Abstract
Plant-growth-promoting bacteria are often used to enhance crop yield and for biological control of phytopathogens. Bacillus sp. CHEP5 is a biocontrol agent that induces systemic resistance (ISR) in Arachis hypogaea L. (peanut) against Sclerotium rolfsii, the causal agent of root and stem wilt. In this work, the effect of the co-inoculation of Bacillus sp. CHEP5 and the peanut nodulating strain Bradyrhizobium sp. SEMIA 6144 was studied on induction of both systemic resistance and nodulation processes. Bradyrhizobium sp. SEMIA 6144 did not affect the ability of Bacillus sp. CHEP5 to protect peanut plants from S. rolfsii by ISR and the priming in challenged-plants, as evidenced by an increment in phenylalanine ammonia-lyase enzyme activity. Additionally, the capacity of Bradyrhizobium sp. SEMIA 6144 to induce nodule formation in pathogen-challenged plants was improved by the presence of Bacillus sp. CHEP5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Plant-growth-promoting bacteria (PGPB) are associated with plants and stimulate their growth either directly or indirectly. PGPB exert a direct effect on plant growth by producing phytohormones or by facilitating the uptake of certain nutrients from the environment, such as nitrogen, phosphorus or iron. On the other hand, indirect promotion occurs when PGPB lessen or prevent the deleterious effect of phytopatogenic organisms by antibiosis, competition for space and nutrients, production of siderophores, and induction of systemic resistance in plants against a broad spectrum of root and foliar pathogens (Podile and Kishore 2006; Lugtenberg and Kamilova 2009).
The induced systemic resistance (ISR) is an indirenct biocontrol mechanism that involves the activation of plant defence state only upon pathogen attack (Kloepper et al. 1992). A common feature of the resistance responses induced by beneficial microorganisms is priming. Primed plants display faster and/or stronger activation of cellular defence responses when they are attacked by pathogens (Pieterse et al. 2000; Paré et al. 2005; Conrath et al. 2006; Pozo et al. 2008). ISR involves a cascade of defence reactions that spread from the site of induction to distant parts of the plant and encompass signal transduction mediated by phytohormones, generation of phytoalexins, oxidative stress protection, enzymes related to plant defence (phenylalanine ammonia-lyase, peroxidase, β-1,3-glucanase), and formation of structural barriers such as wall thickening, callose deposition and accumulation of phenolics (Reymond and Farmer 1998, Verhagen et al. 2004). Considering that the protection mediated by ISR is not specific and is effective against a broad range of phytopathogens (Van Loon et al. 1998), this mechanism becomes promising as an alternative or complement to pesticide application in the integrated disease management.
Among PGPB there is a wide group of microorganisms called rhizobia, which provide nitrogen to legumes by a symbiotic process. They can form nodules on roots of leguminous plants in which they convert N2 into ammonia, which become available as nitrogen source for the plant (Podile and Kishore 2006; Lugtenberg and Kamilova 2009; Fabra et al. 2010).
Peanut (Arachis hypogaea L.) is an economically important legume throughout the world and its productivity is affected, among other environmental stresses, by different diseases. A soil-borne fungal disease that adversely affects peanut yields all over the world’s growing areas is root and stem wilt caused by Sclerotium rolfsii. The extensive use of pesticides could contribute to environmental pollution and to an increase in the production cost. Moreover, beneficial microorganisms, including those that fix atmospheric nitrogen, could be negatively affected by this practice. Previous studies in our laboratory demonstrated that the native isolate Bacillus sp. CHEP5 induces systemic resistance in peanut against the pathogen S. rolfsii and primes the activity of the enzyme phenylalanine ammonia-lyase (PAL) when the plant is challenged by the phytopathogen (Tonelli et al. 2011). On the other hand, Bradyrhizobium sp. SEMIA 6144 is able to establish a symbiotic association with peanut plants, inducing the formation of root nodules (Fabra et al. 2010).
Plants are in constant interplay with many soil microorganisms. There is an appreciable amount of literature on physiological effects on host plant growth during the interaction between only one group of microorganisms and plants (especially rhizobia and legumes). However, enhanced and concentrated efforts are nedeed to obtain a greater clarity about the beneficial effects of different groups of PGPB on plant growth, considering that in the rhizosphere, communication among more than one group of microorganisms and roots are established.
Since the induction of systemic resistance against phytopathogens and the improvement in nitrogen nutrition are both desirable features for peanut growth promotion, it becomes relevant to evaluate the effect of Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144 co-inoculation on this legume. Therefore, the aim of this work was to determine whether the co-inoculation of these microorganisms affects their peanut growth promoting activities.
2 Materials and methods
2.1 Bacterial strains, pathogen and culture conditions
The native biocontrol agent Bacillus sp. CHEP5 and the symbiont Bradyrhizobium sp. SEMIA 6144 (reference strain recommended by Microbiological Resource Center, Porto Alegre, Brasil) were used in this study. Bacillus sp. CHEP5 was cultured at 28°C on Trypticase Soya Broth (TSB) or Agar (TSA) (Britania) media. Bradyrhizobium sp. SEMIA 6144 was cultured at 28°C onYeast Extract Mannitol broth (YEM) or YEM-agar (YEMA) (Vincent 1970). The strains were kept in 20% glycerol at −80°C for long-term storage and in 40% glycerol at −20°C for short-term storage.
The fungal pathogen S. rolfsii was grown on Potato Dextrose Agar (PDA) at room temperature for 7 days. The phytopathogen was kept in 15% glycerol at −20°C for long-term storage.
2.2 Indole acetic acid-like molecule production by Bradyrhizobium sp. SEMIA 6144
Indole acetic acid (IAA)-like molecule production was detected by the method described by Bric et al. (1991). YEMA plates supplemented with l-tryptophan and containing nitrocellulose discs were spot-inoculated with 10 μL bacterial cultures (108 cfu mL−1) and incubated at 28°C for 120 h. Nitrocellulose discs were transferred to test tubes and impregnated with Salkowsky reagent (Gordon and Weber 1951). Appearance of pink colour after 30 min to 3 h of incubation indicated IAA-like molecule production. Uninoculated nitrocellulose discs were used as negative control.
2.3 Simultaneous growth of Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144 on solid media
To evaluate whether the growth of one strain is affected by the presence of the other, the following assays were done: (a) One half of a YEMA plate was streaked with the strain Bradyrhizobium sp. SEMIA 6144 and, at the same time, the other half was streaked with Bacillus sp. CHEP5 culture; (b) Bacillus sp. CHEP5 was streaked earlier (24 h) than Bradyrhizobium sp. SEMIA 6144; (c) Bradyrhizobium sp. SEMIA 6144 was streaked earlier (5 days) than Bacillus sp. CHEP5. The plates were incubated at 28°C.
2.4 Plant material and growth conditions
Seeds of Arachis hypogaea L. (peanut) var. Runner cultivar Granoleico, susceptible to S. rolfsii root and stem wilt, were surface sterilized as described by Vincent (1970). Briefly, the seeds were soaked in 96% ethanol for 30 s followed by 20% H2O2 for 15 min, and then washed six times with sterile distilled water. The surface sterilized seeds were germinated at 28°C in sterilized Petri dishes with one layer of Whatman No.1 filter paper and moist cotton, until the radicle reached approximately 2 cm. Seedlings were sown in plastic cups filled with sterilized quartz sand, watered regularly and supplied with Hoagland medium (Hoagland and Arnon 1950) once a week. Plants were grown under controlled environment (light intensity of 200 mmol m−2 s−1, 16 h day/8 h night cycle, at a constant temperature of 28°C and a relative humidity of 50%).
2.5 Bacterial inoculum preparation
Bacillus sp. CHEP5 was cultured on TSB medium for 24 h, while Bradyrhizobium sp. SEMIA 6144 was cultured on YEM media for 10 days. They were incubated at 28°C until cultures reached an OD620nm= 1 (108–109 cfu mL−1) approximately. The number of viable cells was determined following the methodology described by Somasegaran and Hoben (1994). The cultures were centrifuged at 4000g for 12 min at room temperature and the cells were suspended in 0.85% NaCl sterile solution. For co-inoculation treatments, mix inoculums were prepared in a 1:1 ratio.
2.6 Phytopathogen inoculum preparation
Wet wheat seeds contained in a 50 mL Erlenmeyer flask were autoclaved and then infected with 5 mm diameter S. rolfsii mycelia plugs. The Erlenmeyer was maintained at room temperature until abundant mycelium growth was observed (7–10 days approximately) (Grupta et al. 2002).
2.7 Bioassays to evaluate the induction of systemic resistance
To avoid direct contact between the bacteria and the phytopathogen, the root system of 12-day-old plants growing in pots with quartz sand was separated (without cutting) using the split root system methodology (Tonelli et al. 2011). Each root part was placed in 100 mL glass tubes containing agarized (0.6%) Hoagland medium (Hoagland and Arnon 1950). One of the root parts was inoculated with 3 mL (109 cfu mL−1) of each strain culture (Bradyrhizobium sp. SEMIA 6144 or Bacillus sp. CHEP5) or with a mixture (1:1) of both strains. The root part inoculated with the bradyrhizobial strain was growing in nitrogen-free agarized Hoagland medium. A week later, the other root part was challenged with the pathogen by adding one wheat seed infected with S. rolfsii mycelium (20 mg). Non-pathogenized and non-bacterized control plants were also included. The activity of the enzyme PAL was determined in plants at 24 h and 30 days post-pathogen challenge. At this last time, disease symptoms were recorded and total chlorophyll content and shoot and root dry weights were also analysed.
The experiment was repeated four times with 4 replicates for each treatment.
2.8 BOX –PCR analysis
The fingerprinting analysis was performed in order to confirm that there was no direct contact between Bacillus sp. CHEP5 and S. rolfsii in plants inoculated with the PGPB and challenged with the phytopathogen. Bacteria were isolated from the root half that was challenged with S. rolfsii, as described by Tonelli et al. (2010). Approximately 10–12 colonies from YEMA or TSA plates were selected to obtain DNA template. Total genomic DNA was extracted with Illustra bacteria genomicPrep Mini Spin KitA (GE Healthcare Life Sciences, UK) according to the manufacturer’s protocol. DNA concentration of the samples was approximately 5 ng μL−1. The DNA sequences of BOX primer BOX-AR1 5′-CTACGGCAAGGCGACGCTGACG- 3′ used in this study was described by Versalovic et al. (1994). The BOX-PCR was performed in 25 μL reaction mixture containing 10× PCR buffer, 50 mM MgCl2, 2 mM DNTPs, 50 pmol mL−1 of primer, 1 U of Taq DNA Polymerase (Promega, USA) and 6 μL of DNA template solution. The temperature profile was as follows: initial denaturation at 95°C for 7 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min, extension at 65°C for 8 min and a final extension step at 65°C for 16 min.
PCRs were performed in a Mastercycler gradient block (Eppendorf, Germany). The BOX amplification products in 12 μL sub-samples were separated according to molecular size by horizontal electrophoresis on 1.5% (w/v) agarose gels stained with SYBR Green.
2.9 Determination of PAL (EC 4.3.1.5) activity
Peanut leaves (0.1 g) were homogenized with liquid nitrogen using a mortar and pestle containing appropriate buffer solution (50 mM potassium phosphate and 1 mM EDTA, pH 7.8) and 1% PVP (polyvinylpyrrolidone). The tissue extract was centrifuged at 12000g for 20 min at 4°C. The supernatant was stored at −20°C to be used for enzymatic activity determination.
The protein concentration of the extracts was determined by the method described by Bradford (1976), using bovine albumin (1mg mL−1) as standard.
PAL activity was assayed following the method described by Paynet et al. (1971) by measuring the amount of trans-cinnamic acid formed at 290 nm. The reaction mixture consisted of 100 μL of enzyme extract, 900 μL 6 mM of l-phenylalanine and 500 mM Tris HCl buffer solution (pH 8). The mixture was placed in a water bath at 37°C for 70 min and the reaction was stopped by the addition of 50 μL of 5 N HCl. Trans-cinnamic acid (1 mg ml−1) was used as standard and PAL activity was expressed as ‘μg trans-cinnamic acid min−1 mg protein−1’.
2.10 Total chlorophyll determination
The amount of total chlorophyll was determined by the method described by Arnon (1949). Briefly, 0.1 g of fresh weight peanut leaves was placed in a mortar and the tissue was ground to fine pulp after the addition of 80% acetone. The extract was transfered to a Buchner funnel containing a pad of Whatman filter paper. While filtering the extract, the gridding of the leaves pulp was repeated to adjust the final volume of the filtrate to 10 mL. The chlorophyll content was spectrophotometically determined at 650 and 665 nm. The amount of total chlorophyll was calculated on the basis of μg of chlorophyll per gram of fresh leaf tissue, according to the following equation (Mc Kinney 1938):
2.11 Nodulation tests
Peanut seedlings obtained as described above were sown in sterilized plastic cups filled with quartz sand. Ten-day-old plants were inoculated at the junction between stems and roots (crown) with 4 mL of bacterial culture (109 cfu mL−1) or with the same volume of a mixture (1:1) of Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144. At this time they were also challenged with the pathogen in the plant crown by adding one wheat seed infected with S. rolfsii mycelium (20 mg). Plants were grown under controlled environment, watered regularly and supplied with Hoagland medium (Hoagland and Arnon 1950) once a week. Non-pathogenized and non-bacterized control plants were also included.
The plants were harvested 30 days after inoculation and the number of nodules was determined.
The experiment was repeated four times with 4 replicates for each treatment.
2.12 Statistical analysis
Statistical analysis was performed by subjecting the data to analysis of ANOVA. Statistical significance was determined by the Tukey and LSD tests at p<0.05, using Infostat software (1.0, FCA, UNC, Argentina).
3 Results
3.1 Growth compatibility of Bacillus sp. CHEP5 with Bradyrhizobium sp. SEMIA 6144
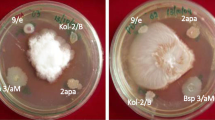
In order to determine whether there is antagonism between Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144, they were streaked at the same time or sequentially on the same Petri plate. Both strains grew optimally under all the conditions evaluated (data not shown), suggesting that there is no antagonistic effect between them.
3.2 Evaluation of the ISR response in peanut plants inoculated with Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144
ISR response was evaluated by using the split root system. Notably, nodules were not formed in roots growing in nitrogen-free medium and inoculated with Bradyrhizobium sp SEMIA 6144. Therefore, the assay was repeated and the plants were grown in Hoagland medium supplemented with nitrogen. Inoculation with each strain or with their mixture reduced disease severity, as the shoot and root dry weights from challenged plants previously inoculated with Bacillus sp. CHEP5, Bradyrhizobium sp. SEMIA 6144 or their mixture were greater than the weights of S. rolfsii -challenged uninoculated plants (table 1). Moreover, the chlorophyll content of challenged plants inoculated with Bacillus sp. CHEP5 and the mixture of the PGPB was higher than the chlorophyll content of uninoculated challenged plants (table 2).
The fingerprinting analysis done with bacterial genomic DNA obtained from the root part challenged with S. rolfsii did not show the typical BOX-profile of Bacillus sp. CHEP5 (figure 1). This result indicated the absence of direct contact between S. rolfsii and Bacillus sp. CHEP5 in plants inoculated with the bacterium before the phytopathogen challenge. Therefore, we confirmed that there was no direct antagonism between the biocontrol bacterium and the phytopathogen and that Bacillus sp. CHEP5 protection against S. rolfsii was mediated by ISR.
The shoot and root dry weights and the chlorophyll content increased in plants inoculated with Bradyrhizobium sp. SEMIA 6144 compared with uninoculated ones. Considering that these plants were growing in the presence of an available nitrogen source, growth promotion could not be attributed to biological nitrogen fixation. The fact that Bradyrhizobium sp. SEMIA 6144 synthetises IAA-like molecules (data not shown) allows us to speculate that they are involved in the growth promotion of plant inoculated with this bradyrhizobial strain.
3.3 PAL activity in peanut plants inoculated with Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144
Peanut PAL activity determined at 30 days after S. rolfsii challenge was higher than at 24 h post challenge. At both times, a significant increase was observed in plants that had previously been inoculated only with Bacillus sp. CHEP5 or with the bacterial mixture, compared with uninoculated pathogen-challenged plants (figures 2 and 3).
PAL enzyme activity in peanut plants inoculated with Bacillus sp. CHEP5 and/or Bradyrhizobium sp. SEMIA 6144, 24 h after S. rolfsii challenge. Values are the mean ± SE (n=16). Different letters indicate significant differences according to the Tukey test (p<0.05). The experiment was repeated four times with 4 replicates for each treatment.
PAL enzyme activity in peanut plants inoculated with Bacillus sp. CHEP5 and/or Bradyrhizobium sp. SEMIA 6144, 30 days after S. rolfsii challenge. Values are the mean ± SE (n=16). Different letters indicate significant differences according to the Tukey test (p<0.05). The experiment was repeated four times with 4 replicates for each treatment.
3.4 Bradyrhizobium sp. SEMIA 6144 symbiotic behaviour in the presence of Bacillus sp. CHEP5 and S. rolfsii
The number of nodules formed was unchanged in coinoculated plants compared with plants inoculated only with Bradyrhizobium sp. SEMIA 6144. As expected, the number of nodules decreased dramatically when the plants were challenged with S. rolfsii. Interestingly, the inoculation with Bacillus sp. CHEP5 reverted this effect, as the number of nodules induced in co-inoculated plants was higher than that obtained in challenged plants inoculated with Bradyrhizobium sp. SEMIA 6144 (table 3).
4 Discussion
We have previously reported that Bradyrhizobium sp. SEMIA 6144 is an effective peanut nodulating strain that promotes plant growth by fixing nitrogen in peanut and that Bacillus sp. CHEP5 induces systemic resistance in this legume (Fabra et al. 2010, Tonelli et al. 2011). Therefore, it becomes relevant to evaluate the impact of the co-inoculation of Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA 6144 on the ISR and the symbiotic interaction in peanut plants.
To attribute a biocontrol agent the ability to induce systemic resistance against phytopathogens, it is important to inoculate the bacterium and the phytopathogen separately in space and time (Van Loon et al. 1998). In this work we used the split root system methodology, since it guarantees the physical separation between the bacteria and the rhizoplane phytopathogen S. rolfsii, as confirmed by the BOX-PCR analysis of the gram-positive bacteria isolated from the root half inoculated only with the fungus.
The evaluation of biocontrol activity mediated by ISR not only involves recording disease incidence and severity but also pathogenesis-related enzymes activity usually determined as ISR markers (Heil and Bostock 2002). Increased activity and accumulation of these proteins depend mainly on the biocontrol agent and the phytopathogen species, as well as on the plant genotype and growth conditions (Madhaiyan et al. 2006). Some of these proteins are involved in the fungal cell wall degradation (Van Loon et al. 2006), but other enzymes associated with ISR such as PAL are related to phytoalexin biosynthesis (Compant et al. 2005). In peanut plants inoculated with Bacillus sp. CHEP5, we have previously determined a significant increment in PAL activity 15 days after S. rolfsii challenge (Tonelli et al. 2011), and an increase in the AhPAL relative transcript amount after 24 h S. rolfsii inoculation (Tonelli et al. 2013). Therefore, in this work PAL activity was evaluated at this last time and also when S.rolfsii-challenged plants showed significant wilt symptoms (30 days after pathogen inoculation).
Since PAL activity was higher in plants inoculated with the biocontrol agent, as compared with those that were challenged only with S. rolfsii, we assume that the plant defence response was primed to be potentiated towards the pathogen infection. Moreover, at 24 h post-pathogen challenge a significant increment of PAL activity was registered in peanut plants inoculated both with Bacillus sp. CHEP5 and with the bacterial mixture. In agreement with our results, Madhaiyan et al. (2006) and Podile and Laxmi (1998) also reported a fast accumulation of PAL enzyme after pathogen challenge in leguminous plants.The fact that PAL activity in peanut plants is increased at short times allows us to assume that this is an early event in the priming phenomenon that characterizes the ISR-mediated protection.
It was interesting to find that, at 24 h post-pathogen challenge, PAL activity was lower in plants inoculated with the mixture of the PGPB than in plants inoculated only with Bacillus sp. CHEP5. The establishment of rhizobia requires mutual recognition with the plant. During the first stages of establishment, rhizobia are initially recognized as potential invaders. In fact, it is known that there are many similarities between the early legume responses to infection by pathogenic and symbiotic organisms (Santos et al. 2001). However, at later stages of the interaction, they are able to modulate plant defence responses to enable successful colonization of host roots (Zaminoudis and Pieterse 2012). Therefore, we suggest that at first stages of the interaction with the plant, Bradyrhizobium sp. SEMIA 6144 modulated the defence response and this could diminish PAL activity even when Bacillus sp. CHEP5 was present. However, the increase in PAL activity in plants inoculated with the mixture of the PGPB was restored at 30 days post-pathogen challenge. Although finding reasons for this response is beyond the scope of this work, it became evident that in the rhizosphere the different populations of the microbial consortium and the plant roots detect each other and respond to each other’s presence.
At 30 days post-pathogen challenge, PAL activity in plants inoculated with Bradyrhizobium sp. SEMIA 6144 reached values similar to those in plants inoculated with the bacterial mixture. The fact that PAL activity was higher in plants inoculated with the PGPB as compared with pathogen-challenged plants confirms that the presence of Bradyrhizobium sp. SEMIA 6144 did not affect the ability of Bacillus sp. CHEP5 to induce ISR against S. rolfsii.
Comparison between shoot and root dry weights and chlorophyll content from challenged-PGPB inoculated plants and from challenged plants reveals that the inoculation with one or both bacteria reduced wilt severity and improved growth and physiological state of pathogen-treated peanut plants. Considering that Bradyrhizobium sp. SEMIA 6144 was not able to induce nodule formation in the system used in this study (split root), plant growth promotion cannot be attributed to nitrogen fixation, but to other growth promoting mechanisms such as IAA production. It has been demonstrated that increased root proliferation, related to bacterial IAA biosynthesis, enhances plant mineral uptake (Spaepen et al. 2007). This could place the plant in a better fitness state and condition to face the phytopathogen challenge and, as a consequence, diminish the pathogen’s deleterious effect. In this study, the root dry weight of Bradyrhizobium sp. SEMIA 6144-inoculated plants was higher compared to uninoculated ones.
In addition to enhanced total chlorophyll content and PAL activity, the increase in shoot and root dry weight of co-inoculated and pathogen-challenged peanut plants suggests that Bradyrhizobium SEMIA sp. 6144 does not affect the ability of Bacillus sp. CHEP5 to induce systemic resistance against S. rolfsii.
To assess whether Bacillus sp. CHEP5 affects the capacity of Bradyrhizobium sp. SEMIA 6144 to nodulate S. rolfsii-challenged peanut plants, a nodulation assay was done in the presence of the biocontrol agent. Bacillus sp. CHEP5 inoculation reversed the harmful effect of S. rolfsii on the number of nodules formed, indicating that the symbiotic interaction in pathogen-challenged plants is not affected and, in fact, is improved by the presence of Bacillus sp. CHEP5. Similar results have been recently reported in peanut plants inoculated with Bradyrhizobium spp. and the biocontrol agents Serratia marcescens and/or Trichoderma harzianum (Badawi et al. 2011).
In summary, data obtained in this work indicate that the presence of Bradyrhizobium sp. SEMIA 6144 does not affect Bacillus sp. CHEP5 ISR-mediated protection of peanut plants against S. rolfsii. Moreover, the inoculation of both PGPB strains, in the presence or absence of S. rolfsii, promotes plant growth and increases the number of nodules formed by Bradyrhizobium sp. SEMIA 6144. It is possible that, as has been proposed, the biocontrol agent alters host plant metabolism and/or produces antimicrobial compounds that favour competition abilities of the rhizobial strain (Podile and Kishore 2006; Verma et al. 2010).
Living in a community is thought to generate robustness to environmental stresses and to promote stability for the members of a consortium over time. Communities might be more capable of resisting invasion by other species than monocultures (Brenner et al. 2008). The results obtained in this work demonstrated that microbial consortia constituted by PGPB may outperform the beneficial effects achieved by each strain (pure cultures). This is an attractive trait that can be used to increase peanut plant production.
References
Arnon DI 1949 Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15
Badawi FSF, Biomy AMM and Desoky AH 2011 Peanut plant growth and yield as influenced by co-inoculation with Bradyrhizobium and some rhizo-microorganisms under sandy loam soil conditions. Ann. Agric. Sci. 56 1–9
Bradford M 1976 A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72 248–254
Brenner K, You L and Arnold FH 2008 Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 26 483–489
Bric JM, Bostock RM and Silverstone SE 1991 Rapid in situ assay for indolacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 5 535–538
Compant S, Duffy B, Nowak J, Clément C and Ait Barka E 2005 Minireview: Use of plant growth promoting bacteria for biocontrol of plant disease: principles, mechanisms of action and future prospects. Appl. Environ. Microbiol. 71 4951–4959
Conrath U, Beckers G, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, et al. 2006 Priming: getting ready for battle. Mol. Plant Microbe. Interact. 19 1062–1071
Fabra A, Castro S, Taurian T, Angelini J, Ibañez F, Dardanelli M, Tonelli ML, Bianucci E, et al. 2010 Interaction among Arachis hypogaea L. (peanut) and beneficial soil microorganisms: how much is it known? Crit. Rev. Microbiol. 36 179–194
Gordon SA and Weber RP 1951 Colorimetric estimation of indolacetic acid. Plant Physiol. 26 192–195
Grupta CP, Dubey RC and Maheshwari DK 2002 Plant growth enhancement and suppression of Macrophomina phaseolina causing charcoal rot of peanut by fluorescent Pseudomonas. Biol. Fertil. Soils 35 399–405
Heil M and Bostock R 2002 Induced systemic resistance (ISR) against pathogens in the context of induced plant defenses. Ann. Bot. 89 503–512
Hoagland DR and Arnon DI 1950 The water culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 347 1–39
Kloepper J, Tuzun S and Kúc J 1992 Proposed definitions related to induced disease resistance. Biocontrol. Sci. Technol. 2 349–351
Lugtenberg B and Kamilova F 2009 Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 63 541–556
Madhaiyan M, Suresh Reddy BV, Anandham R, Senthilkumar M, Poonguzhali S, Sundaram SP and Sa T 2006 Plant-growth promoting Methylobacterium induces defence responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr. Microbiol. 53 270–276
Mc Kinney G 1938 Some absorption spectra of leaf extract. Plant Physiol. 13 128–140
Paré PW, Farag MA, Krishnamachari V, Zhang H, Ryu CM and Kloepper JW 2005 Elicitors and priming agents initiate plant defense responses. Photosynth. Res. 85 149–159
Paynet M, Martin C and Girand M 1971 Activité phenylalanine ammonia lyase et hypersensibilite au virus de la mosaique du tabac. Académie des Sciences, Paris 273 537–539
Pieterse C, Van Pelt J, Ton J, Parchmann S, Mueller M, Buchala A, Meâ Traux JP and Van Loon L 2000 Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis thaliana requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57 123–134
Podile AR and Kishore K 2006 Plant growth-promoting rhizabacteria; in Plant-associated bacteria (ed) SS Gnanamanickam (Springer) pp 195–230
Podile AR and Laxmi VDV 1998 Seed bacterization with Bacillus subtilis AF 1 increases phenylalanine ammonia-lyase and reduces the incidence of fusarial wilt in Pigeonpea. J. Phytopathol. 146 255–259
Pozo MJ, Van Der Ent S, Van Loon LC and Pieterse CM 2008 Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance en Arabidopsis thaliana. New Phytol. 180 511–523
Reymond P and Farmer E 1998 Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 5 404–411
Somasegaran P and Hoben H 1994 Quantifying the growth of rhizobia; in Handbook for rhizobia: Methods in legume rhizobia technology (New York: Springer) Section 3, pp 382–390
Santos R, Herouart D, Sigaud S, Touati D and Puppo A 2001 Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14 86–89
Spaepen S, Vanderleyden J and Remans R 2007 Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31 425–448
Tonelli ML, Taurian T, Ibáñez F, Angelini J and Fabra A 2010 Selection and in vitro characterization of biocontrol agents to protect peanut plants against fungal pathogens. J. Plant Pathol. 92 73–82
Tonelli ML, Furlán A, Taurian T, Castro S and Fabra A 2011 Peanut priming induced by biocontrol agents. Physiol. Mol. Plant Pathol. 75 100–105
Tonelli ML, Ibañez F, Taurian T, Argüello J and Fabra A 2013 Analysis of a phenylalanine ammonia-lyase gene sequence from Arachis hypogaea L. and its transcript abundance in induced systemic resistance against Sclerotium rolfsii. J. Plant Pathol. 95 191–195
Van Loon LC, Bakker PHM and Pieterse JCM 1998 Systemic Resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36 453–483
Van Loon L, Rep M and Pieterse C 2006 Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44 135–162
Verhagen BWM, Glazebrook J, Zhu T, Chang HS, Van Loon LC and Pieterse CMJ 2004 The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant Microbe. Interact. 17 895–908
Verma JP, Yadav J, Tiwari KN and Lavakush Singh V 2010 Impact of plant growth promoting rhizobacteria on crop production. Int. J. Agricul. Res. 5 954–983
Versalovic J, Schneider M, de Bruijn F and Lupski J 1994 Genomic fingerprint of bacteria using repetitive secuence based PCR (rep-PCR). Meth. Mol. Cell. Biol. 5 25–30
Vincent JM 1970 A manual for the practical study of root nodule bacteria; in International Biological Programme Handbook n° 15 (Oxford: Blackwell Scientific Publications Ltd.) pp 73–97
Zamioudis C and Pieterse CMJ 2012 Modulation of host immunity by beneficial microbes. MPMI 25 139–150
Acknowledgements
This study was financially supported by the SECyT-UNRC, CONICET, Ministerio de Ciencia y Tecnología de Córdoba, ANPCyT and Fundación Maní Argentino grants. The authors are grateful to Verónica L Muñoz, MA in Applied Linguistics, for editing the language aspects of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: DURGADAS P KASBEKAR
MS received 04 March 2014; accepted 05 August 2014
Corresponding editor: Durgadas P Kasbekar
[Figueredo MS, Tonelli ML, Taurian T, Angelini J, Ibañez F, Valetti L, Muñoz V, Anzuay MS, Ludueña L and Fabra A 2014 Interrelationships between Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA6144 in the induced systemic resistance against Sclerotium rolfsii and symbiosis on peanut plants. J. Biosci. 39 1–9] DOI 10.1007/s12038-014-9470-8
Rights and permissions
About this article
Cite this article
Figueredo, M.S., Tonelli, M.L., Taurian, T. et al. Interrelationships between Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA6144 in the induced systemic resistance against Sclerotium rolfsii and symbiosis on peanut plants. J Biosci 39, 877–885 (2014). https://doi.org/10.1007/s12038-014-9470-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12038-014-9470-8