Abstract
Extracellular vesicle (EV)–encapsulated circRNAs have the potential role in affecting brain disorders. However, the role of circ_0000075 in cerebral ischemic injury remains unclear. Here, we tried to investigate the mechanism of bone marrow mesenchymal stem cell (BMSC)–derived EVs carrying circ_0000075 in the control of cerebral ischemic injury. Initially, a mouse model with cerebral ischemic injury was induced by middle cerebral artery occlusion (MCAO), followed by the determination of circ_0000075 expression. Then, neurons were isolated and subjected to oxygen–glucose deprivation/reperfusion. BMSCs were isolated for extraction of EVs. The correlation among circ_0000075, microRNA (miR)-218-5p, and Smad ubiquitination regulatory factor 2 (SMURF2) was detected with their roles in cerebral ischemic injury analyzed in vivo and in vitro. circ_0000075 was down-regulated in MCAO mice and engineered RVG-EVs were internalized by neurons to up-regulate circ_0000075 expression. Treatment of RVG-circ_0000075-EVs reduced brain tissue damage, increased neuronal count, and significantly curtailed apoptosis rate, suppressing cerebral ischemic injury in vitro and in vivo. miR-218-5p was targeted by circ_0000075 in neurons, which promoted SMURF2 expression. A negative correlation between SMURF2 and transcriptional regulator Yin Yang 1 (YY1) was identified. In vitro experiments further proved that circ_ 00,000 75 could down-regulate the expression of YY1 through SMURF2, and finally relieving cerebral ischemic injury. Collectively, engineered EVs delivered circ_0000075 into brain tissues and increased circ_0000075 expression, which down-regulated miR-218-5p and up-regulated SMURF2, thus alleviating cerebral ischemic injury.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ischemic stroke is caused by hypoxic-ischemic encephalopathy and acute cerebrovascular accident and can contribute to damages in cognitive and motor function, neurodegenerative disorders, and even acute death [1]. During cerebral ischemia, disturbances in cerebral blood flow occur, which may induce cell necrosis and apoptosis [2]. It has been reported that cerebral ischemia is the leading cause of death and disability in the world and cerebral ischemic injury shows a great correlation to neuron death [3]. Moreover, neuroprotection is one of the main therapeutic strategies currently adopted to treat the cerebral ischemic injury, but most neuroprotective agents result in poor efficacy or severe toxicity/side effects [4]. Therefore, exploration of the molecular mechanism underlying neuroprotection in cerebral ischemic injury is necessitated for developing new therapeutic agents.
Mesenchymal stem cells (MSCs) possess potential therapeutic effects on a paracrine mode through extracellular vesicles (EVs), which might be an alternative to cell therapy [5]. Of note, the systematical administration of MSC-EVs is of therapeutic efficacy on hypoxia–ischemia-induced injury [6]. EVs carry cargo proteins, nucleic acids (RNA and DNA), and metabolites [7], while circular RNAs (circRNAs) are newly investigated noncoding RNAs, which are abundant in the brain, so it is inferred that circRNAs may be engaged in the processes of cerebral physiology and pathology [8]. Although hsa_circ_0000075 participates in the atrial fibrillation pathogenesis [9], its correlation with cerebral ischemic injury is largely unknown. It has been shown that microRNA (miR)-218-5p expression is elevated in the plasma of patients with acute ischemic stroke and that down-regulation of miR-218-5p significantly inhibits the release of inflammatory cytokines and may serve as a novel biomarker of acute ischemic stroke progression [10, 11], which indicated a possible relationship between miR-218-5p and cerebral ischemic injury. Additionally, the correlation between miR-218-5p and nerve injury has been mentioned recently, as evidenced by the elevation of miR-218-5p is able to reduce the damage of dopaminergic neurons in rats with Parkinson’s disease [12]. In further exploration, another target, SMURF2, caught our attention. SMURF2 has been found to contribute to recovery in patients with stroke [13]. Importantly, SMURF2 regulates the EZH2/AKT/GSK3β signaling pathway and plays a protective role in ischemic reperfusion brain injury [14]. Moreover, SMURF2 has been shown to degrade YY1 transcription factor (YY1) and inhibit its protein expression through its E3 ubiquitin ligase function [15] while YY1 can promote the development of cerebral ischemic injury [16]. However, the mechanism by which BMSC-EVs play a neuroprotective role through the circ_0000075/miR-218-5p/SMURF2 axis has not been fully elucidated. Therefore, this study aims to investigate the mechanism by which BMSC-EVs alleviated cerebral ischemic injury by regulating the circ_0000075/miR-218-5p/SMURF2 axis both in vitro and in vivo in an attempt to provide a better option for the treatment of cerebral ischemic injury in the future.
Materials and Methods
Experimental Animals
This study was ratified by the Animal Ethics Committee of the First Affiliated Hospital of Nanchang University and all procedures were carried out per the Guide for the Care and Use of Laboratory Animals. Adult male C57BL/6 J mice (8–10 weeks old; weighing 22–25 g) were procured from the Institute of Laboratory Animal Research, Chinese Academy of Medical Sciences (China) and housed in a temperature-controlled room under a 12 h light/dark cycle.
Establishment of Middle Cerebral Occlusion (MCAO) Mouse Model
Under isoflurane anesthesia, focal ischemia for 45 min was induced by the MCAO method using a silicone-coated monofilament nylon thread (SMC23B104PK100, RWD Life Science, Shenzhen, China) [17, 18]. During surgery, the rectal temperature was kept at 37 ± 0.5 °C using a heating pad. Sham-operated mice were anesthetized and operated in the absence of MCAO. All mice were placed in postoperative cages and kept warm and undisturbed for at least 2 h for observation.
Assessment of Neurological Function
The foot fault test was performed to assess motor function and the rota-rod test was performed to assess balance and coordination. The detailed experimental procedures were implemented based on previously published methods [19].
In the adhesive removal test, the tape adhered to the palmar surface of the forepaw and the time required to touch and remove the tape on the first attempt was recorded.
Additionally, the Morris water maze (MWM) test was performed with a 120-cm-diameter pool [20]. Tests were implemented on non-consecutive days to reduce possible confusion due to motor learning.
RNA Extraction and Sequencing
Mice were dissected, and the infarct area of modeled mice and the area corresponding to the infarct in sham-operated mice were dissected to collect the brain tissues. Total RNA was isolated utilizing Trizol reagent (15,596,026, Invitrogen, Thermo Fisher Scientific, Waltham, MA). RNA sample concentrations were measured by OD260/280 employing a Nanodrop ND-1000 spectrophotometer (1011U, Nanodrop, Thermo Fisher Scientific). RNA concentrations were assayed using the Qubit RNA Analysis Kit. Total RNA samples meeting the requirements as RNA integrity index (RIN) ≥ 7.0 and 28S:18S ratio ≥ 1.5 were used for subsequent experimentations.
Sequencing libraries were prepared and sequenced by CapitalBio Technology (Beijing, China). About 5 μg RNA was used per sample. Briefly, ribosomal RNA (rRNA) was eliminated from total RNA with the help of the Ribo-Zero™ magnetic kit (MRZE706, Epicentre Technologies, Madison, WI). For the removal of linear RNA, total RNA was detached with RNase R (Epicentre Technologies). The NEB Next Ultra RNA Library Preparation Kit (#E7775, NEB, Ipswich, MA) for Illumina was adopted for the construction of libraries for sequencing. The RNA was then fragmented into fragments of approximately 300 base pairs (bp) in length in NEB Next First Strand Synthesis Reaction Buffer (5 ×). The first strand complementary DNA (cDNA) was synthesized using reverse transcriptase primers and random primers, and the synthesis of the second strand cDNA was completed in Second Strand Synthesis Reaction Buffer in dUTP Mix (10 ×). cDNA fragments were end-repaired, including the addition of ployA tails and the ligation of sequencing junctions. After ligation of the Illumina sequencing junction, the second strand of cDNA was detached using USER Enzyme (#M5508, NEB) to construct a strand-specific library. The library DNA was amplified, while the library was purified and enriched by polymerase chain reaction (PCR). Libraries were then identified by Agilent 2100 and quantified using the KAPA library quantification kit (KK4844, KAPA Biosystems). Finally, paired-end sequencing was processed on an Illumina NextSeqCN500 sequencer.
Quality Control and Comparison
The quality of paired-end reads of raw sequencing data was checked using FastQC software v0.11.8. Pre-processing of raw data was performed using Cutadapt software 1.18, where the Illumina sequencing junctions and poly(A) tail sequences were removed. Reads with more than 5% N content were removed by Perl scripts, while 70% of reads with base mass above 20 were extracted using FASTX Toolkit software 0.0.13. Paired-end sequences were repaired using BBMap software. Finally, the filtered fragments of high-quality reads were compared to the mouse reference genome (mm9) by bwa software (0.7.12) [21].
Bioinformatics Analysis
The CIRI software was used to identify reverse splicing events, and sequencing data that could not be directly compared to the reference genome were used for subsequent circRNA analysis. The identified circRNAs were annotated via the circRNA annotation file on the circBase website. Differential analysis of circRNAs was performed using the R software limma package (v3.32.10), where the screening threshold for significant differentially expressed circRNA screening was |logFoldChange|> 1 and p value < 0.05. The miRanda algorithm, the starBase (v2.0) database, and MicroT-CDS database were applied to predict the miRNAs interacting with circRNAs and the downstream target genes of miRNAs. The target binding sites were analyzed through the starbase database and the RNA22 database. GSE84216, a miRNA sequencing dataset, and GSE16561, a gene microarray dataset, associated with stroke, were then obtained through the GEO database, and used to validate the downstream regulatory genes of circRNAs and the trends of their impact factors in stroke samples. DisGeNET database was introduced to retrieve cerebral ischemic stroke–related genes.
Cell Processing and Treatment
Primary hippocampal neurons were isolated from mouse embryos at embryonic day 17.5. Hippocampal bodies were dissected and cut into pieces. The tissues were detached with 0.25% EDTA-free trypsin at 37 °C for 20 min and gentle shook at 5-min intervals for removal of tissue fragments. Then, cells were suspended in DMEM replenished with 20% FBS and seeded in culture dishes pre-coated with poly-L-lysine (50 μg/mL, Sigma-Aldrich) at 37 °C for 4 h. After adhesion, neurons were washed with PBS to eliminate FBS and incubated in a neural basal medium appended to 1% Glutamax and 2% B27 (Gibco) at 37 °C in a 5% CO2 humidified chamber. Neurons were identified by immunofluorescence detection of MAP2 using rabbit anti-MAP2 (A17409, 1: 200, Abclonal, Woburn, MA) expression.
Lentivirus harboring circ_0000075 or short hairpin RNA (shRNA) against circ_0000075 (sh-circ_0000075), GNSTM-RVG-lysosome-associated membrane glycoprotein 2b (Lamp2b)-HA lentiviral plasmid, miR-218-5p mimic, SMURF2 overexpression plasmid, and SMURF2 knockdown plasmid were all provided by RiboBio (Guangzhou, China). Interference sequence was designed specifically for reverse splice site and lentivirus harboring sh-circ_0000075 was therefore constructed [22]. The detailed grouping is displayed in Table S1.
Neurons or 293 T cells (CRL-3216, ATCC) were transfected with miR-218-5p mimic or infected with circ_0000075 overexpressing lentiviral plasmids following the instructions of Lipofectamine 3000 (L3000001, Thermo). The detailed grouping is displayed in Table S1.
Cell Viability Assay
Primary neuronal death was assessed by lactate dehydrogenase (LDH) release in the culture medium. The release of LDH in the supernatant of cultured cells was tested employing a LDH assay kit (Beyotime). The percentage of cell death was calculated: cytotoxicity (%) = LDH release (OD492)/maximum (OD492). Cell viability was quantified utilizing a cell counting kit (CCK-8; Dojindo, Japan). A total of 1 × 105 neurons were seeded in 96-well plates, treated, and then incubated with CCK-8 reagent at 37 °C for 3 h. OD450 values following the different treatment protocols were measured using a Bio-Rad680 microplate meter (Bio-Rad680, Bio-Rad, Hercules, CA).
RT-qPCR
Total RNA was extracted from EVs using the Exosomal RNA Isolation Kit (NGB-58000, Norgen Biotek), while the total RNA was extracted from tissues and cells using the MiRneasy kit (QIAGEN, Germantown, MD). For miRNA detection, miRNA cDNA containing PolyA tail was obtained by means of PolyA tailing reaction detection kit (B532451, Sangon, Shanghai, China), followed by RT-qPCR using miRcute miRNA qPCR Detection (FP401, ANGEN Biotech, Beijing, China). circRNAs and mRNAs were reversely transcribed using HiScript Q RT SuperMix (R123-01, Vazyme, Nanjing, China), followed by RT-qPCR using AceQ qPCR SYBR Green Master Mix (Q141-02, Vazyme). U6 was served as the normalizer for miRNA while results of circRNAs and mRNAs were normalized to β-actin. Data analysis was conducted using 2∆∆Ct and the primer sequences are exhibited in Table S2.
Fluorescence In SituHybridization (FISH)
A 6-FAM-labelled specific circ_0000075 FISH probe (GeneSeed, Guangzhou, China) was designed and used for the experiment. Adherent cells were fixed in 4% PFA, washed with PBS and then detached with proteinase K (Sangon, Shanghai, China) for 5 min. After PBS washing, cells were fixed in 1% PFA and then serially dehydrated in 70%, 85%, and 100% alcohol. Dilute probe drops of hybridization solution were placed on cell slides, and the cells were denatured at 73 °C for 3 min and hybridized overnight in the dark at 42 °C. A total of 50% formamide/2 × SSC was pre-warmed to 43 °C while 0.1% NP40/2 × SSC was pre-warmed to 37 °C. The sections were stained by DAPI and images were acquired with the help of a confocal laser scanning microscope (LSM 510 META, Carl Zeiss AG).
Western Blot Analysis
Cellular proteins were collected and lysed in radio immunoprecipitation assay (RIPA; P0013B, Beyotime) containing a protease inhibitor (A8260, Solarbio). Protein quantification was performed by bicinchoninic acid assay method (AR0197, Boster, Wuhan, China). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to the polyvinylidene fluoride membranes, which were blocked with 5% skimmed milk powder for 1 h. The membranes were incubated with diluted primary antibodies against Lamp2b (ab18529, 1: 1000, Abcam), CD63 (ab217345, 1: 1000, Abcam), GM130 (ab52649, 1: 2000, Abcam), Alix (ab88388, 1: 1000, Abcam), SMURF2 (ab53316, 1: 1000, Abcam), YY1 (A12928, 1: 1000, Abclonal), and β-actin (AC026, 1: 50,000, Abclonal, internal reference). The following day, the membranes were incubated with secondary antibodies of peroxidase-conjugated AffiniPure Goat Anti-Rabbit Immunoglobulin G (IgG) (H + L) (#111,035,003, Jackson ImmunoResearch, West Grove, PA) for 1 h. The membranes were exposed to luminescent solution (Thermo Fisher Scientific) followed by color development. Analysis was performed using Image J software.
Dual-Luciferase Reporter Gene Assay
The correctly constructed circ_0000075 wild-type (WT) and circ_0000075 mutant-type (MUT) reporter plasmids were co-transfected with miR-218-5p mimic or mimic-NC into 293 T cells using Lipofectamine 3000. After 24 h, the luciferase activity was measured using a dual luciferase reporter assay system (E1910, Promega). The binding capacity of miR-218-5p to SMURF2 was assayed using the same method. miR-218-5p mimic or mimic-NC was transfected with the luciferase reporter plasmid pGL3-Luciferase Reporter vector containing the WT SMURF2-3'UTR or MUT SMURF2-3'UTR into 293 T cells.
RNA Pull Down
Biotinylated miR-218-5p mimic RNA or control RNA (RiboBio) was transfected with cells at a final concentration of 50 nM, and cells were collected after 48 h, followed by the addition of 0.7 mL lysis buffer and 10-min incubation on ice. Cell lysis buffer was incubated with streptavidin-coated magnetic beads (Life Technologies, Carlsbad, CA) for 10 min at 10,000 × g to pull down the biotin-coupled RNA complexes. The abundance of circ_0000075 and SMURF2 in the binding region was detected by RT-qPCR.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
Cells were lysed with RIPA lysis buffer and RIP assay was performed with the Magna RIP™ Kit (Millipore, Billerica, MA). Argonaute2 antibody (Anti-Ago2) (ab186733, 1: 50, Abcam) and normal IgG antibody (Anti-IgG) (ab172730, 1: 100, Abcam) were incubated with magnetic beads (Millipore) at 4 °C for 1 h. Then, a portion of the cell lysis buffer was used as a NC (Input) and another portion of the cell lysis buffer was incubated with magnetic beads at 4 °C overnight. After purification, RT-qPCR was processed to quantify the target binding ability of circ_0000075 to miR-218-5p.
Isolation and Identification of Mouse BMSCs
Mouse BMSCs were isolated and cultured. Specifically, the C57BL/6 mice (6–8 week old) were euthanized, and their hind limbs were removed and rinsed in 70% ethanol, followed by collection of bone marrow. Cell mass was removed using a 70-mm filter. Cells were cultured in Dulbecco’s modified eagle medium (DMEM; Gibco by Life technologies, Grand Island, NY) replenished with 10% FCS penicillin (100 IU/mL; Gibco by Life technologies) and streptomycin (100 µg/mL) at 37 °C in an incubator with 5% CO2. After 3 h, the un-adhered cells were removed and the medium was renewed every 3–4 days. Upon 80% confluence, the mouse BMSCs were passaged and cultured using 0.25% trypsin. The mouse BMSCs at passage 3 were involved in the identification of surface markers.
For identification of surface marker of mouse BMSCs, 1 × 104 cells were incubated with primary antibodies (all from BD Bioscience, San Diego, CA) of PE anti-mouse CD44 (1: 100, 553,134), PE anti-mouse CD29 (1: 100, 562,801), PE anti-mouse CD31 (1: 100, 561,073), and fluorescein isothiocyanate anti-mouse CD45 (1: 100, 553,080) at 4 °C for 30 min. Cells were further incubated for 30 min with specific secondary antibodies protected from light, followed by the determination of the expression of surface markers in mouse BMSCs by FACScan flow cell flow system (Becton Dickinson, San Diego, CA).
Differentiation ability of mouse BMSCs was detected. The mouse BMSCs were cultured in osteogenic differentiation medium (HUXUC-90021, Cyagen) for 14 days, adipogenic differentiation medium (HUXUC-90031, Cyagen) for 21 days, or chondrogenic differentiation medium (HUXUC-9004, Cyagen) for 21 days. Subsequently, the differentiated cells were fixed in 4% paraformaldehyde (PFA), and then identified by alkaline phosphatase staining and 2% Alizarin Red (A5533, Sigma) staining for osteogenic differentiation. Oil Red O staining (O0625, Sigma) was performed for adipogenic differentiation detection, and 1% Alcian Blue solution (A5268, Sigma) for chondrogenic differentiation detection.
Engineered EVs
Mouse BMSCs were infected with GNSTM-RVG-Lamp2b-HA lentiviral plasmid at 60% confluence. After 6 h, cells were infected with circ_0000075 lentiviral plasmid. After 24 h of infection, the double-positive BMSCs were screened. The culture medium for double-positive cells was renewed with EV-free serum culture medium for another 48-h incubation to induce the release of EVs. RVG-EVs enriched with circ_0000075 were harvested from the culture medium.
Isolation and Identification of EVs
BMSCs overexpressing RVG-Lamp2b or circ_0000075 were subjected to differential centrifugation for extraction of circ_0000075-EVs, RVG-circ_0000075-EVs, and RVG-vector-EVs. Cell conditioned medium was collected and centrifuged at 300 × g for 5 min and 2000 × g for 20 min. The conditioned medium was then filtered through a filter (microporous, 0.22 μm) and centrifuged twice at 10,000 × g for 1.5 h. Subsequently, the obtained EVs were resuspended with phosphate buffer saline (PBS), fixed with 1% glutaraldehyde, and then stained with 1% phosphotungstic acid. Morphology of EVs was observed employing a JEM-2100 transmission electron microscope (TEM; JEOL, Tokyo, Japan), while the size distribution and concentration of EVs were analyzed by Zetaview. Western blot analysis was performed to quantify the expression of markers of RVG-Lamp2b-modified EVs: Lamp2b, CD63, Alix, and GM130 (negative marker).
Internalization of EVs
EVs were labeled using the Dil-membrane Evs Labeling Purification Kit (EXOPDIL10-1, RenGenbio, Liaoning, China). Specifically, 5 μL of DiO labeling dye was added to 50 μL of reaction buffer and stirred well until the dye was completely dissolved. Then, 50 μL EVs were added to the dye mixture and incubated for 30 min at 37 °C. A total of 100 µL EVs were carefully added to the top of the elution column and eluted by centrifugation at 50 × g for 90 s. Dil-EVs were co-cultured with neurons for 24 h, washed with PBS and fixed in 4% PFA. Uptake was then observed by fluorescence microscopy. Reverse transcription quantitative PCR (RT-qPCR) was performed after RNase A treatment of BMSC conditioned medium to detect expression of circ_0000075 and verify that circ_0000075 was encapsulated in EVs. To exclude that the expression of circ_0000075 was endogenously induced, the expression of circ_0000075 was quantified by RT-qPCR after neurons were treated with actinomycin D (0.1 μM, Apexbio, Boston, MA).
Animal Model Processing and Treatment
To study the effect of circ_0000075 on cerebral ischemic injury, 2 μL of LV-circ_0000075 was injected into the right ventricle of mice. One day after injection, a MCAO model was established and a series of behavioral tests were carried out to comprehensively assess the dysfunction of the mice before and 28 days after MCAO. Another part of the mice was taken, and brain tissues were dissected at day 7 for quantification of circ_0000075 expression. Mice were treated with LV-vector or LV-circ_0000075.
To determine the targeting effect of RVG-circ_0000075-EVs on brain tissues, Dil-labeled RVG-circ_0000075-EVs (12 mg/kg) were injected into the tail vein and 12 h later, 3D fluorescence imaging was performed to observe the aggregation of EVs and mouse tissues were then taken for immunofluorescence detection. Mice were treated with circ_0000075-EVs or RVG-circ_0000075-EVs.
To study the effect of RVG-circ_0000075-EVs on brain damage repair in MCAO mice, differently treated EVs (RVG-circ_0000075-EVs, RVG-EVs, or circ_0000075-EVs) were injected into mice via tail vein, followed by the establishment of a MCAO model one day later. Then, brain tissues were harvested on day 7 to observe the brain damage. Mice were treated with sham operation, MCAO, RVG-circ_0000075-EVs, or RVG-vector-EVs, with ten mice per treatment.
Pathohistological Observations
Serial frozen sections of the brain, liver, spleen, heart, lung, and kidney were prepared (14 mm in thickness). After incubation with 3% bovine serum albumin and 0.3% Triton X-100 in PBS for 1 h, sections were incubated with primary antibodies (Abcam, Cambridge, UK) against rabbit anti-Iba1 (ab178846, 1: 2000) and NeuN (ab177487, 1: 3000) overnight at ambient temperature. Corresponding secondary antibodies of Alexa Fluor 488 (ab150073, Abcam) were added at ambient temperature for another 2-h incubation. 4′,6-diamidino-2-phenylindole (DAPI) staining of cell nuclei was performed, while terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was performed in the light of the kit instructions (C1088, Beyotime, Shanghai, China). The percentage of TUNEL+ cells to DAPI+ nuclei was calculated. Hematoxylin–eosin (H&E) staining (C0105, Beyotime) was processed to observe morphological changes in brain tissues. Quantitative analysis was performed by a blind method.
OGD/R-Treated Neurons
Neurons were exposed to glucose-free Earl’s solution [5.4 mmoL/L KCl, 116.4 mmoL/L NaCl, 0.8 mmoL/L MgSO4, 1.8 mmoL/L CaCl2, 26.2 mmoL/L NaHCO3, 2.6 mmol/L NaH2PO4, and 20.1 mmol/L HEPES (pH = 7.4)] and incubated at 37 °C in 5% CO2 and 95% N2 (OGD) for 2 h. The OGD was then terminated by the addition of Earl’s solution with 5.6 mmol/L glucose. Cells were continued to be incubated in 5% CO2 and 95% air for 12 h. Neurons were treated with LV or EVs for 6 h and then the medium was renewed with the standard medium. After another 72-h treatment, cells received OGD/R treatment.
5-Triphenyltetrazolium Chloride (TTC) Staining
The brains of mice were cut into 1-mm coronal sections, 6 sections in total. Sections were incubated with TTC staining solution (G3004, Solarbio, Beijing, China) at 37 °C for 15 min and then fixed overnight in 10% formalin solution. The sections were imaged with Image-Pro Plus 6.0 software and the volume of the infarct area was quantified.
Modified Neurological Severity Scoring (mNSS)
A mNSS system ranging from 0 to 14 was applied, where 0 indicates normal and 14 indicates the highest degree of neurological deficit. Locomotor testing was used to observe the bending and twisting of the limbs of the mice by holding up the tail (0–3 points). The posture of walking on the floor (0–3 points) was also calculated. In the balance test, mice were placed on a beam. The degree of neurological deficit was evaluated based on whether the mice could maintain balance on the beam, whether the limbs fell off the beam and whether they could pass the beam (0–6 points). Sensory and reflex tests examined the auricular and corneal reflexes, respectively (0–2 points). The higher score was indicative of a more severe injury.
Statistical Analysis
The SPSS 19.0 statistical software (IBM, Armonk, NY) was used for data analysis. Each group of experiments was repeated at least three times independently, and the measurement data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. Unpaired t-test was performed for data in normal distribution and homogeneous variance. Data normality was tested using the Kolmogorov–Smirnov method, in which normally distributed data were compared employing one-way analysis of variance. A value of p < 0.05 indicates a statistically significant difference.
Results
Low Expression of circ_0000075 Is Detected in the Brain Tissues of MCAO Mice
By sequencing and analyzing circRNAs in brain tissues from MCAO mice and sham-operated mice, we obtained circRNAs that were significantly differentially expressed in brain tissue samples (Fig. 1A–B). To determine the protective circRNAs in cerebral ischemic injury, the top 5 down-regulated circRNAs were subjected to RT-qPCR in brain tissues of MCAO mice and sham-operated mice. It was found that the expression of circ_0000075 was distinctly lower in the MCAO mice compared with the sham-operated mice while no significant difference was witnessed regarding the expression of the other 4 circRNAs (Fig. 1C–D). The above results suggest that the expression of circ_0000075 is diminished in stroke.
Low expression of circ_0000075 in MCAO mice. A Heat map of differentially expressed circRNAs in ischemic tissues of mice in the MCAO group (n = 2) and normal brain tissues in the sham group (n = 2). B Volcano map of differentially expressed circRNAs in ischemic tissues of mice in the MCAO group (n = 2) and normal brain tissues in the sham group (n = 2). C RT-qPCR of top 5 down-regulated circRNAs expression from sequencing analysis in the blood of mice in the MCAO group and sham group. n = 8; * p < 0.05 vs. the sham group. D RT-qPCR of top 5 down-regulated circRNAs from sequencing analysis in the brain tissues of mice in the MCAO group and the sham group. n = 8; * p < 0.05 vs. the sham group
Exogenous circ_0000075 Attenuates Cerebral Ischemic Injury in MCAO Mice
To further investigate the effect of circ_0000075 on cerebral ischemic injury, a lentiviral vector-mediated overexpression of circ_0000075 was constructed (Fig. S1A). LV-circ_0000075 was injected intracranially into mice 1 day prior to MCAO modeling. Sensory-motor function in mice before and 28 days after MCAO was assessed comprehensively by a series of behavioral tests (Fig. S1B–E). Mice injected with LV-circ_0000075 showed significantly less sensorimotor deficits, better neural recovery, and significantly less foot faults, as well as spent longer time to use the rota-rod, and less time to remove the tape. In addition, MWM test revealed that mice injected with LV-circ_0000075 had significantly less avoidance latency during the learning phase and spent more time in the target quadrant during the memory phase after MCAO modeling, indicating improved long-term learning and memory behavior (Fig. S1F–G). Similarly, RT-qPCR results clarified that the expression of circ_0000075 in mouse brain tissues was elevated after LV-circ_0000075 injection (Fig. S1H). The above results suggest that exogenous overexpression of circ_0000075 can reduce cerebral ischemic injury in MCAO mice.
Engineered RVG-EVs Are Internalized by Neurons to Up-regulate circ_0000075 Expression
To further study circ_0000075, we need to manipulate its expression in cells. Due to the in vivo toxicity of viral vectors, EVs, as cell-secreted vesicles, are the best choice as a gene drug delivery vector [23]. Mouse BMSCs were chosen for isolation of engineered EVs. Mouse BMSCs were isolated, followed by identification of the surface markers of MSCs using flow cytometry, results of which showed high expression of CD29 (97%) and CD44 (99%) as well as low expression of CD31 (5%) and CD45 (5%) (Fig. S2A). In addition, the BMSCs were tested for their ability to differentiate in a targeted manner, and the results showed that the cells we isolated and cultured were capable of osteogenic, lipogenic, and chondrogenic differentiation (Fig. S2B). The above results indicate that we successfully isolate and extract BMSCs.
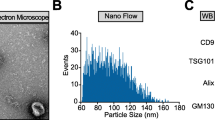
To explore the role of circ_0000075 in stroke pathology, circ_0000075-EVs targeting the brain were developed and BMSCs were treated with lentiviral plasmids encoding GNSTM-RVG-Lamp2b-HA and circ_0000075. As a control, lentivirus harboring sh-circ_0000075 was constructed and used to treat BMSCs, followed by EV purification (Fig. 2A). EVs were isolated from the supernatant of cultured cells and the isolated EVs were characterized by Zetaview, TEM, and Western blot analysis (Fig. 2B–D). Also, RVG-Lamp2b-HA was expressed in BMSCs and was integrated into BMSC-derived EVs (Fig. 2D). circ_0000075 expression was elevated in the BMSCs treated with RVG-circ_0000075-EVs, yet lower in presence of RVG-sh-circ_0000075-EVs, suggesting that circ_0000075 was integrated into BMSC-derived EVs (Fig. 2E).
Engineered EVs can transfer circ_0000075 to neurons. A Schematic diagram of obtaining RVG-circ_0000075-EVs. B Zetaview analysis of the particle size distribution and concentration of EVs. C, TEM observation of EVs; bar = 100 nm. D, Western blot detection of Lamp2b, CD63, GM130, and Alix expression in BMSCs and EVs. E RT-qPCR of circ_0000075 expression in EVs. * p < 0.05 vs. the RVG-vector-EV group. F Fluorescence microscopy observation of EV internalization; Dil-labeled EVs (red) (bar = 25 μm). G RT-qPCR of circ_0000075 expression in neurons. * p < 0.05 vs. the RVG-vector-EV group. H RT-qPCR of circ_0000075 expression after co-treatment of BMSC-CM with RNase A or RNase A + TritonX-100. * p < 0.05 vs. the control group (no treatment). I Expression of circ_0000075 in neurons after actinomycin D treatment. * p < 0.05 vs. the RVG-circ_0000075-EVs + DMSO group. The cell experiment was repeated 3 times independently
To determine that RVG-circ_0000075-EVs could be internalized by neurons, we extracted mouse neurons (Fig. S2C) which were co-incubated with Dil-labelled RVG-circ_0000075-EVs or RVG-sh-circ_0000075-EVs. The fluorescence microscopy revealed red fluorescence around the nuclei of neurons, which indicated that neurons internalized RVG-circ_0000075-EVs and RVG-sh-circ_0000075-EVs (Fig. 2F). The treatment of RVG-circ_0000075-EVs significantly promoted the expression of circ_0000075 in neurons when compared with treatment of RVG-vector-EVs while no significant difference was witnessed regarding circ_0000075 expression in response to RVG-sh-circ_0000075-EVs as EVs-depletion (Fig. 2G). After the conditioned medium of BMSCs was treated with RNase A, RNA was extracted, and the expression of circ_0000075 was essentially the same in BMSCs treated with RNase A compared to the control, but circ_0000075 expression was undetectable in BMSCs after treatment of both RNase A and TritonX-100 (Fig. 2H).
To further exclude that the expression of circ_0000075 was endogenously induced, neurons were treated with actinomycin D. RT-qPCR assay revealed that there was no significant difference in the expression of circ_0000075 between the actinomycin D-treated neurons and control, indicating that the expression of circ_0000075 was not endogenously induced (Fig. 2I).
The above results suggest that RVG-circ_0000075-EVs can be internalized by neurons, which in turn up-regulates circ_0000075 expression in neurons.
RVG-circ_0000075-EVs Target Brain Tissues and Thus Reduce Cerebral Ischemic Injury
To determine the effect of RVG-circ_0000075-EVs on cerebral ischemic injury in vivo, the targeting of RVG-circ_0000075-EVs to brain tissues was first explored. After intravenous injection of Dil-labeled RVG-circ_0000075-EVs, IVIS revealed that the brain tissues from the mice treated with RVG-circ_0000075-EVs had stronger fluorescence intensity (Fig. 3A), and circ_0000075 expression was increased in the brain tissues of mice treated with RVG-circ_0000075-EVs (Fig. 3B), suggesting that RVG-circ_0000075-EVs targeted brain tissues to increase circ_0000075 expression. Moreover, the aggregation of EVs in brain tissues of mice treated with RVG-circ_0000075-EVs was increased (Fig. 3C).
RVG-circ_0000075-EVs target brain tissues. A EV targeting observed by IVIS 12 h after intravenous injection of 12 mg/kg circ_0000075-EVs or RVG-circ_0000075-EVs. B RT-qPCR of circ_0000075 expression in brain, lung, heart, spleen, kidney, and liver of mice. * p < 0.05 vs. the circ_0000075-EV group. C Immunofluorescence of EV (Dil, red) distribution in mouse brain, lung, heart, spleen, kidney, and liver; bar = 25 μm. D Immunofluorescence of EV distribution in neurons and microglia. Neurons (NeuN); microglia cells (Iba-1); EVs (Dil); bar = 25 μm. n = 3
To determine the internalization of RVG-circ_0000075-EVs by cells from brain tissues, immunofluorescence assay was performed on brain tissues (Fig. 3D), and the results showed that EVs were detected in both neurons and microglia, indicating that RVG-circ_0000075-EVs were internalized by cells from brain tissues.
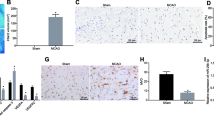
To determine the effect of RVG-circ_0000075-EVs on cerebral ischemic injury, RVG-circ_0000075-EVs were injected via the tail vein 1 day before MCAO modeling, and brain tissues were taken on the 7th day to observe brain injury. As revealed by RT-qPCR, MCAO significantly decreased circ _0000075 expression when compared with sham operation while RVG-circ_0000075-EVs significantly increased circ _0000075 expression in brain tissues (Fig. 4A). TTC staining showed that the infarct area was larger in the MCAO mice, which was reduced after RVG-circ_ 0,000,075-EV treatment (Fig. 4B). In addition, water content was enhanced in the MCAO mice, yet reduced after RVG-circ_0000075-EV treatment (Fig. 4C). Further evaluation of the neuroprotective effect, as shown in Fig. 4D, revealed a significant betterment in mNSS after RVG-circ_0000075-EV treatment. Moreover, in MCAO mice, we found severe brain tissue damage, decreased neuronal count, and increased cell apoptosis. However, after the treatment of RVG-circ_0000075-EVs, alleviated brain tissue damage, increased neuronal count, and significantly reduced apoptosis rate were observed (Fig. 4E–G).
RVG-circ_0000075-EVs reduce cerebral ischemic injury in MCAO mice. A RT-qPCR of circ_0000075 expression in brain tissues. B TTC staining to observe the infarct area. C Detection of brain water content after MCAO to measure the severity of brain injury. D mNSS to assess the neurological function of mice on day 7 after MCAO. E H&E staining of coronal sections of mouse brain after MCAO; bar = 50 μm. F NeuN staining of neurons in mice after MCAO. G TUNEL staining to observe neuronal apoptosis in mice after MCAO; n = 8. * p < 0.05 vs. the sham group; # p < 0.05 vs. the MCAO group; & p < 0.05 vs. the RVG-vector-EV group
The above results suggest that RVG-circ_0000075-EVs can target brain tissues and up-regulate the expression of circ_0000075 in brain tissues, thus relieving cerebral ischemic injury.
RVG-circ_0000075-EVs Inhibit Neuronal Apoptosisin Vitro
To further investigate the molecular mechanism of RVG-circ_0000075-EVs affecting neurons in vitro, we constructed an in vitro neuronal OGD/R model and found that circ_0000075 expression was reduced in cells treated with OGD/R. circ_0000075 expression was not significantly different regarding circ_0000075 expression in presence of RVG-vector-EVs, while RVG-circ_0000075-EV treatment significantly up-regulated circ_0000075 expression in neurons after OGD/R and no significantly difference was induced by RVG-sh-circ_0000075-EV treatment where circ_0000075 expression was similar to that after exposure to OGD/R (Fig. 5A).
RVG-circ_0000075-EVs reduce neuronal injury and apoptosis after OGD/R. A RT-qPCR of circ_0000075 expression in each group of cells after OGD/R treatment. B CCK-8 assay to detect cell survival. C TUNEL staining to detect apoptosis; bar = 25 μm. D LDH release to assess neuronal injury. * p < 0.05 vs. the control group; # p < 0.05 vs. the RVG-vector-EV group. The cell experiment was repeated 3 times independently
In addition, neuronal survival was detected by CCK-8 (Fig. 5B), apoptosis by TUNEL staining (Fig. 5C), and cell damage by LDH assay (Fig. 5D). The results showed that OGD/R treatment decreased cell survival, increased neuronal apoptosis, and promoted LDH release. No significant difference was detected in OGD/R-treated cells following treatment of RVG-vector-EVs. However, RVG-circ_0000075-EV treatment brought about contrary results. After exposure to OGD/R, RVG-sh-circ_0000075-EV treatment led to no significant difference.
These results suggest that RVG-circ_0000075-EVs can attenuate OGD/R-induced neuronal damage, increase neuronal survival, and inhibit neuronal apoptosis in vitro.
circ_0000075 Promotes SMURF2 Expression by Binding to miR-218-5p
To further investigate the molecular mechanisms by which circ_0000075 affects neuronal injury, FISH for circ_0000075 localization in neurons revealed that circ_0000075 was mainly expressed in the cytoplasm (Fig. 6A). circ_0000075 may be able to inhibit the binding between miRNA and its target mRNA as a miRNA sponge through ceRNA mechanism, thereby controlling cerebral ischemic injury. Therefore, 33 differentially expressed miRNAs were screened out from microarray dataset GSE84216 (10 sham-operated mice and 10 MCAO mice), among which 19 miRNAs were up-regulated (Fig. S3A). We used the miRanda algorithm to predict their targeting relation with circ_0000075, revealing miR-218-5p, miR-183-5p and miR-34c-5p as 3 putative up-regulated miRNAs that bind to circ_0000075 (Fig. S3B).
Targeted regulatory relationship among circ_0000075/miR-218-5p/SMURF2. A FISH detection of circ_0000075 localization in neurons; bar = 25 μm. B The upper panel indicates the target binding site of human circ_0000075 with miR-218-5p predicted by the starBase database, and the lower panel indicates the target binding site of mouse circ_0000075 with miR-218-5p predicted by the RNA22 database. C RIP to detect the enrichment of circ_0000075. * p < 0.05 vs. the mimic NC group. D The upper panel indicates the target binding site of human miR-218-5p with SMURF2 predicted by the RNA22 database, and the lower panel indicates the target binding site of mouse miR-218-5p with SMURF2 predicted by the starBase database. E Dual luciferase reporter gene assay to verify binding of circ_0000075 to miR-218-5p and SMURF2 to miR-218-5p. * p < 0.05 vs. the mimic NC group. F Biotin-coupled miR-218-5p mimic for pull down assay to detect circ_0000075 and SMURF2 enrichment. * p < 0.05 vs. the Biotin-NC or LV-NC group. G RT-qPCR to detect the expression of circ_0000075 and miR-218-5p in each group of cells. * p < 0.05 vs. the mimic NC + LV-NC group; # p < 0.05 vs. the miR-218-5p mimic + LV-NC group. H Western blot analysis to detect the protein expression of SMURF2 in each group of cells. * p < 0.05 vs. the mimic NC + LV-NC group; # p < 0.05 vs. the miR-218-5p mimic + LV-NC group. I RT-qPCR to detect the expression of miR-218-5p in neurons treated with RVG-circ_0000075-EVs. * p < 0.05 vs. the RVG-vector-EV group. J Western blot analysis to detect the protein expression of SMURF2 in neurons after treatment with RVG-circ_0000075-EVs. * p < 0.05 vs. the RVG-vector-EV group. The cell experiment was repeated 3 times independently
Previous evidence has documented the critical role of miR-218-5p in acute ischemic stroke [11]. Thus, miR-218-5p was selected for subsequent analysis. The target binding site of human (the upper panel in Fig. 6B) and mouse (the lower panel in Fig. 6B) circ_0000075 to miR-218-5p was predicted using the starBase database and the RNA22 database. To determine that circ_0000075 targets and binds to miR-218-5p, we performed RIP experiments as shown in Fig. 6C, and circ_0000075 was specifically enriched in miR-218-5p-transfected cells.
Next, target genes of miR-218-5p were predicted in starBase and MicroT-CDS database, results of which were further intersected with cerebral ischemic injury–related genes retrieved from DisGeNET database. In total, 24 target genes were identified in the intersection, among which SMURF2 drew our attention (Fig. S3C). Then, the RNA22 database and starBase database predicted the human (the upper panel in Fig. 6D) and mouse (the lower panel in Fig. 6D) binding sites between miR-218-5p and SMURF2 (Fig. 6D). Therefore, we further investigated the effect of circ_0000075 on development of cerebral ischemic injury via miR-218-5p/SMURF2.
To verify the interaction of circ_0000075, miR-218-5p, and SMURF2, the results of the dual luciferase reporter gene assay showed that transfection of miR-218-5p mimic significantly reduced the luciferase activity of WT circ_0000075 and SMURF2, but did not affect that of MUT circ_0000075 and SMURF2 (Fig. 6E). RNA pull-down assays were performed using biotin-coupled miR-218-5p mimic to detect the binding activity of circ_0000075 and SMURF2 with miR-218-5p in neurons (Fig. 6F). Enrichment of circ_0000075 and SMURF2 was found in miR-218-5p, but overexpression of circ_0000075 in cells resulted in reduced enrichment of SMURF2 on miR-218-5p. Transfection of miR-218-5p mimic in neurons significantly increased miR-218-5p expression and suppressed SMURF2 expression. When circ_0000075 was also overexpressed, miR-218-5p expression was reduced and circ_0000075 and SMURF2 expression was up-regulated (Fig. 6G-H). Similarly, when neurons were treated with RVG-circ_0000075-EVs, the expression of miR-218-5p was reduced and the expression of SMURF2 was increased (Fig. 6I–J). The above results suggest that in neurons, circ_0000075 binds to miR-218-5p to modulate SMURF2 expression.
circ_0000075 Ameliorates Neuronal Injury by Regulating the miR-218-5p/SMURF2 Axis
The role of circ_0000075/miR-218-5p/SMURF2 axis in cerebral ischemic injury was subsequently investigated. miR-218-5p expression was up-regulated and SMURF2 mRNA expression was down-regulated in the brain tissues of MCAO mice (Fig. 7A), the protein expression of SMURF2 showed the same expression trend. However, in mice treated with circ_0000075 and RVG-circ_0000075-EVs, miR-218-5p expression was down-regulated and SMURF2 mRNA expression was up-regulated in brain tissues (Fig. 7B, C), the protein expression of SMURF2 showed the same expression trend. It is thus evident that circ_0000075 may affect cerebral ischemic injury through mediation on miR-218-5p/SMURF2 expression in the mouse model.
circ_0000075/miR-218-5p/SMURF2 ameliorates neuronal injury. A RT-qPCR for miR-218-5p and SMURF2 expression in brain tissues of MCAO mice (n = 8). * p < 0.05 vs. the sham group. B RT-qPCR and Western blot analysis for miR-218-5p and SMURF2 expression in brain tissues of mice treated with circ_0000075 (n = 8). * p < 0.05 vs. the LV-NC or RVG-vector-EV group. C RT-qPCR and Western blot analysis for miR-218-5p and SMURF2 expression in brain tissues of mice treated with RVG-circ_0000075-EVs (n = 8). * p < 0.05 vs. the LV-NC or RVG-vector-EV group. D RT-qPCR and Western blot analysis to detect miR-218-5p and SMURF2 expression in OGD/R-treated neurons. * p < 0.05 vs. the control group. E LDH release to assess neuronal damage. * p < 0.05 vs. the mimic NC + Vector or mimic NC + RVG-vector-EV group. # p < 0.05 vs. the mimic NC + circ_0000075 or mimic NC + RVG-circ_0000075-EVs or miR-218-5p mimic + Vector group. The cell experiment was repeated 3 times independently
Furthermore, the role of miR-218-5p/SMURF2 was verified in cells treated with OGD/R. It was found that OGD/R treatment increased the expression of miR-218-5p and reduced the mRNA and protein expression of SMURF2 (Fig. 7D). When neurons were treated with miR-218-5p inhibitor, cell survival was promoted and apoptosis was suppressed (Fig. S4A–D), and consistent results were obtained when SMURF2 was overexpressed in neurons (Fig. S5A–D).
We further performed circ_0000075 or RVG-circ_0000075-EV treatment while overexpressing miR-218-5p in neurons and found that overexpression of miR-218-5p attenuated the effects of circ_0000075 and RVG-circ_ 0,000,075-EVs on neuronal injury. Similarly, after both overexpression of miR-218-5p and SMURF2, it was found that SMURF2 ameliorated the exacerbating effect of overexpression of miR-218-5p on neuronal injury (Fig. 7E).
The above results suggest that circ_0000075 can regulate the expression of miR-218-5p, which in turn affects the expression of SMURF2, to alleviate the neuronal injury induced by OGD/R treatment.
circ_0000075 May Up-regulate SMURF2 to Regulate the YY1 and Thereby Relieve Cerebral Ischemic Injury
Analysis of the GEO database (GSE16561) showed a significant negative correlation between SMURF2 and YY1 expression (Fig. 8A), with YY1 expression significantly higher in the cerebral ischemic injury samples from GSE16561 than that in control samples (Fig. 8B). The above results suggest that SMURF2 may attenuate cerebral ischemic injury by affecting the YY1 axis.
SMURF2 regulates YY1 expression in neurons. A Correlation analysis of SMURF2 and YY1 in microarray GSE16561. B Expression of YY1 in microarray GSE16561. C Western blot analysis of YY1 expression in brain tissues of MCAO mice. * p < 0.05 vs. the sham group; n = 8. D Western blot analysis of YY1 protein expression in OGD/R-treated neurons. * p < 0.05 vs. the control group. E Western blot analysis of YY1 protein expression in neurons after overexpression of SMURF2. * p < 0.05 vs. the vector group. F Western blot analysis for YY1 protein expression in neurons after knockdown of SMURF2 and CHX treatment. G Western blot analysis for YY1 expression in brain tissues of circ_0000075- and RVG-circ_0000075-EV-treated mice. * p < 0.05 vs. the LV-NC group; # p < 0.05 vs. the LV-circ_0000075 group. H Detection of LDH release to assess neuronal damage. I CCK8 was performed for detection of cell viability. J TUNEL was used to detect cell apoptosis in each group. * p < 0.05 vs. the LV-NC + sh-NC + sh-NC group or the RVG-vector-EVs + sh-NC + sh-NC group; # p < 0.05 vs. the circ_0000075 + sh-NC + sh-NC group or the RVG-circ_0000075-EVs + sh-NC + sh-NC group; and p < 0.05 vs. the LV-circ_0000075 + sh-SMURF2 + sh-NC group or the RVG-circ_0000075-EVs + sh-SMURF2 + sh-NC group. The cell experiment was repeated 3 times independently
To quantify the expression of YY1 in cerebral ischemic tissues, Western blot examined YY1 expression in brain tissues of MCAO mice, and the results showed that YY1 expression was up-regulated in the MCAO mice (Fig. 8C). Also, YY1 protein expression was up-regulated after OGD/R treatment of neurons (Fig. 8D), and that overexpression of SMURF2 in neurons significantly inhibited YY1 protein expression (Fig. 8E). Knockdown of SMURF2 and treatment with CHX enhanced YY1 protein stability (Fig. 8F). Treatment of circ_0000075 and RVG-circ_0000075-EVs down-regulated YY1 protein expression in brain tissues of mice while addition of sh-SMURF2 reversed the changing tendency induced by circ_0000075 and RVG-circ_0000075-EVs (Fig. 8G).
To further demonstrate the role of SMURF2/YY1 axis, neurons were treated with circ_0000075 or RVG-circ_0000075-EVs, followed by knockdown of SMURF2 and knockdown of YY1. Treatment of circ_0000075 or RVG-circ_0000075-EVs reduced the release of intracellular LDH, while knockdown of SMURF2 inhibited this effect, but the release of LDH was still reduced following knock down of YY1 and knockdown of SMURF2 (Fig. 8H). CCK8 and TUNEL assays revealed that treatment of circ_0000075 or RVG-circ_0000075-EVs enhanced cell viability and reduced cell apoptosis, while sh-SMURF2 treatment led to opposite trends; cell viability was enhanced and cell apoptosis was reduced upon sh-SMURF2 and sh-YY1 treatment (Fig. 8I, J).
The above results suggest that circ_0000075 may alleviate cerebral ischemic injury in brain tissues by increasing SMURF2 expression to regulate downstream YY1 expression.
Discussion
Ischemic stroke attributes to a deficiency in blood and oxygen in focal or complete brain and sometimes results in irreversible damage to the cerebrum [24]. Moreover, cerebral ischemic injury leads to apoptosis and even damage or death of a significant proportion of neurons, yet the underlying mechanisms remain largely unclear [25]. Previous study showed that MSC-EVs suppressed neuroinflammation induced by hypoxia–ischemia in vivo [26]. Herein, this study attempts to study the effect of BMSC-EVs on neurons after cerebral ischemic injury as well as the molecular mechanism. Consequently, engineered EVs carried exogenous circ_0000075 increased the expression of circ_0000075 in brain tissues, while circ_0000075 promoted the expression of SMURF2 by down-regulating miR-218-5p in neurons, which reduced the stability of YY1 protein, thereby alleviating cerebral ischemic injury (Fig. 9).
Molecular mechanism by which BMSC-derived EVs deliver circ_0000075 to brain tissues to affect cerebral ischemic injury. circ_0000075 binds to miR-218-5p in neurons, thus attenuating the targeting inhibition of SMURF2 by miR-218-5p and up-regulating the expression of SMURF2, which increases neuronal survival after cerebral ischemic injury by decreasing YY1 protein stability, promoting LEF1 protein expression
As our RNA-seq analysis and experimental results shown that we detected poorly expressed circ_0000075 in MCAO mice. The published work demonstrated that transient MCAO modeling was able to alter the profile of circRNAs [27]. Furthermore, circRNAs have regarded as a potent therapeutic target for ischemic stroke due to its high stability and evolutionary conservation in the brain tissues [28]. Similar to our finding, circDLGAP4 was under-expressed in cerebral ischemia and overexpression of circDLGAP4 attenuated neurological deficits and decreased infarct areas in MCAO model [29]. However, the effects of circ_0000075 on cerebral ischemic injury were barely reported in previous literature. In our study, we further found that engineered EVs increased circ_0000075 expression, which reduced brain tissue damage, increased neuronal count, and curtailed cell apoptosis rate. Since the in vivo toxicity of viral vectors limits their use, EVs, as a cell-secreted vesicle, are the best choice as a gene drug delivery vehicle [30]. Standard bioengineered EV is capable of leading EVs toward clinical success for stroke therapy [31].
Our further analysis revealed that circ_0000075 could bind to miR-218-5p, while loss of function of miR-218-5p increased SMURF2 expression. Recent research has documented that circRNAs can sponge miRNA, modulate splicing and transcription, and modify parental gene expression [32]. Of note, circRNAs can modulate gene expression through a circRNA-miRNA-gene pathway in OGD/R-induced neuronal injury [33]. Similar to the molecular mechanism our study investigated, mmu_circRNA_40806 is a potential sponge for mmu-miR-20a-3p in cerebral ischemia [34]. Moreover, miR-218-5p down-regulation has been demonstrated to ameliorate nitric oxide levels and suppress neuronal cell apoptosis [11], which was consistent with our results that decreased expression of miR-218-5p by circ_0000075 participated in the inhibition of cell apoptosis and promotion of cell survival. Notably, SMURF2 bears great responsibility in diverse biological pathways, including cell cycle, cell cytoskeletal remodeling, and limiting TGF-β signaling pathway [35]. Furthermore, Yu et al. unfolded that SMURF2 was capable of accelerating functional recovery in rats after ischemic stroke through inducing neuron differentiation [13]. As our study found, SMURF2 ameliorated the exacerbating effect of overexpression of miR-218-5p on neuronal injury.
Furthermore, SMURF2 up-regulation decreased YY1 protein level, but not mRNA level, and that Smurf2 enhanced the poly-ubiquitination of YY1 [36]. Accordingly, previous data have pinpointed the elevation of YY1 in MCAO mice and OGD-treated neurons [16]. In our study, we also identified a negatively correlation between SMURF2 and YY1. In addition, we demonstrated that the treatment of circ_0000075 and RVG-circ_0000075-EVs down-regulated YY1 protein expression in brain tissues of mice, which relieved cerebral ischemic injury.
Conclusions
Together, the findings of the current study have demonstrated that circ_0000075 binds with miR-218-5p to enhance SMURF2 expression, resulting in reduced YY1. Up-regulated circ_0000075 attenuated neuronal deficits, decreased infarct areas, and ameliorated brain damage. Thus, circ_0000075 delivered by engineered BMSC-EVs may be important both as a target for therapeutic interventions in cerebral ischemic injury and as a biomarker for disease activity.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
Wei K, Wang P, Miao CY (2012) A double-edged sword with therapeutic potential: an updated role of autophagy in ischemic cerebral injury. CNS Neurosci Ther 18(11):879–886
Singh N, Sharma G, Mishra V (2012) Hypoxia inducible factor-1: its potential role in cerebral ischemia. Cell Mol Neurobiol 32(4):491–507
Chen SD, Yang DI, Lin TK, Shaw FZ, Liou CW, Chuang YC (2011) Roles of oxidative stress, apoptosis, PGC-1alpha and mitochondrial biogenesis in cerebral ischemia. Int J Mol Sci 12(10):7199–7215
Chen W, Sun Y, Liu K, Sun X (2014) Autophagy: a double-edged sword for neuronal survival after cerebral ischemia. Neural Regen Res 9(12):1210–1216
Kaminski N, Koster C, Mouloud Y, Borger V, Felderhoff-Muser U, Bendix I, Giebel B, Herz J (2020) Mesenchymal stromal cell-derived extracellular vesicles reduce neuroinflammation, promote neural cell proliferation and improve oligodendrocyte maturation in neonatal hypoxic-ischemic brain injury. Front Cell Neurosci. 14:601176
Ophelders DR, Wolfs TG, Jellema RK, Zwanenburg A, Andriessen P, Delhaas T, Ludwig AK, Radtke S et al (2016) Mesenchymal stromal cell-derived extracellular vesicles protect the fetal brain after hypoxia-ischemia. Stem Cells Transl Med 5(6):754–763
Saheera S, Jani VP, Witwer KW, Kutty S (2021) Extracellular vesicle interplay in cardiovascular pathophysiology. Am J Physiol Heart Circ Physiol 320(5):H1749–H1761
Yang J, Chen M, Cao RY, Li Q, Zhu F (2018) The Role of Circular RNAs in cerebral ischemic diseases: ischemic stroke and cerebral ischemia/reperfusion injury. Adv Exp Med Biol. 1087:309–325
Zhang PP, Sun J, Li W (2020) Genome-wide profiling reveals atrial fibrillation-related circular RNAs in atrial appendages. Gene. 728:144286
Jin F, Xing J (2017) Circulating pro-angiogenic and anti-angiogenic microRNA expressions in patients with acute ischemic stroke and their association with disease severity. Neurol Sci 38(11):2015–2023
Zhu H, Wang X, Chen S (2020) Downregulation of MiR-218–5p protects against oxygen-glucose deprivation/reperfusion-induced injuries of pc12 cells via upregulating N-myc Downstream regulated gene 4 (NDRG4). Med Sci Monit 26:e920101
Ma X, Zhang H, Yin H, Geng S, Liu Y, Liu C, Zhao J, Liu Y et al (2021) Up-regulated microRNA-218–5p ameliorates the damage of dopaminergic neurons in rats with Parkinson’s disease via suppression of LASP1. Brain Res Bull. 166:92–101
Yu YL, Chou RH, Shyu WC, Hsieh SC, Wu CS, Chiang SY, Chang WJ, Chen JN et al (2013) Smurf2-mediated degradation of EZH2 enhances neuron differentiation and improves functional recovery after ischaemic stroke. EMBO Mol Med 5(4):531–547
Liu H, Sun S, Liu B (2021) Smurf2 exerts neuroprotective effects on cerebral ischemic injury. J Biol Chem 297(2):100537
Ramkumar C, Cui H, Kong Y, Jones SN, Gerstein RM, Zhang H (2013) Smurf2 suppresses B-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat Commun. 4:2598
Liu W, Guo Q, Zhao H (2018) Oxidative stress-elicited YY1 potentiates antioxidative response via enhancement of NRF2-driven transcriptional activity: a potential neuronal defensive mechanism against ischemia/reperfusion cerebral injury. Biomed Pharmacother. 108:698–706
Zhao H, Tao Z, Wang R, Liu P, Yan F, Li J, Zhang C, Ji X et al (2014) MicroRNA-23a-3p attenuates oxidative stress injury in a mouse model of focal cerebral ischemia-reperfusion. Brain Res. 1592:65–72
Fan Y, Xiong X, Zhang Y, Yan D, Jian Z, Xu B, Zhao H (2016) MKEY, a peptide inhibitor of CXCL4-CCL5 heterodimer formation, protects against stroke in mice. J Am Heart Assoc 5(9):e003615
Hong M, Kim M, Kim TW, Park SS, Kim MK, Park YH, Sung YH, Shin MS (2020) Treadmill exercise improves motor function and short-term memory by enhancing synaptic plasticity and neurogenesis in photothrombotic stroke mice. Int Neurourol J 24(Suppl 1):S28-38
Li F, Geng X, Huber C, Stone C, Ding Y (2020) In search of a dose: the functional and molecular effects of exercise on post-stroke rehabilitation in rats. Front Cell Neurosci. 14:186
Brooks MJ, Rajasimha HK, Roger JE, Swaroop A (2011) Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes. Mol Vis. 17:3034–3054
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 7:11215
Martinez Escude, de Castilla P, Tong L, Huang C, Sofias AM, Pastorin G, Chen X, Storm G, Schiffelers RM et al (2021) Extracellular vesicles as a drug delivery system: a systematic review of preclinical studies. Adv Drug Deliv Rev. 175:113801
Chen C, Chu SF, Liu DD, Zhang Z, Kong LL, Zhou X, Chen NH (2018) Chemokines play complex roles in cerebral ischemia. Neurochem Int. 112:146–158
Radak D, Katsiki N, Resanovic I, Jovanovic A, Sudar-Milovanovic E, Zafirovic S, Mousad SA, Isenovic ER (2017) Apoptosis and Acute Brain Ischemia in Ischemic Stroke. Curr Vasc Pharmacol 15(2):115–122
Xin D, Li T, Chu X, Ke H, Liu D, Wang Z (2021) MSCs-extracellular vesicles attenuated neuroinflammation, synapse damage and microglial phagocytosis after hypoxia-ischemia injury by preventing osteopontin expression. Pharmacol Res. 164:105322
Mehta SL, Pandi G, Vemuganti R (2017) Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke 48(9):2541–2548
Wang Q, Liu X, Zhao J, Zhu R (2020) Circular RNAs: novel diagnostic and therapeutic targets for ischemic stroke. Expert Rev Mol Diagn 20(10):1039–1049. https://doi.org/10.1080/14737159.2020.1826313
(2020) Erratum: Bai et al., “Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity”. J Neurosci 40(44):8601
Zhang Y, Liu Y, Liu H, Tang WH (2019) Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 9:19
Khan H, Pan JJ, Li Y, Zhang Z, Yang GY (2021) Native and bioengineered exosomes for ischemic stroke therapy. Front Cell Dev Biol. 9:619565
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K et al (2015) Circular RNA: A new star of noncoding RNAs. Cancer Lett 365(2):141–148
Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ (2016) Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun 471(1):52–56
Liu C, Zhang C, Yang J, Geng X, Du H, Ji X, Zhao H (2017) Screening circular RNA expression patterns following focal cerebral ischemia in mice. Oncotarget 8(49):86535–86547
Dong S, Liu J, Li L, Wang H, Ma H, Zhao Y, Zhao J (2019) The HECT ubiquitin E3 ligase Smurf2 degrades mu-opioid receptor 1 in the ubiquitin-proteasome system in lung epithelial cells. Am J Physiol Cell Physiol 316(5):C632–C640
Jo DH, An H, Chang DJ, Baek YY, Cho CS, Jun HO, Park SJ, Kim JH et al (2014) Hypoxia-mediated retinal neovascularization and vascular leakage in diabetic retina is suppressed by HIF-1alpha destabilization by SH-1242 and SH-1280, novel hsp90 inhibitors. J Mol Med (Berl) 92(10):1083–1092
Author information
Authors and Affiliations
Contributions
Yue Liu and You-Ping Li conceived and designed research. Li-Min Xiao and Li-Ke Chen performed experiments. Su-Yue Zheng and Er-Ming Zeng interpreted results of experiments. Yue Liu and Chun-Hua Xu drafted paper. You-Ping Li edited and revised manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This study was ratified by the Animal Ethics Committee of the First Affiliated Hospital of Nanchang University and all procedures were carried out per the Guide for the Care and Use of Laboratory Animals.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflicts of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary File 1
(DOCX 21 kb)

Supplementary Fig. 1
Effect of exogenous circ_0000075 on cerebral ischemic injury in MCAO mice. A, The overexpression efficiency of circ_0000075 assayed within BMSCs. B, Foot fault test from 1 day before to 28 days after MCAO. C, Rota-rod test from 1 day before to 28 days after MCAO. D, E, Adhesive removal from 1 day before to 28 days after MCAO. F, Time to locate the mice to the platform (escape latency) measured 22 - 26 days after MCAO. G, Spatial memory assessed by measuring the time spent in the target quadrant during platform removal 27 days after MCAO. H, RT-qPCR of circ_0000075 expression in mouse brain tissues 7 days after MCAO; n = 8. * p < 0.05 vs. the LV-NC group. (PNG 210 kb)

Supplementary Fig. 2
Extraction and identification of BMSCs and neurons. A, Flow cytometry of surface markers of BMSCs. B, Induction of multifunctional differentiation of BMSCs. i: Alizarin red cytochemical staining for osteogenic differentiation, scale bar = 50 μm; ii: Oil red O cytochemical staining for adipogenic differentiation; iii: Alcian Blue cytochemical staining for chondrogenic differentiation, scale bar = 50 μm. C, Primary Neuronal MAP-2 staining; red: MAP-2, blue: Hoechst (bar = 25 μm). All experiments were repeated 3 times independently. (PNG 717 kb)

Supplementary Fig. 3
Target miRNAs of circ_0000075 and target genes of miRNA. A, Volcano map of differentially expressed miRNAs in MCAO mice (n = 10) and sham-operated mice (n = 10). B, Venn diagram displaying the intersection of up-regulated miRNAs in cerebral ischemic injury and target miRNAs of circ_0000075. C, Venn diagram displaying the intersection of target genes of miR-218-5p and cerebral ischemic injury-related genes. (PNG 329 kb)

Supplementary Fig. 4
Effect of down-regulated miR-218-5p expression on neuronal damage after OGD/R. A, RT-qPCR to detect miR-218-5p expression in each group of cells after OGD/R treatment. B, CCK-8 assay to detect cell survival. C, TUNEL staining to detect apoptosis. D, LDH release to assess neuronal injury. * p < 0.05 vs. the OGD/R + inhibitor NC group. The cell experiment was repeated 3 times independently. (PNG 82 kb)

Supplementary Fig. 5
Effect of SMURF2 overexpression on neuronal damage after OGD/R. A, Western blot assay to detect SMURF2 expression in each group of cells after OGD/R treatment. B, CCK-8 assay for cell survival. C, TUNEL staining for apoptosis. D, LDH release to assess neuronal damage. * p < 0.05 vs. the OGD/R + vector group. The cell experiment was repeated 3 times independently. (PNG 126 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Li, YP., Xiao, LM. et al. Extracellular Vesicles Derived from Bone Mesenchymal Stem Cells Carrying circ_0000075 Relieves Cerebral Ischemic Injury by Competitively Inhibiting miR-218-5p and Up-regulating E3 Ubiquitin Ligase SMURF2. Mol Neurobiol 60, 2801–2818 (2023). https://doi.org/10.1007/s12035-022-03192-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03192-9