Abstract
Diabetic neuropathic pain (DNP) is a common complication of diabetes, and its complicated pathogenesis, as well as clinical manifestations, has brought great trouble to clinical treatment. The spinal cord is an important part of regulating the occurrence and development of DNP. Spinal microglia can regulate the activity of spinal cord neurons and have a regulatory effect on chronic pain. P2Y12 receptor is involved in DNP. P2Y14 and P2Y12 receptors belong to the Gi subtype of P2Y receptors, but there is no report that the P2Y14 receptor is involved in DNP. Closely related to many human diseases, the dysregulation of long noncoding RNA (lncRNA) has the effect of promoting or inhibiting the occurrence and development of diseases. The aim of this research is to investigate the function of the spinal cord P2Y14 receptor in type 2 DNP and to understand the function as well as the possible mechanism of lncRNA-UC.25 + (UC.25 +) in rat spinal cord P2Y14 receptor–mediated DNP. Our results showed that P2Y14 shRNA can reduce the expression of P2Y14 in DNP rats, thereby restraining the activation of microglia, decreasing the expression of inflammatory factors and the level of p38 mitogen–activated protein kinase (p38 MAPK) phosphorylation. At the same time, UC.25 + shRNA can downregulate the expression of the P2Y14 receptor, reduce the release of inflammatory factors, and diminish the p38 MAPK phosphorylation, indicating that UC.25 + can alleviate spinal cord P2Y14 receptor–mediated DNP. The RNA immunoprecipitation result showed that UC.25 + enriched signal transducers and activators of transcription1 (STAT1) and positively regulated its expression. The chromatin immunoprecipitation result indicated that STAT1 combined with the promoter region of the P2Y14 receptor and positively regulated the expression of the P2Y14 receptor. Therefore, we infer that UC.25 + may alleviate DNP in rats by regulating the expression of the P2Y14 receptor in spinal microglia via STAT1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus (DM) is a universal chronic illness. The rising morbidity and mortality rate have seriously affected the health and quality of life of patients. The latest ninth edition statistics of the International Diabetes Federation in 2019 show that diabetes has become a global epidemic, and the number of patients suffering from diabetes is increasing with the prevalence rate of 9.3% (approximately 463 million) [1, 2]. Diabetic neuropathic pain (DNP) develops from peripheral neuropathy. The main features are hyperalgesia, tactile pain, secondary pain, and spontaneous pain. The pathogenesis and clinical manifestations of DNP are relatively complex, and it is even difficult to relieve pain symptoms in patients by comprehensive treatment measures [3]. Therefore, the research on the pathogenesis and prevention of diabetic neuropathic pain is particularly urgent.
The pathogenesis of DNP involves not only the peripheral but the central nervous systems. As one part of the central nervous system, the spinal cord plays significant roles on the regulation of the occurrence and development of DNP [4]. The traditional treatments for pain mainly focus on neurons, and the clinical analgesic effect of a large number of neuron-targeted therapeutic drugs is not quite satisfactory, which boost the exploration of glial cells, a new hot spot in pain research. Research has shown that glial cells, such as microglia and astrocytes, can regulate the activity of neurons in the spinal cord and have a significant regulatory effect on the pathological changes in chronic pain [5]. Glial cell activation is observed in a variety of neuropathic pain models. The activated glial cells release multiple inflammatory factors, which enhance the excitability of neurons and promote the spread of the pain [6]. As immunocompetent cells, like macrophages, glia cells participate in the function of the immune barrier under normal circumstances. When the peripheral or central nervous system suffers from diabetes-related injuries, microglia are rapidly activated and undergo morphological and functional changes [7]. Activated microglia produce and secrete a series of pro-inflammatory factors, which increase sensitivity and affect synaptic transmission, leading to pathological pain. Therefore, the reason why the current pain treatment effect is not ideal may be the incorrect selection of the target, and glial cells may be a more suitable target [8].
As an energy donor for normal cellular metabolism, intracellular adenosine triphosphates (ATPs) play an important biological role. Inflammation as well as tissue damage can cause intracellular ATP release. Besides, the extracellular ATPs can also act as signal molecules and participate in signal transduction by binding to purinergic receptors located on the cell membrane [9]. Purinergic receptors include P1 receptors and P2 receptors. P2 receptors include ligand-gated ion channel type P2X receptors and G protein-coupled P2Y receptors. P2Y receptors include eight subtypes (P2Y1, 2, 4, 6, 11, 12, 13, 14). Research has shown that in the rat model of neurogenic neuropathic pain, the expression of P2Y14 receptors in spinal microglia is significantly increased, lasting at least 2 weeks. The application of P2Y14 receptor antisense oligonucleotide can significantly relieve pain and inhibit the expression of P2Y14 receptor, which proves that P2Y14 receptor can participate in neuropathic pain [7]. So, it can be speculated that P2Y14 receptors may participate in the development of DNP.
LncRNA is a DNA transcription product over 200 nucleotides in length, which can be found in the nucleus or cytoplasm and is stably expressed in tissues and cells. With the development of whole-genome sequencing technology and transcriptome sequencing, lncRNA has also received more attention [10]. LncRNA is not only an intermediary between DNA and protein, but also an important participant in cell function, which can regulate gene expression in a variety of ways from different levels, including epigenetic level, transcription level, and post-transcriptional processing [11]. Recent research has shown that the dysregulation of lncRNA is going hand in hand with numerous human diseases, and has the effect of promoting or inhibiting the development of diseases [12]. The abnormal expression of lncRNA was closely related to DM [13]. Therefore, the expression of individual lncRNA may be suitable for disease diagnosis and prognostic evaluation. LncRNA UC.25 + (UC.25 +) is one of the ultra-conserved orthologous regions, which is 100% identity between rats, mice, and humans [14]. Our microarray result revealed the expression of UC.25 + was raised in the DM rats. However, there is no report on the function of UC.25 + .
In this research, a rat model of DNP was built to explore whether spinal microglia P2Y14 receptor is involved in DNP and whether UC.25 + affects the P2Y14 receptor–mediated DNP via the speculated mechanism.
Materials and Methods
Animal Model
In order to define that the P2Y14 receptor participates in DNP, an animal model of DNP was established. Adult male Sprague–Dawley (SD) rats (430,726,220,100,076,156) weighing between 180 and 220 g were purchased from Changsha Tianqin Biotechnology Co., Ltd., Hunan Province. All experimental operations have been reviewed and authorized by the Medical Laboratory Animal Ethics Committee of Nanchang University. Before the rats were sacrificed, 3 ml/kg 10% hydrous chloric acid was injected intraperitoneally for anesthesia to relieve the pain of the rats. Before establishing the model, the rats were fed with normal diet in a clean environment for 1 week to fully adapt to the environment and reduce the influence of environmental differences on this experiment. After 1 week of normal diet, the rats were randomly divided into Control group (n = 12) and DNP group (n = 50). Rats in the control group were fed with normal diet, while DNP group was fed with high-lipid and high-glucose diet, including 77.8% normal diet, 10% lard, 10% sugar, 2% cholesterol, and 0.2% cholic acid sodium. After a high-lipid and high-sugar diet for a month, the rats in the DNP group were fasted for 8–10 h and injected intraperitoneally with streptozotocin 35 mg/kg. After 7 days, the fasting blood sugar ≥ 7.8 mmol/L and the obvious neuropathic pain symptoms were used as the criteria of the DNP model. The rats in the normal group were left untreated, and the tail vein blood glucose test showed normal. Finally, 36 rats were randomly chosen from the modeled rats and divided into three groups: DNP group (DNP group), DNP + P2Y14 shRNA treatment group (DNP + P2Y14 shRNA), and DNP + scramble shRNA as negative control treatment group (DNP + NC shRNA), with 12 rats in each group.
In order to explore the role and possible mechanism of lncRNA-UC.25 + in rat spinal cord P2Y14 receptor–mediated DNP, the animal model was established again, and the establishment method was the same as above. The test was divided into four groups: control group (Control), diabetic neuropathologic pain group (DNP group), DNP + lncRNA-UC.25 + shRNA treatment group (DNP + UC.25 + shRNA), and DNP + scramble shRNA treatment group (DNP + NC shRNA). There were 12 rats in each group.
Intrathecal Injection
According to the instructions of the animal transfection kit of Beijing Engreen Biotechnology Co., Ltd., P2Y14 shRNA, lncRNA-UC.25 + shRNA and NC shRNA plasmid were transfected into rats by intrathecal injection at a dose of 5 µg. The absorbent cotton soaked in ether was placed in the centrifuge tube, and the mouth and nose of the prone rats were placed in the tube. The needle was inserted vertically along the middle of the L5–L6 spinous process, and there was an obvious sense of breakthrough when entering the subarachnoid space. Wagging the tail was a sign of successful intrathecal injection. Injected the plasmid. The rats were put in an observation cage to wake up until they resumed normal activities. No treatment for control group and DNP group.
Cell Experiment
Select human umbilical vein endothelial cells (HUVEC) for the experiment. The cultural conditions were endothelial cell medium (ECM), 5% fetal bovine serum, 1% penicillin and streptomycin, 1% growth factor, and the cultural environment was 37 °C and 5% CO2. HUVEC cells were divided into control group and high-glucose group. The glucose concentration in the high-glucose group and the control group were 33.3 mmol/L and 5.5 mmol/L. In order to explore the relationship between UC.25 + and the transcription factor, signal transducer and activator of transcription 1 (STAT1), the cells were set up in the following groups: control group (Control), UC.25 + overexpression group (UC.25 + OE) and vector plasmid group (Vector) and high-glucose cell group (HG), high-glucose plus UC.25 + low-expression group (HG + UC.25 + shRNA), and high-glucose plus scramble shRNA group (HG + NC shRNA). Meanwhile, set up the following groups: control group (Control), STAT1 overexpression group (STAT1 + OE), vector plasmid group (Vector), and high-glucose cell group (HG), high-glucose plus STAT1 low-expression group (HG + STAT1 shRNA), and high-glucose plus scramble shRNA group (HG + NC shRNA) to detect the effect of STAT1 on P2Y14 receptor. STAT1 (Gene ID: 6772; NCBI Reference Sequence: NM_007315.3), and UC.25 + were inserted into the pCDNA3.1 plasmid, respectively. The overexpression plasmid (1000 ng/μl) was transfected into cells by FuGENE6.
Measurement of the Thermal Withdrawal Latency and Mechanical Withdrawal Threshold
Each group of rats was placed in a transparent plastic box and allowed to acclimate to the environment for 30 min. The electromechanical pain meter was used for testing, adjusting the relevant parameters of the instrument in advance, and then stimulating the center of the left hind plantar of the rat with a test needle with a force sensor. The intensity is gradually increased until the rat feels pain and lifts the left hind foot. The displayed data at this time is recorded as the rat’s mechanical hyperalgesia threshold. Place each group of rats in a transparent plastic box on a horizontal glass table. After the start of the test, turn on the tungsten lamp with a thermal light source and aim at the center of the left hind plantar of the rat to give thermal stimulation. Until the rat feels pain and lifts the left hind foot, the value recorded at this time is the thermal hyperalgesia threshold. Repeat the measurement 6 times for each rat, and take the average of the measurement results.
Quantitative Real-Time PCR
The separated lumbar 4–6 (L4–6) spinal cord is washed with pre-cooled phosphate buffer saline (PBS); it was placed in a homogenizer that had been treated with ribozyme-free water in advance. Collect total RNA with TRIZOL total RNA reagent and use Transgeen Reverse Transcription Kit for RNA reverse transcription. Then, add the complementary DNA (cDNA) template obtained by reverse transcription according to the Promega’s Real-time PCR kit, and use the step-one plus real-time fluorescent quantitative PCR instrument for experiment. All primers are from Beijing Shenggong Biological Co., Ltd. Primer sequence is as follows: P2Y14 Forward: 5′-GCATTGTGCTCGTATTTGTCG -3′, Reverse: 5′-CTAAACGGCTGGCATAAGAAG-3′; UC.25 + Forward: 5′-TGGTCAAAAGCAAAACAAG-3′, Reverse: 5′-TATGCAAGAAAAGGCAGAG-3′; STAT 1 Forward: 5′-GCCAAAGGAAGCACCAGAGCC-3′, Reverse: GAGCCCACTATCCGAGACACC-3′; Rat β-actin Forward: 5′-GCTCTCTTCCAGCCTTCCTT-3′, Reverse: 5′-CTTCTGCATCCTGTCAGCAA-3′; Homo β-actin Forward: 5′-CAAGAGATGGCCACGGCTGCT-3′, Reverse: 5′-TCCTTCTGCATCCTGTCGGCA-3′. Real-time quantitative PCR was performed with SYBR® Green Master Mix by an ABI PRISM® 7500 (Applied Biosystems, Inc., Foster City, CA, USA). Individual experiments were performed on each sample and repeated three times. The quantification of gene expression was calculated by the 2−ΔΔCt method in comparison with respective levels of β-actin mRNA.
Western Blotting
Freshly isolated L4-6 spinal cord tissue was washed in pre-cooled PBS, and put into the radio immunoprecipitation assay (RIPA) lysis solution. The mixture was thoroughly ground and decomposed on ice for 30 min, followed by centrifugation to obtain supernatant. The protein in the supernatant was separated by polyacrylamide gel electrophoresis, and the separated protein was transferred to the polyvinylidene fluoride (PVDF) membrane. The protein-containing PVDF membrane was incubated with 5% nonfat dried milk for 1 h, and then incubated with anti-rabbit P2Y14 (1:500, APR026, Alomone Labs), OX42 (1:1000, AB1211, Abcam), interleukin-1 beta (IL-1β) (1:500, AF5103, Affinity), tumor necrosis factor-alpha (TNF-α) (1:500, PB0082, Boster), p38 mitogen-activated protein kinase (p38 MAPK) (1:800, 8690, Cell Signaling Technology), and phosphorylated p38 (p-p38) MAPK (1:800, 4511, Cell Signaling Technology) overnight at 4 °C. After three washes with triethanolamine-buffered saline-tween (TBST), the membranes were incubated in secondary antibody with horseradish peroxidase (1:2000, Beijing Zhongshan Biotechnology Co.) for 1 h. Put the membranes into the gel imaging system, and the enhanced chemiluminescence (ECL) luminescent solution was added to expose the image. The results were recorded and stored. The integrate optical density (IOD) analysis of protein results was analyzed by Image-Pro Plus software, and the expression of target protein in each group was standardized by β-actin.
Immunofluorescence
The isolated L4-6 spinal cord was rinsed with pre-cooled PBS and dehydrated overnight in an eppendorf (EP) tube containing 20% sucrose solution. The spinal cord was embedded by optimum cutting temperature (OCT) and sectioned with a frozen microtome. First rinse the sections with PBS, then soak them in 0.3% Triton X-100 PBS solution for 30 min, and incubate with 10% normal goat serum to block non-specific staining. The sections were incubated overnight with anti-rabbit P2Y14 receptor (1:200, APR026, Alomone Lab) and anti-mouse OX42 (1:200, AB1211, Abcam). After washing the next day, the sections were incubated with goat anti-rabbit tetramethyl rhodamin isothiocyanate (TRITC) (1:200, Zhongshan Biotech Co.) and goat anti-mouse fluorescein isothiocyanate (FITC) (1:200, Zhongshan Biotech Co.). The images of the dorsal horns of the spinal cord were captured by fluorescence microscopy (Olympus DP72, Japan).
Bioinformatics Technology Predicts the Transcription Factor STAT1 of the P2Y 14 Receptor
The transcription factors of P2Y14 were searched and screened through UCSC, GENECARDS and other databases, and multiple prediction results showed that transcription factor STAT1 might regulate the expression of P2Y14 receptor. Finally, the sequence of binding sites with higher STAT1 score was searched through JASPAR website and subsequent validation experiments were carried out.
RNA Immunoprecipitation
The interaction between UC.25 + and STAT1 protein was detected by RNA immunoprecipitation (RIP) experiment. HUVEC cells were cultured and divided into two groups: Control group: co-transfected with two plasmids pcDNA3.0–12 × MS2bs and pcDNA3.0-Flag-2 × MS2; Experimental group: co-transfected with two plasmids pcDNA3.0-UC.25 + -12 × MS2bs and pcDNA3.0-Flag-2 × MS2. After plasmid transfection, the cells were cultured for 48 h. After the cells were cross-linked by UV and lysed, collected the supernatant, added ANTI-FLAG M2 magnetic beads eluted with the eluent I, and turned over overnight. The supernatant was collected as the output group, and the remaining magnetic beads were continuously eluted through eluent I and eluent II. Finally, the obtained supernatant was subjected to a Western blot experiment to detect the protein bound to UC.25 + .
Chromatin Immunoprecipitation
In order to explore the binding of the transcription factor STAT1 to the DNA fragments in the promoter region of the P2Y14 receptor, a ChIP assay kit was used, and experiments was performed according to the instructions. HUVEC cells were cross-linked with 1% formaldehyde and chromatin was disrupted by an ultrasonicator (UH-250A, Beijing, China). STAT1 antibody (1:50, 14,994, Cell Signaling Technology) was used to immunoprecipitate cross-linked protein-DNA complexes. After purification, the obtained DNA was amplified by PCR. The PCR primers were designed, and the sequences were Homo STAT1 290 bp, Sense: 5′-TCCAGCAACCTGATGTAA-3′; Antisense: 5′-ACACTTGGGAGTAGAGGG-3′.
Statistical Analysis
The experimental results were analyzed by SPSS 21, and the results of each group were expressed as mean ± SEM. The results of each group were compared by one-way analysis of variance (ANOVA) combined with least significant difference (LSD). Two-way ANOVA was performed to compare 2 points between groups for TWL and MWT. Paired t-test was used for comparison between the two groups. p < 0.05 was considered to be statistically significant.
Results
P2Y 14 Receptor Mediates DNP
Determination of MWT and TWL
The MWT and TWL were measured to define the changes of pain behavior. The data indicated that MWT and TWL in DNP group were significantly lower than those in Control group after STZ injection (p < 0.01); after intrathecal injection of P2Y14 shRNA, compared with the DNP group, the MWT and TWL of DNP + P2Y14 shRNA group were obviously raised, indicating that P2Y14 shRNA can alleviate pain behavior in DNP rats (p < 0.01). There was no significant difference in MWT and TWL between DNP + NC shRNA and DNP group (p > 0.05) (Fig. 1).
Expression of P2Y14 Receptor and OX42 in Rat Spinal Cord
Compared with the Control group, the expression of P2Y14 receptor protein and mRNA in the DNP group were obviously raised (p < 0.01); after intrathecal injection of P2Y14 shRNA, the expression of P2Y14 receptor protein and mRNA in DNP + P2Y14 shRNA group were obviously lower than those in DNP group (p < 0.01); there was no significant difference between the DNP + NC shRNA group and the DNP group (p > 0.05) (Fig. 2A, B). The co-expression of P2Y14 receptor and microglial marker OX42 was detected by immunofluorescence in the control group (Fig. 2C). The results indicated that P2Y14 receptor was co-localized with OX42, and P2Y14 receptor could be expressed in spinal microglia. Compared with Control group, the expression level of OX42 in DNP group was significantly raised (p < 0.01); after P2Y14 shRNA treatment, the expression level of OX42 was obviously reduced than that in the DNP group (p < 0.01). Compared with the DNP group, the expression of OX42 was not obviously and statistically different in the DNP + NC shRNA group (p > 0.05) (Fig. 2D).
The expression of P2Y14 receptor and OX42 in the spinal cord. A Relative expression of P2Y14 mRNA. B The expression levels of P2Y14 proteins and the bar histograms of P2Y14 proteins analysis results. C The co-expression of P2Y14 receptor and OX42 in the spinal cord was detected by immunofluorescence double labeling. The red signal means that P2Y14 is stained with TRITC. The green signal means that OX42 is stained with FITC. The yellow signal means the P2Y14 and OX42 double staining. Scale bar, 50 μm. D The expression of OX42 is detected by Western blot. The results are expressed as the mean ± SEM. n = 12 in each group. **p < 0.01 vs. Control group; ##p < 0.01 vs. DNP group. All data were statistically analyzed by one-way ANOVA
P2Y14 shRNA Downregulated the Expression of TNF-α, IL-1β, and p-p38 Protein in the Spinal Cord
Western blot was used to detect the expression of pro-inflammatory factors and p-p38 protein of each group. The results indicated that the expression levels of TNF-α and IL-1β protein in the DNP group were obviously increased than those in the Control group (p < 0.01); compared with the DNP group, after intrathecal injection of P2Y14 shRNA, the expression of TNF-α and IL-1β in DNP + P2Y14 shRNA group were obviously reduced (p < 0.01); there was no significant difference between the DNP + NC shRNA group and the DNP group (p > 0.05) (Fig. 3A, B). The expression of p38 of rats was not statistically different in each group (F = 3.798, p = 0.058). Compared with the Control group, the phosphorylation level of p38 in the DNP group was obviously raised (p < 0.01); after intrathecal injection of P2Y14 shRNA, the phosphorylation level of p38 was obviously lower than that in the DNP group (p < 0.01); the phosphorylation level of p38 was not significantly different between the DNP + NC shRNA group and the DNP group (p > 0.05) (Fig. 3C).
P2Y14 shRNA downregulated the expression levels of TNF-α, IL-1β, and p-p38 protein in the spinal cord. A Expression of IL-1β. B Expression of TNF-α. C The expression levels of p38 and p-p38. The results are expressed as the mean ± SEM using one-way ANOVA. n = 12 rats per group. **p < 0.01 vs. Control group; ##p < 0.01 vs. DNP group. All data were statistically analyzed by one-way ANOVA
The Effect and Mechanism of lncRNA-UC.25 + on P2Y14 Receptor–Mediated DNP
Determination of MWT and TWL
In order to evaluate the pain behavior of rats, the MWT and TWL were measured for each group. After successfully modeling, these data indicated that the TWL and MWT in the DNP group were obviously lower than those in the Control group (p < 0.01); after intrathecal injection of UC.25 + shRNA, the TWL and MWT of the DNP + UC.25 + shRNA group were obviously higher than those in the DNP group (p < 0.01); there were no significant difference in TWL and MWT between the DNP group and the DNP + NC shRNA group (p > 0.05). These results indicated that UC.25 + shRNA can reduce hyperalgesia threshold in DNP group rats (Fig. 4).
Expression of UC.25 + , P2Y14 Receptor, and OX42
Compared with Control group, the expressions of UC.25 + , P2Y14 receptor, and OX42 in DNP group were obviously raised (p < 0.01); after intrathecal injection of UC.25 + shRNA, the expressions of UC.25 + , P2Y14 receptor, and OX42 in DNP + UC.25 + shRNA group were obviously decreased than those in the DNP group (p < 0.01); there was no significant difference in the expression of UC.25 + , P2Y14 receptor, and OX42 between DNP + NC shRNA group and DNP group (p > 0.05) (Fig. 5A, B, C, and D).
The expression of UC.25 + , P2Y14 receptor, and OX42. A Relative expression of UC.25 + . B Relative expression of P2Y14 mRNA. C The expression levels of P2Y14 proteins. D The expression of OX42 The results are expressed as the mean ± SEM.n = 12 in each group. **p < 0.01 vs. Control group; ##p < 0.01 vs. DNP group. All data were statistically analyzed by one-way ANOVA
UC.25 + shRNA Downregulated the Expression of TNF-α、IL-1β, and p-p38 Protein in the Spinal Cord
The expression levels of inflammatory factors and p-p38 protein were detected. Those data showed that the protein expressions of TNF-α and IL-1β in DNP group were obviously higher than those in Control group (p < 0.01); after treatment with UC.25 + shRNA, the expression levels of TNF-α and IL-1β in DNP + UC.25 + shRNA group were obviously lower than those in DNP group (p < 0.01); there was no significant difference in the expression levels of IL-1β and TNF-α between the DNP + NC shRNA group and the DNP group (p > 0.05) (Fig. 6A, B). There was no significant difference in the protein expression of p38 MAPK among the groups (F(3, 8) = 3.622, p = 0.065); the expression of p-p38 MAPK in DNP group was obviously higher than that in Control group (p < 0.01); after UC.25 + shRNA treatment, the expression of p-p38 MAPK in DNP + UC.25 + shRNA group was obviously lower than that in DNP group (p < 0.01); there was no significant difference in the expression of p-p38 MAPK between DNP + NC shRNA group and DNP group (p > 0.05) (Fig. 6C).
UC.25 + shRNA downregulated the expression of TNF-α, IL-1β, and p-p38 protein in the spinal cord. A Expression of IL-1β. B Expression of TNF-α. C The expression levels of p38 and p-p38. The results are presented as the mean ± SEM. n = 12 rats per group. **p < 0.01 vs. Control group; ##p < 0.01 vs. DNP group. All data were statistically analyzed by one-way ANOVA
Determine the Interaction Between lncRNA and STAT1 and Between STAT1 and P2Y14 Receptor
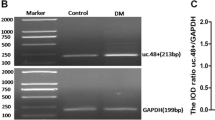
RIP results indicated that compared with the Control group, the enrichment of STAT1 protein in the experimental group was obviously increased (p < 0.01), indicating that UC.25 + could bind to STAT1, and there was obvious interaction between them. Flag-Tag (DYKDDDDK) as an internal reference (Fig. 7A). ChIP results indicated that the Input group containing the total DNA of the sample had obvious expression; compared with the Control group, the STAT1 group was specifically expressed at 290 bp, indicating that STAT1 can specifically bind to the promoter of P2Y14 gene, and there is an obvious interaction between them (Fig. 7B).
Determination of interaction. AThe interaction between UC.25 + and STAT1 protein. The results are expressed as the mean ± SEM of three independent experiments. **p < 0.01 vs. Control group. B The interaction between STAT1 protein and P2Y14 receptor。 M: DNA marker; Iuput: total DNA; Control: no antibody; STAT1: STAT1 antibody. Paired t-test was used for statistics
Regulation of UC.25 + on STAT1
HUVEC cells were cultured in high-glucose and normal environments respectively, and UC.25 + was overexpressed in normal cells and low-expressed in high-glucose cells. The effect of UC.25 + on STAT1 was detected by real-time quantitative PCR and Western blot. Results showed that UC.25 + was successfully overexpressed in HUVEC cells. Compared with Control group, the expression of UC.25 + in UC.25 + OE group transfected with UC.25 + overexpression plasmid was obviously increased (p < 0.01) (Fig. 8A). Meanwhile, the expression of STAT1 protein was obviously increased (p < 0.01) (Fig. 8B); UC.25 + was successfully low-expressed in high-glucose HUVEC cells. Compared with HG group, the expression of UC.25 + in HG + UC.25 + shRNA group transfected with UC.25 + low-expression plasmid was obviously decreased (p < 0.01) (Fig. 8C), and the expression of STAT1 protein was also obviously decreased (p < 0.01) (Fig. 8D), indicating that UC.25 + can positively regulate the expression of STAT1 in HUVEC cells.
The expression of STAT1 in HUVEC after UC.25 + overexpression and silence. A, C Relative expression of UC.25 + . B, D Expression of STAT1 proteins. The results are displayed as mean ± SEM of three independent experiments. **p < 0.01 vs. Control group (or HG group). All data were statistically analyzed by one-way ANOVA
Regulation of STAT1 on P2Y14 Receptor
To clarify the regulatory effect of STAT1 on P2Y14 receptor, HUVEC cells were cultured again in normal and high-glucose environments. STAT1 was overexpressed in normal cells and low-expressed in high-glucose cells. The effect of STAT1 on P2Y14 receptor was detected by real-time quantitative PCR and Western blot. The results showed that STAT1 was successfully overexpressed in HUVEC cells. Compared with Control group, the expression of STAT1 in STAT1 OE group transfected with STAT1 overexpression plasmid was obviously increased (p < 0.01) (Fig. 9A), and the expression of P2Y14 protein was also obviously increased (p < 0.01) (Fig. 9B). STAT1 was successfully low-expressed in high glucose HUVEC cells. Compared with the HG group, the expression of STAT1 in the HG + STAT1 shRNA group transfected with the STAT1 low-expression plasmid was obviously decreased (p < 0.01) (Fig. 9C). At the same time, the expression level of P2Y14 protein was obviously decreased (p < 0.01) (Fig. 9D), suggesting that the transcription factor STAT1 can positively regulate the expression of P2Y14 receptor in HUVEC cells.
The expression of P2Y14 receptor in HUVEC after STAT1 overexpression and silence. A, C Relative expression of STAT1 mRNA. B, D Expression of P2Y14 proteins. The results are displayed as mean ± SEM of three independent experiments. **p < 0.01 vs. Control group (or HG group). All data were statistically analyzed by one-way ANOVA
Discussion
Diabetes can bring about various injuries in the body thanks to its complications, which include DNP caused by peripheral neuropathy. The symptoms of DNP are obvious, including hyperalgesia, touch-induced pain, spontaneous pain, and hyperalgesia. Most of them can develop into chronic pain, lasting for several years [15]. Thus, it is essential to study the pathogenesis and pretreatment of DNP. In this study, P2Y14 shRNA and UC.25 + shRNA treatment increased the mechanical hyperalgesia and thermal hyperalgesia thresholds of rats, indicating that P2Y14 shRNA and UC.25 + shRNA can alleviate the pain behavior of DNP rats.
P2Y14 receptor can perform specific physiological functions by modulating different signal transduction pathways in cells [16]. Studies have confirmed that in neuropathic pain models, the expression of P2Y14 receptors was obviously increased, and the application of P2Y14 antisense locked nucleotides can relieve the pain [7]. The upregulation of P2Y12 receptors in satellite glial cells of dorsal root ganglion can be increase mechanical or thermal hyperalgesia in diabetic rats, thereby mediating the DNP process [17]. The activation of P2Y13 receptors in spinal microglia can promote the transmission of pain in diabetic rats, and the application of P2Y13 receptor–specific antagonist MRS2211 can significantly relieve pain. P2Y12, P2Y13, and P2Y14 receptors belong to Gi/o-coupled receptors with strong sequence homologies, and it has been proved that P2Y12 and P2Y13 receptors exist in spinal microglia [18]. Therefore, we speculated that the P2Y14 receptor in spinal cord may participate in the development of DNP. The data showed that the expression of P2Y14 receptor protein and mRNA in the DNP group was remarkably increased than that in the Control group. P2Y14 shRNA treatment could reduce the expression of P2Y14 receptor. Combined with the results of rat pain behavior, it indicated that the abnormal pain in rats was related to the upregulation of P2Y14 receptor. Inhibiting the expression of P2Y14 receptor could alleviate the pain behavior of rats, implying that P2Y14 receptor may participate in the DNP process. At the same time, UC.25 + shRNA treatment could reduce the expression level of P2Y14 receptor and alleviate the pain behavior of DNP rats, indicating that UC.25 + may affect the transmission of pain in rats through P2Y14 receptor.
Microglia are extremely sensitive to the change in the surrounding environment and can respond quickly. When the nerves are injured or noxiously stimulated, microglia are rapidly activated, releasing a large number of cytokines to act on neurons and alter the synaptic effects [19]. Furthermore, it is found that inhibiting the activation of microglia can effectively relieve the pain [20]. OX42 (CD11b monoclonal antibody) is a marker for identifying the activation of microglia [21]. Immunofluorescence showed that OX42 and P2Y14 receptors were co-expressed in microglia, indicating that P2Y14 receptors can be expressed in microglia. Western blot result showed that the expression of OX42 was increased obviously during DNP, indicating that microglia were activated in diabetic rats after nerve injury. P2Y14 shRNA treatment reduced the OX42 expression. The activation of microglia is accompanied by the upregulation of P2Y14 receptors. The inhibition of P2Y14 receptors diminishes the activation of microglia and reduces the transmission of pain. UC.25 + shRNA treatment reduced the expression of OX42 and P2Y14 receptor, suggesting that UC.25 + may decrease the activation of microglia by inhibiting the expression of P2Y14 receptor, thereby alleviating DNP.
Diabetes is a low-grade systemic inflammatory state [22]. When peripheral nerves are injured, the secretion of pro-inflammatory cytokines increases. The upregulated cytokines will further enhance inflammatory signals and promote the development of pathological pain [23]. The activation of microglia can also promote the secretion of inflammatory factors. This study found that the expression levels of IL-1β and TNFα in the DNP group were obviously higher than those in the control group, indicating that DNP rats released more inflammatory factors and produced a stronger inflammatory response; P2Y14 shRNA treatment can downregulate the elevated expression of inflammatory factors induced by DNP, indicating that the P2Y14 receptor is related to the release of inflammatory factors. UC.25 + shRNA suppressed the P2Y14 expression, suggesting that UC.25 + may reduce the inflammatory response by inhibiting the P2Y14 receptor.
Mitogen-activated protein kinases (MAPKs) participate in cell signal transduction. Studies have shown that during inflammatory pain, the activation of p38 MAPK can promote the synthesis and release of inflammatory factors, and p38 MAPK inhibitors can relieve pain; activation of the P2Y12 receptor in spinal microglia can over-release inflammatory factors and activate the p38 MAPK pathway, and participate in maintenance of various types of pain [18]. Our data indicated that the phosphorylation of p38 MAPK was obviously raised during DNP, and P2Y14 shRNA or UC.25 + shRNA treatment inhibited its phosphorylation, indicating that downregulated expression of P2Y14 receptor or UC.25 + could relieve the phosphorylation of p38 MAPK.
Recent studies have verified that lncRNAs can participate in the development of diseases [24]. The dysregulation of lncRNAs may be associated with neuropathic pain [25]. Our results indicated that UC.25 + shRNA inhibited the expression of P2Y14 receptor, alleviated the DNP process.
In order to further investigate the potential mechanism of UC.25 + regulating the P2Y14 receptor, the screening results through multiple databases showed that the P2Y14 promoter region contained the response element of STAT1, suggesting that the transcription factor STAT1 may participate in the regulation of P2Y14 receptor expression. In this study, the RIP results showed that UC.25 + can enrich STAT1, indicating that there is an interaction between UC.25 + and STAT1. The overexpression or low-expression of UC.25 + in HUVEC cells respectively increased or decreased the expression of STAT1, suggesting that UC.25 + could positively regulate the expression of STAT1. In addition, ChIP results showed that STAT1 could specifically bind to the response element of P2Y14 gene promoter. Similarly, overexpression or low-expression STAT1 in HUVEC cells enhanced or reduced the expression of P2Y14, implying that STAT1 could positively regulate the expression of P2Y14 receptor. In summary, UC.25 + can enrich STAT1 and positively regulate the expression of STAT1. As a transcription factor of the P2Y14 receptor, STAT1 positively regulates the expression of the P2Y14 receptor, thereby promoting the occurrence and development of P2Y14 receptor–mediated DNP.
Data Availability
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N et al (2019) Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract 157:107843. https://doi.org/10.1016/j.diabres.2019.107843
Li Y, Teng D, Shi X, Qin G, Qin Y, Quan H et al (2020) Prevalence of diabetes recorded in mainland China using 2018 diagnostic criteria from the American Diabetes Association: national cross sectional study. BMJ 369:m997. https://doi.org/10.1136/bmj.m997
Duehmke RM, Derry S, Wiffen PJ, Bell RF, Aldington D, Moore RA (2017) Tramadol for neuropathic pain in adults. Cochrane database of systematic reviews 6:CD003726. https://doi.org/10.1002/14651858.CD003726.pub4
Wang D, Couture R, Hong Y (2014) Activated microglia in the spinal cord underlies diabetic neuropathic pain. Eur J Pharmacol 728:59–66. https://doi.org/10.1016/j.ejphar.2014.01.057
Tsuda M, Inoue K (2016) Neuron-microglia interaction by purinergic signaling in neuropathic pain following neurodegeneration. Neuropharmacology 104:76–81. https://doi.org/10.1016/j.neuropharm.2015.08.042
Rahman MH, Jha MK, Suk K (2016) Evolving insights into the pathophysiology of diabetic neuropathy: implications of malfunctioning glia and discovery of novel therapeutic targets. Curr Pharm Des 22(6):738–757. https://doi.org/10.2174/1381612822666151204001234
Kobayashi K, Yamanaka H, Yanamoto F, Okubo M, Noguchi K (2012) Multiple P2Y subtypes in spinal microglia are involved in neuropathic pain after peripheral nerve injury. Glia 60(10):1529–1539. https://doi.org/10.1002/glia.22373
Ji RR, Berta T, Nedergaard M (2013) Glia and pain: is chronic pain a gliopathy? Pain 154(Suppl 1):S10-28. https://doi.org/10.1016/j.pain.2013.06.022
Franke H, Verkhratsky A, Burnstock G, Illes P (2012) Pathophysiology of astroglial purinergic signalling. Purinergic Signal 8(3):629–657. https://doi.org/10.1007/s11302-012-9300-0
Weirick T, Militello G, Muller R, John D, Dimmeler S, Uchida S (2016) The identification and characterization of novel transcripts from RNA-seq data. Brief Bioinform 17(4):678–685. https://doi.org/10.1093/bib/bbv067
Melissari MT, Grote P (2016) Roles for long non-coding RNAs in physiology and disease. Pflugers Arch 468(6):945–958. https://doi.org/10.1007/s00424-016-1804-y
Li Z, Li X, Chen X, Li S, Ho IHT, Liu X et al (2019) Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif 52(1):e12528. https://doi.org/10.1111/cpr.12528
Li G, Sheng X, Xu Y, Jiang H, Zheng C, Guo J et al (2017) Co-expression changes of lncRNAs and mRNAs in the cervical sympathetic ganglia in diabetic cardiac autonomic neuropathic rats. J Neurosci Res 95(8):1690–1699. https://doi.org/10.1002/jnr.24000
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS et al (2004) Ultraconserved elements in the human genome. Science 304(5675):1321–1325. https://doi.org/10.1126/science.1098119
Schreiber AK, Nones CF, Reis RC, Chichorro JG, Cunha JM (2015) Diabetic neuropathic pain: Physiopathology and treatment. World J Diabetes 6(3):432–444. https://doi.org/10.4239/wjd.v6.i3.432
von Kugelgen I, Hoffmann K (2016) Pharmacology and structure of P2Y receptors. Neuropharmacology 104:50–61. https://doi.org/10.1016/j.neuropharm.2015.10.030
Jia T, Rao J, Zou L, Zhao S, Yi Z, Wu B et al (2017) Nanoparticle-encapsulated curcumin inhibits diabetic neuropathic pain involving the P2Y12 receptor in the dorsal root ganglia. Front Neurosci 11:755. https://doi.org/10.3389/fnins.2017.00755
Tatsumi E, Yamanaka H, Kobayashi K, Yagi H, Sakagami M, Noguchi K (2015) RhoA/ROCK pathway mediates p38 MAPK activation and morphological changes downstream of P2Y12/13 receptors in spinal microglia in neuropathic pain. Glia 63(2):216–228. https://doi.org/10.1002/glia.22745
Eyo UB, Peng J, Murugan M, Mo M, Lalani A, Xie P et al (2016) Regulation of physical microglia-neuron interactions by fractalkine signaling after status epilepticus. eNeuro. 3(6) https://doi.org/10.1523/ENEURO.0209-16.2016
Inoue K, Tsuda M (2018) Microglia in neuropathic pain: cellular and molecular mechanisms and therapeutic potential. Nat Rev Neurosci 19(3):138–152. https://doi.org/10.1038/nrn.2018.2
Chamera K, Trojan E, Szuster-Gluszczak M, Basta-Kaim A (2020) The potential role of dysfunctions in neuron-microglia communication in the pathogenesis of brain disorders. Curr Neuropharmacol 18(5):408–430. https://doi.org/10.2174/1570159X17666191113101629
Poreba M, Rostoff P, Siniarski A, Mostowik M, Golebiowska-Wiatrak R, Nessler J et al (2018) Relationship between polyunsaturated fatty acid composition in serum phospholipids, systemic low-grade inflammation, and glycemic control in patients with type 2 diabetes and atherosclerotic cardiovascular disease. Cardiovasc Diabetol 17(1):29. https://doi.org/10.1186/s12933-018-0672-5
Zhu MD, Zhao LX, Wang XT, Gao YJ, Zhang ZJ (2014) Ligustilide inhibits microglia-mediated proinflammatory cytokines production and inflammatory pain. Brain Res Bull 109:54–60. https://doi.org/10.1016/j.brainresbull.2014.10.002
Peng H, Zou L, Xie J, Wu H, Wu B, Zhu G et al (2017) lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Mol Neurobiol 54(1):511–523. https://doi.org/10.1007/s12035-015-9632-1
Jiang BC, Sun WX, He LN, Cao DL, Zhang ZJ, Gao YJ (2015) Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain 11:43. https://doi.org/10.1186/s12990-015-0047-9
Funding
This study was supported by grants from the National Natural Science Foundation of China (81860217, 82160163, 81560219, 81870574, and 81200853), and a grant from the Key Research and Development programs of Jiangxi Province (20192BBH80017).
Author information
Authors and Affiliations
Contributions
Research project conception: Guilin Li, Shangdong Liang. Execution of experimental procedures: Baoguo Wu, Congfa Zhou, Zehao Xiao, Gan Tang. Data analysis: Hongmin Guo, Zihui Hu, Qixing Hu, Hao Peng, Lingzhi Pi, Zhihua Zhang, Miaomiao Wang, Taotao Peng, Jiaqi Huang. Interpretation of results: Baoguo Wu, Congfa Zhou, Hongmin Guo, Zihui Hu. Draft of the manuscript: Baoguo Wu, Gan Tang, Shangdong Liang, Guilin Li. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Animal experiments were approved by the Institutional Animal Investigation Committee of the Nanchang University and were performed in accordance with the Guidelines for Animal Experiments at Nanchang University.
Consent to Participate
No human subjects were involved in this research, so consent to participate is not relevant.
Consent for Publication
All authors have approved this manuscript and consented to its submission for publication.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Baoguo Wu and Congfa Zhou are both first authors.
Rights and permissions
About this article
Cite this article
Wu, B., Zhou, C., Xiao, Z. et al. LncRNA-UC.25 + shRNA Alleviates P2Y14 Receptor–Mediated Diabetic Neuropathic Pain via STAT1. Mol Neurobiol 59, 5504–5515 (2022). https://doi.org/10.1007/s12035-022-02925-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02925-0