheat. Nat. Biotechnol. 38 582–585
Listgarten J, Weinstein M, Kleinstiver BP, Sousa AA, Joung JK, Crawford J and Fusi N 2018 Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2 38
Liu D, Chen X, Liu J, Ye J and Guo Z 2012 The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaportheoryzae and salt tolerance. J. Exp. Bot. 63 3899–3912
Liu H, Wei Z, Dominguez A, Li Y, Wang X and Qi LS 2015 CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 31 3676–3678
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA and Jaenisch R2016 Editing DNA methylation in the mammalian genome. Cell 167 233–247.e17
Liu Y, Gao Y, Gao Y and Zhang Q 2019 Targeted deletion of floral development genes in Arabidopsis with CRISPR/Cas9 using the RNA endoribonuclease Csy4 processing system. Hortic. Res. 6 99
Lowe K, Wu E, Wang N, et al. 2016 Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28 1998–2015
Lu Y and Zhu JK 2017 Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 10 523–525
Lu Y, Ye X, Guo R, Huang J, Wang W, Tang J, Tan L, Zhu JK, Chu C and Qian Y 2017 Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10 1242-5
Lusser M, Parisi C, Plan D and Rodriguez-Cerezo E 2011 New plant breeding techniques: state-of-the-art and prospects for commercial development. In Report EUR 24760, (Technical, J.R.C, ed)
Lyzenga WJ, Harrington M, Bekkaoui D, Wigness M, Hegedus DDand Rozwadowski KL 2019 CRISPR/Cas9 editing of three CRUCIFERIN C homoeologues alters the seed protein profile in Camelina sativa. BMC Plant Biol. 19 292
Ma JK, Drake PM and Christou P 2003 The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 4 794–805
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R and Xie Y 2015 A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8 274–1284
Makarova KS and Koonin EV 2015 Annotation and classification of CRISPR/Cas systems. Methods Mol. Biol. 1311 47–75
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R and Nagamangala KC 2016 DNA-free genetically edited grapevine and apple protoplast using crispr/cas9 ribonucleoproteins. Front Plant Sci. 7 1904
Mao Y, Botella JR, Liu Y and Zhu JK 2019 Gene editing in plants-progress and challenges. Nat. Sci. Rev. 12(8) 047–1059
Mao Y, Zhang H, Xu N, Zhang B, Gao F and Zhu JK 2013 Application of the CRISPR/Cas system for efficient genome engineering in plants. Mol. Plant 6 2008–2011
Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR and Zhu JK 2016 Development of germ-line-specific CRISPR/Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14 519–532
Marino ND, Zhang JY, Borges AL, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP and Bondy-Denomy J 2018 Discovery of widespread type I and type V CRISPR/Cas inhibitors. Science 362 240–242
Mei Y, Beernink BM, Ellison EE, Konečná E, Neelakandan AJ, Voytas D. FandWhitham SA 2019 Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct. 3 1–16
Meng X, Yu H, Zhang Y, Zhuang F, Song X, Gao S, Gao C and Li J 2017 Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 10 1238–41
Mercx S, Smargiasso N, Chaumont F, Pauw ED, Boutry M and Navarre C 2017 Inactivation of the b(1,2)-xylosyltransferase and the a(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 8 00403
Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ and Stupar RM 2015 CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and amodified Cas9enzyme. GM Crops Food 6 243–252
Miki D, Zhang W, Zeng W Feng Z and Zhu JK 2018 CRISPR/ Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9 1967
Minkenberg B, Xie K and Yang Y 2017 Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 89 636–648
Misra S and Mohanty D 2017 Psychobiotics: a new approach for treating mental illness? Crit. Rev. Food Sci. Nutr. 59 1230–1236
Miyamoto T, Takada R, Tobimatsu Y, Takeda U, Suzuki S, Yamamura M, Osakabe K, Osakabe, Y, Sakamoto MUT 2019 OsMYB108 loss-of-function enriches p-coumaroylated and tricin lignin units in rice cell walls. Plant J. 98 975–987
Mohanty D, Chandra A and Tandon R 2016 Germline transformation for crop improvement In: Molecular Breeding for Sustainable Crop Improvement, Vol. 2, eds VR Rajpal SR Rao and SN Raina (Cham: Springer International Publishing)
Mok BY, Moraes MHD, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, Mougous JD and Liu DR 2020 A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. https://doi.org/10.1038/s41586-020-2477-4
Moon SB, Kim DY, Ko J and Kim Y 2019 Recent advances in the CRISPR genome editing tool set. Exp. Mol. Med. 51 130
Moradpour M and Abdulah SNA 2020 CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol. J. 18 32–44
Naim F, Dugdale B, Kleidon J, Brinin A, Shand K, Waterhouse P and Dale J 2018 Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27 451–460
Najera VA, Twyman RM, Christou P and Zhu C 2019 Applications of multiplex genome editing in higher plants. Curr. Opin. Biotechnol. 59 93–102
Negishi K, Kaya H, Abe K, Hara N and Saika Hand Toki S 2019 An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol. J. 17 1476–1478
Nekrasov V, Staskawicz B, Weigel D, Jones JD and Kamoun S 2013 Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 691
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z and Kondo A 2016 Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 16 353
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F and Nureki O 2014 Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156 935–49
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F and Nureki O 2019 Engineered CRISPR/Cas9 nuclease with expanded targeting space. Science 361 1259–1262
Nishitani C, Hirai N, Komori S, Wada M, Okada K, Okada K, Osakabe K, Yamamoto Tand Osakabe Y 2016 Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6 314811
O’BrienAand Bailey TL 2014 O’GT-Scan: identifying unique genomictargets. Bioinformatics 30 26732675
Østerberg JT, Xiang W, Olsen LI, Edenbrandt AK, Vedel SE, Christiansen A, Landes X, Andersen MM, Pagh P, Sandøe P, Nielsen J, Christensen SB, Thorsen BJ, Kappel K, Gamborg C and Palmgren M 2019 Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci. 22 373–384
Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L and Lu G 2016 CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6 24765
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF and Doudna JA 2020 CRISPR/CasΦ from huge phages is a hypercompact genome editor. Science 369 333–337
Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL and Davidson AR 2016 Naturally occurring off-switches for CRISPR/Cas9. Cell 167 1829–1838
Pawluk A, Davidson AR and Maxwell KL 2018 Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 16 12–17
Peterson BA, Haak DC, Nishimura MT, Teixeira PJ, James SR, Dangl JL and Nimchuk ZL 2016 Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PloS One 11 p.e0162169
Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al‐Shareef, S and Mahfouz MM 2015 RNA‐guided transcriptional regulation in planta via synthetic dC as9‐based transcription factors. Plant Biotechnol. J. 13 578–589
Qi W, Zhu T, Tian Z, Li C and Zhang WSR 2016 High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 16 58
Raman NM, Eswaran M, Bharti J, Motalb KFA, Verma R, Kaul R and Kaul T 2019 Ushering in CRISPR/Cas mediated genome engineering for crops. Sch. Acad. J. Biosci. 7 313–320
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y and Zhang F 2013 Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154 1380–1389
Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan, NJ and Bondy-Denomy J 2017 Inhibition of CRISPR/Cas9 with bacteriophage proteins. Cell 168 150–158
Rees HA and Liu DR 2018 Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19 770–788
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X and Lin HZH 2019 Cas9-NG greatly expands the targeting scope of the genome editing toolkit by recognizing NG and other a typical PAMs in rice. Mol. Plant 12 1015-1026.
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, Lin H and Zhou H 2018 Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant 11 623–626
Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S and Liang Z 2016 ‘CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.)’ Sci. Rep. 6 32289
Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji, JY, Li, JB and Ni JQ 2014 Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9 1151–1162
Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME and Lippman ZB 2017 Engineering quantitative trait variation for crop improvement by genome editing. Cell 171 470–480
Romer P, Recht S and Lahaye T 2009 A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. USA 106 20526–20531
Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K and Federici F 2014 Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166 455–469
Sandhya D, Jogam P, Allini VR, Abbagani S and Alok A 2020 The present and potential future methods for delivering CRISPR/Cas9 components in plants. J Genet. Eng. Biotechnol. 18 25
Sauer NJ, Narvaez-Vasquez J, Mozoruk J, Miller RB, Warburg ZJ, Woodward MJ, Mihiret YA, Lincoln TA, Segami RE, Sanders SL, Walker KA, Beetham PR, Schöpke CR and Gocal GF 2016 Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170 1917–28
Schindele A, Dorn A and Puchta H 2020 CRISPR/Cas brings plant biology and breeding into the fast lane. Curr. Opin. Biotechnol. 61 714
Schiml S, Fauser F and Puchta H 2014 The CRISPR/C as system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80 1139–1150
Sedeek KEM, Mahas A and Mahfouz M 2019 Plant Genome Engineering for Targeted Improvement of Crop Traits. Front. Plant Sci. 10 114
Seonghee L, Cheolmin Y, Kevin F and Vance M 2018 Whitaker CRISPR gene editing in strawberry. The Institute of Food and Agricultural Sciences (IFAS)
Shan QW, Zhang Y, Chen KL, Zhang K and Gao CX2015 Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13 791–800
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL and Gao C 2013 Targeted genome modification of crop plants using a CRISPR/Cas system. Nat. Biotechnol. 31 686–688
Sharma S and Petsalaki E 2018 Application of CRISPR/Cas9 based genome-wide screening approaches to study cellular signalling mechanisms. Int. J. Mol. Sci. 19 933
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H and Habben JE 2017 ARGOS 8 variants generated by CRISPR/Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15 207–216
Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB and Ertl DS 2003 The maize low phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 131 507–515
Shimatani Z, Kashojiya S, Takayama M, Terada R, ArazoeT, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T and Kondo A 2017 Targeted base editing in rice and tomato using a CRISPR/Cas cytidine deaminase fusion. Nat. Biotechnol. 35 440–446
Shen C, Que Z, Xia Y, Tang N, Li D, He R, He, R and CM 2017 Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 60 539–547
Shen L, Wang C, Fu Y, Wang J, Liu Q, Zhang X, Yan C, Qian Q and Wang K 2018 QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 60 89–93
Shin J, Jiang F, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE and Doudna, JA 2017 Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3 1701620
Silva NV and Patron NJ 2017 CRISPR-based tools for plant genome engineering. Emerg. Top Life Sci. 1 135–149
Singh R, Kuscu C, Quinlan A and Qi YAM 2015 Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucl. Acid. Res. 43 e118e118
Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez, J and M and Lippman ZB 2016 Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49 162
Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, Natarajan R, Kelkar M, De A, Choudhary B and Raghavan SC 2012 An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151 1474–87
Steinert J, Schiml S and Puchta H 2016 Homology-based double strand break-induced genome engineering in plants. Plant Cell Rep. 35 1429–1438
Stemmer M, Thumberger T, Keyer MDS, Wittbrodt J and Mateo JL 2015 CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PloS One 10 e0124633
Sternberg SH, Redding S, Jinek M, Greene EC and Doudna JA 2014 DNA interrogationby the CRISPR RNA-guided endonuclease Cas9. Nature 507 62
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura, I and Kohchi T 2014 CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55 475–481
Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL and Gu Z 2015 Self-assembled DNA nanoclews for the efficient delivery of CRISPR/Cas9 for genome editing. Angew. Chem. Int. Ed Engl. 54 12029–12033
Sun Y Jiao G, Liu Z, Zhang X, Li J, Guo X, Du W, Du J, Francis F, Zhao Y and Xia L 2017 Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8 298
Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y and Xia L 2016 Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9 628–631
Svitashev S, Schwartz C, Lenderts B, Young JK and Cigan AM 2016 Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7 13274
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC and Cigan AM 2015 Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169 931–945
Symington LS and Gautier J 2011 Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45 247–271
Tang L, Mao B, Li Y, Lv Q, Zhang L, Chen C, He H, Wang W, Zeng X, Shao Y, Pan Y, Hu Y, Peng Y, Fu X, Li H, Xia S and Zhao B 2017a Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating Indica rice without compromising yield. Sci. Rep. 7 14438
Tang X, Liu G, Zhou J, Ren Q, You Q, Tian L, Xin X, Zhong Z, Liu B, Zheng X and Zhang D 2018 A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genom. Biol. 19 84
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y and Qi Y 2017b A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3 17103
Tashkandi M, Ali Z, Aljedaani F, Shami A and Mahfouz MM 2018 Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Sci. Plant Signal. Behav. 362 236–239
Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q and Jin W and Tian F 2019 Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365 658–664
Tingting L, Di F, Lingyu R, Yuanzhong J, Rui L and Keming L 2015 Highly efficient CRISPR/Cas9 mediated targeted mutagenesis of multiple genes in Populus. YiChuan 37 1044–1052
Tripathi NJ, Ntui VO, Ron M, Muiruri SK, Britt A and Tripathi L 2019 CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Comm. Biol. 2 https://doi.org/10.1038/s42003-019-0288-7
Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V and Aryee MJ 2015 GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR/Cas nucleases. Nat. Biotechnol. 33 187
Turcotte MM, Araki H, Karp DS, Poveda K and Whitehead SR 2017 The ecoevolutionary impacts of domestication and agricultural practices on wild species. Phil. Trans. R. Soc. B. 372 20160033
Upadhyay SK, Kumar J, Alok A and Tuli R 2013 RNA-guided genome editing for target gene mutations in wheat. G3: Genes, Genomes, Genetics 3 2233–2238
Vartak SV and Raghavan SC 2015 Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J. 282 4289–94
Wada N, Ueta R, Osakabe Y and Osakabe K 2020 Precision genome editing in plants: state of the art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 20 234
Waltz E 2016 Gene-edited CRISPR mushroom escapes US regulation. Nat. Biotechnol. 532 293
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG and Zhao K 2016 Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11 e0154027
Wang M, Lu Y, Botella JR, Mao Y, Hua K and Zhu JK 2017 Gene targeting by homology-directed repair in rice using a gemini virus based CRISPR/Cas9 system. Mol. Plant 10 1007–1010
Wang H, Wu Y, Zhang Y, Yang J, Fan W, Zhang H, Zhao S, Yuan L and Zhang P 2019b CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20 4702
Wang M, Mao Y, Lu Y, Tao X and Zhu JK 2019c Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10 1011–1013
Wang M, Wang Z, Mao Y, Lu Y, Yang R, Tao X and Zhu JK 2019a Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. 17 1697–1699
Wang S, Zhang S, Wang W, Xiong X, Meng F and Cui X 2015b Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 34 1473–1476
Wang T, Guan C, Guo J, Liu B, Wu Y, Xie Z, Zhang Cand Xing X 2018b Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 9 2475
Wang W, Simmonds J, Pan Q, Davidson D, He F, Battal A, Akhunova A, Trick HN, Uauy C and Akhunov E 2018a Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 131 2463–2475
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C and Qiu JL 2014 Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 947–951
Wang ZP, Xing HL, Dong L, Zhang HY, Han, CY, Wang, XC and Chen QJ 2015a Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genom. Biol. 16 144
Watters KE, Fellmann C, Bai HB, Ren SM and Doudna JA 2018 Systematic discovery of natural CRISPR/Cas12a inhibitors. Science 362 236–239
Weinstock DM and Jasin M 2006 Alternative pathways for the repair of RAG-induced DNA breaks. Mol. Cell Biol. 26 131–139
Wolt JD 2017 Safety, security, and policy considerations for plant genome editing. Progr. Mol. Biol. Translational Sci. 149 1873–1877
Wolt JD, Wang K and Yang B 2016 The regulatory status of genome‐edited crops. Plant Biotechnol. J. 14 510–518
Wolter F, Klemm J and Puchta H 2018 Efficient in planta gene targeting in Arabidopsis using egg-cell specific expression of the Cas9 nuclease of S. aureus. Plant 94 735–746
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S and Kim JS 2015 DNA-free genome editing in plants with preassembled CRISPR/Cas9 ribonucleoproteins. Nat. Biotechnol. 33 1162
Wu Y, Xu W, Wang F, Zhao S, Feng F, Song J, Zhang C and Yang J 2019 Increasing cytosine base editing scope and efficiency with engineered Cas9-PmCDA1 fusions and the modified sgRNA in rice. Front. Genet. 10 379
Wu J, Chen C, Xian G, Liu D, Lin L, Yin S, Sun Q, Fang Y, Zhang H and Wang Y 2020 Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 1–3
Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G and Zhang B 2014 CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30 8 1180–1182
Xie K and Yang Y 2013 RNA-guided genome editing in plants using a CRISPR/Cas system. Mol. Plant 6 1975–1983
Xie K, Minkenberg B and Yang Y 2015 Boosting CRISPR/Cas9 multiplex editingcapability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112 3570–3575
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC and Chen QJ 2014 A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14 327
Xing Y and Zhang Q 2010 Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61 421–442
Xu Z, Xu X, Gong Q, Li Z, Li Y, Wang S, Yang Y, Ma W, Liu L, Zhu B, Zou L and Chen G 2019 Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12 1434–1446
Xu P, Zhang Y, Kang L, Roossinck MJ and Mysore KS 2006 Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142 429–440
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC and Yang JB 2015 Generation of inheritable and ‘transgene clean’ targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5 11491
Xu R, Qin R, Li H, Li D, Li L, Wei P and Yang J 2017 Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 15 713–717
Yadava P, Abhishek A, Singh R, Singh I, Kaul T, Pattanayak A and Agrawal PK 2017 Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci. 7 1949
Yan W, Chen D and Kaufmann K 2016 Efficient multiplex mutagenesis by RNA-guided Cas9 and its use in the characterization of regulatory elements in the AGAMOUS gene. Plant Methods 12 23
Yang H, Wu JJ, Tang T, Liu KD and Dai C 2017 CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7 7489
Yang H and Patel DJ 2017 Inhibition mechanism of an anti-CRISPR suppressor AcrIIA4 targeting SpyCas9. Mol. Cell 67 117–1275
Ye M, Peng Z, Tang D, Yang Z, Li D, Xu Y, Zhang C and Huang S 2018 Generation of self-compatible diploid potato by knockout of S-RNase. Nat. Plants 4 651–654
Yin K, Gao C and Qiu J 2017 Progress and prospects in plant genome editing. Nat. Plants 3 17107
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY and Land Liu Y 2015 A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 5 14926
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La RM, Xie M, Ding S and Qi LS 2015 Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16 142–147
Yu Q, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo, Yan P, Wang Q and Asmutola P 2017 CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7 11874
Yunyan F, Jie Y, Fangquan W, Fangjun F, Wenqi L, Jun W, Yang X, Jinyan Z and Weigong Z 2019 Production of two elite glutinous rice varieties by editing Wx Gene. Rice Sci. 26 118–124
Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, Gennaro EMD, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott, DA, Severinov K, van der Oost and Zhang F 2017 Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35 31–34
Zhang F, LeBlanc C, Irish VF and Jacob Y 2017b Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36 1883–1887
Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D and Gao C 2018 Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36 894–898
Zhang J, Zhang H, Botella JR and Zhu JK 2017a Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. Integr. Plant Biol. 60 369–375
Zhang P, Du H, Wang J, Pu Y, Yang C, Yan R, Yang H, Cheng H and Yu D 2019 Multiplex CRISPR/Cas9-mediated metabolic engineering increases soybean isoflavone content and resistance to soybean mosaic virus. Plant Biotechnol. J. https://doi.org/10.1111/pbi.13302.
Zhang Y, Liang J, Zong Y, Wang Y, Liu J, Chen K, Qiu JL and Gao C 2016 Efficient and transgenefree genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7 12617
Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li WX, Mao L, Chen B, Xu Y, Li X and Xie C 2016 An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 6 23890
Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Yand Zhang Y 2019 Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 12 1027–1036
Zhong Z, Zhang Y, You Q, Tang X, Ren Q, Liu S, Yang L, Wang Y, Liu X, Liu B, Zhang T, Zheng X, Le Y, Zhang Y and Qi Y 2018 Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol. Plant 11 999–1002
Zhou J, Peng Z, Long J, Sosso D, Liu B, Eom JS, Huang S, Liu S, Vera Cruz C, Frommer WB, White FF and Yang B 2015a Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82 632–643
Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, Wei Y, Hu X, Zeng R, Li, Y Zhou H, Guo F and Yang H 2019 Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571 275
Zhou X, Jacobs TB, Xue LJ, Harding SA and Tsai CJ 2015b Exploiting SNPs for biallelic CRISPR mutations in the out crossing woody perennial Populus reveals 4 coumarate:Co Aligase specificity and redundancy. New Phytol. 208 298–301
Zhu Y, Gao A, Zhan Q, Wang Y, Feng H, Liu S, Gao G, Serganov A and Gao P 2019 Diverse mechanisms of CRISPR/Cas9 inhibition by Type IIC anti-CRISPR proteins. Mol. Cell 74 296–309
ZKBS (German Biosafety Commission) 2012 Position statement of the ZKBS on new plant breeding techniques. http://www.bvl.bund.de/DE/06_Gentechnik/04_Fachmeldungen/2013/2013_08_01_Fa_neue_Techniken_Pflanzenzuechtung.html?nn=1471850.
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D and Gao C 2017 Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35 438–440
Zsögön A, Cermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF and Kudla J 2018 De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36 1211–1216
Zuo E, Sun Y, Wei W, Yuan T, Ying W and Sun H 2019 Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364 289–292
Zuo J and Li J 2014 Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48 99–118
Acknowledgements
The authors are thankful to ICAR-National Agricultural Science Fund (NASF – Grant F. No. NASF/GTR-7025/2018-19) for funding this research. They also take this opportunity to thank the ICGEB core funds and Department of Biotechnology, for providing supportive funding for research at ICGEB, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection: Genetic Intervention in Plants: Mechanisms and Benefits.
Rights and permissions
About this article
Cite this article
Kaul, T., Sony, S.K., Verma, R. et al. Revisiting CRISPR/Cas-mediated crop improvement: Special focus on nutrition. J Biosci 45, 137 (2020). https://doi.org/10.1007/s12038-020-00094-7
Published:
DOI: https://doi.org/10.1007/s12038-020-00094-7