Abstract
18F-fluorodeoxyglucose (FDG) positron emission tomography (PET) and functional magnetic resonance imaging (fMRI) estimate brain activities from different aspects, including regional glucose uptake (rGU) by 18FDG-PET, regional cerebral blood flow (rCBF) by arterial spin labeling, and dynamic changes of deoxyhemoglobin by blood oxygenation level-dependent (BOLD) functional magnetic resonance imaging (fMRI). However, the relationships between them remain incompletely understood. In the current study, twenty-four subjects (14 males, 10 females) were recruited and investigated the correlation among rGU, rCBF, and BOLD fMRI-derived metrics reflecting the neural activity, including amplitude of low-frequency fluctuation (ALFF), regional homogeneity (ReHo), and degree centrality (DC) by hybrid PET/fMRI. Correlation analyses were performed across subject and across space at both voxel level and region level, considering partial volume effects by adjusting for gray matter volume. Each pair of metrics showed significant across-space correlations. rGU against ReHo showed the highest mean correlation coefficients. rGU had higher correlations with three resting-state (RS) fMRI metrics than did ASL-rCBF. However, the across-subject correlations were not significant among functional modalities (rGU, rCBF, and RS-fMRI BOLD data) at either voxel level or region level even with a liberal threshold, except for significant across-subject correlation between RS-fMRI metrics (ALFF, ReHo, and DC). These comprehensive findings from hybrid PET/MR might provide complementary information to reveal the underlying mechanisms of the brain activity and open new perspective to interpret pathologic conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hybrid scans of positron emission tomography (PET) and magnetic resonance imaging (MRI) are becoming increasingly popular for clinical diagnosis. PET and functional MRI (fMRI) are two of the most frequently used imaging techniques for non-invasively mapping the human brain function. Both 18F-fluorodeoxyglucose (FDG)-PET and fMRI estimate brain activities from different aspects, such as energy consumption, low-frequency fluctuation, and regional cerebral blood flow (rCBF). However, the complex relationships among energy consumption, rCBF, and other fMRI metrics remain unknown.
FDG-PET is the most commonly used technique to assess regional glucose uptake (rGU), which reflects cellular activity by measuring glucose uptake. One method used in fMRI is referred to as blood oxygenation level-dependent (BOLD) imaging, which reflects the oxygen absorption by measuring the ratio changes of oxygenated to deoxygenated hemoglobin [1]. Resting-state functional MRI (RS-fMRI; i.e., without specific cognitive task during scanning [2]), has been widely used to investigate functional activities, including amplitude of low-frequency fluctuation (ALFF) [3], fractional ALFF (fALFF) [4] of every single voxel, regional homogeneity (ReHo) for measuring local synchronization of nearest neighboring voxels [5], and degree of centrality (DC) for measuring global number of functional connections between a given voxel with all voxels in the brain [6]. The vast majority of FDG-PET studies have analyzed only the local activity, and a few FDG-PET studies have investigated metabolic connectivity [7, 8]. Although several studies have correlated local glucose metabolism (e.g., rGU) and RS-fMRI functional connectivity [9, 10], correlation between FDG-PET and fMRI-derived metrics (e.g., fALFF, ReHo, and DC) is more straight-forward. For example, it has been consistently reported that the spatial patterns of rGU and fMRI-derived metrics were very similar, but the voxel-level correlation across subjects was very low [11,12,13].
Arterial spin labeling (ASL) is a perfusion-based functional imaging, which reflects tissue perfusion by magnetically labeled water protons in the blood as an endogenous contrast agent. Both BOLD signals and ASL-rCBF are based on hemodynamic changes. BOLD is sensitive to changes in deoxyhemoglobin [1], whereas ASL is sensitive to changes in rCBF [14]. Although the scanning sequences of ASL permit time series analyses as in BOLD signal analysis (e.g., ALFF and ReHo) [15], the most popular metric for ASL is the mean rCBF over a period of time (e.g., 10 min), just as rGU for PET analysis. A few concurrent PET-ASL studies have reported that certain brain diseases showed similar yet distinct abnormal patterns of rGU and ASL [16].
Although some efforts have been made, further gaps need to be filled. Considering various physiological processes may change between these imaging sessions, simultaneous multimodal information acquisition is the major challenge to investigate the relationship between PET and fMRI. The advanced hybrid PET/MRI solves this problem by offering simultaneous glucose metabolism and RS-fMRI under the same physiological condition, which further provide optimal spatial and temporal registration of both modalities [11].To the best of our knowledge, no concurrent multimodal study has simultaneously collected PET, ASL, and BOLD fMRI data and investigated the complex mutual relationships to elucidate the underlying neurovascular coupling.
In the current study, we simultaneously collected rGU, ASL, and BOLD fMRI data from a group of healthy participants and aimed to investigate across-space correlation as well as across-subject correlation of the voxel-wise metrics between each pair of functional modalities. In addition, we compared the relative values between each pair of functional modalities. We speculated that the three techniques have similar spatial distribution with regional variability in the healthy subjects. These results will help us better understand the pathophysiology of brain disorders, especially when multiple functional imaging modalities are recorded.
Materials and Methods
Subjects
Twenty-four subjects (14 males, 10 females, age 21 ~ 60 years, average age 44.5 ± 8.2 years, fasting venous glucose 6.0 ± 0.7 mmol/L, body mass index 25.2 ± 2.8 kg/m2, all right-handed) were recruited among those referred to whole-body PET/MRI scan for tumor screening in Xuanwu Hospital Capital Medical University and finally proven to be healthy. The inclusion criteria included no brain abnormalities apart from age-related leukoaraiosis, no history of psychiatric or serious medical disorders, and no contradictions to MRI scanning. Before undergoing the current study, written informed consent was obtained from all subjects. The study was approved by the ethics committee of Xuanwu Hospital.
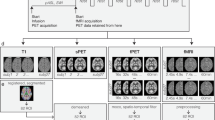
Image Acquisition
One session, including a PET scan, an ASL scan, a BOLD scan, and a T1 scan, was conducted in an integrated GE Signa PET/MR scanner designed with a multi-ring LBS detector block embedded into a 3-T magnetic resonance scanner with a 19-channel phase-array head coil. Subjects were required to fast for at least 6 h before scanning. Subjects who had a fasting venous glucose >140 mg/dL were excluded. The injection dosage of 18F-FDG for each subject was 3.7 MBq/kg × body weight. With a bolus injection of FDG tracer, we simultaneously started PET/fMRI data acquisition. The total time is 1 h. Static PET images were reconstructed from the list-mode data acquired 40–50 min post-injection. During PET acquisition, the following MRI sequences were run sequentially: BOLD, ASL, diffusion weighted images (DWI), diffusion tensor images (DTI), T1-MPRAGE, T2 FLAIR, and T2-FSE. All scanning were performed in a dimmed environment to avoid unnecessary influence. During the scanning, subjects were instructed to keep their eyes open, avoid focusing their minds on anything, and keep their heads as still as possible.
FDG-PET Scanning
FDG-PET images were scanned using the following parameters: 10 min list-mode acquisition, 89 slices (gap = 0 mm) covering the whole brain; field of view (FOV) = 250 × 250 mm2, matrix = 192 × 192; voxel size = 1.82 × 1.82 × 2.78 mm3; scatter and attenuation correction, reconstructed with a time of flight, ordered subset-expectation maximization (TOF-OSEM) algorithm (8 iterations, 32 subsets), and post-filtered with isotropic full-width half-maximum (FWHM) gaussian kernel of 3 mm.
BOLD Scanning
BOLD data were obtained by gradient-echo echo-planar imaging (EPI) sequence with parameters of TR = 2000 ms, TE = 30 ms, FOV = 224 × 224 mm2, matrix = 64 × 64, pixel size = 3.5 × 3.5 mm2, 33 interleaved slices with thickness = 3.6 mm, 0.8 mm gap between slices, and 240 timepoints. The total scan time was 8 min.
ASL Scanning
A pseudo-continuous ASL (pCASL) perfusion imaging sequence was utilized with the following parameters: single-shot gradient-echo EPI in combination with parallel imaging (SENSE factor 2.0), TR = 4852 ms, TE = 10.7 ms, matrix = 64 × 64, FOV = 240 × 240 mm2, voxel size = 1.88 × 1.88 × 4 mm3, 32 slices, slice thickness/gap = 4/0 mm, post-spin labeling delay = 2025 ms, and scanning time = 4 min and 24 s. Thirty-two pairs of control/labeled images were acquired and averaged. An EPI M0 image was obtained separately with the same geometry and the same imaging parameters as the pCASL without labeling, for measurement of the magnetization of arterial blood and for co-registration purposes.
3D-T1 Scanning
T1-weighted, three-dimensional (3D) images were acquired with the following parameters: TR = 1900 ms, TE = 3.24 ms, FOV = 256 × 256 mm2, matrix = 256 × 256, 192 sagittal slices, slice thickness = 1 mm with no gap, and scanning time = 5 min and 21 s.
The clinical routine scanning sequences, including T1, T2, FLAIR, and DWI, were also scanned. No visible structural abnormalities were found.
Image Processing
DPABI [17], SPM12 (https://www.fil.ion.ucl.ac.uk/spm), Analysis of Functional NeuroImages (AFNI [18]), and MATLAB 2014b (Mathworks Inc.) were used for data processing. Considering all data acquired during the same session in the hybrid scanner, the individual imaging data are co-registered. Multimodal imaging normalization are described in our previous studies [19]. Briefly, PET, BOLD, and ASL data were first co-registered to a 3D, high-resolution T1 image. The co-registered T1 images were then segmented by means of a new circular segment and normalization step implemented in SPM12. The transformation parameters derived from MRI spatial normalization were then applied to the co-registered PET, BOLD, and CBF images for spatial normalization to standard template of the Montreal Neurological Institute (MNI). Imaging processing is detailed as follows.
ASL-rCBF
The ASL images were transferred to AW 4.7 workstation (Signa, GE) for further CBF calculation. CBF imaging processing was performed by SPM12, including (1) co-registering 3D-T1 image to co-registered M0 image using a process of 12 parameter affine transformations and rigid-body transformation, (3) spatial normalization to MNI standard space using transformation parameters derived from MRI spatial normalization, (4) resampling to 3-mm isotropic voxels, and (5) spatial smoothing (FWHM = 6 mm).
FDG-PET
SPM12 was used to process FDG-PET images, including (1) co-registering 3D-T1 image to FDG-PET image using a process of 12 parameter affine transformations and rigid-body transformation, (2) spatial normalization to the MNI standard space by the transformation parameters derived from MRI spatial normalization, (3) resampling to 3-mm isotropic voxels, and (4) spatial smoothing (FWHM = 6 mm).
BOLD fMRI
DPABI was used to process BOLD fMRI images. The preprocessing included (1) discarding the first 10 time points for the instability of the initial MRI signal and subject’s adaptation to inherent scanner noise, (2) slice timing, (3) head motion correction (one subject was excluded from the present study due to head motion more than 2.5 mm of maximum displacement in the x, y, or z translation or 2.5° of angular motion during the entire fMRI scan), (4) co-registering individual T1 image of the remaining 23 subjects to the individual averaged fMRI image, (5) spatial normalization, (6) resampling to 3-mm isotropic voxels, (7) removing the linear trend within the time series, and (8) regressing out Friston-24 head motion parameters.
After preprocessing, three metrics (fALFF, ReHo, and DC) were calculated as follows:
-
The fALFF quantifies the amplitude of low-frequency oscillations of each single voxel. It is the fraction of the amplitude within the low-frequency range (0.01–0.1 Hz) to that of the entire detectable frequency range [4].
-
ReHo estimates the degree of local synchronization among the fMRI time series of neighboring voxels. The ReHo calculation is performed as in [5]. It is the Kendall’s coefficient of concordance (KCC) of the time series of every set of 27 neighboring voxels. The KCC value was given to the central voxel.
-
DC represents the functional strength of a given voxel within all voxels in the brain. The DC calculation is performed as in [20, 21]. Specifically, a threshold of r > 0.25 was used, and the number of positive correlations was counted and then assigned to each voxel.
-
All metrics were calculated within a frequency band of 0.01–0.1 Hz. Of note, spatial smoothing (FWHM = 6 mm) was performed before fALFF calculation, but after ReHo and DC calculations [17].
Intersection Mask
An intersection mask was made to constrain further analyses. As shown in Fig. 1, it was made from two masks, one named GM05 mask and one named FOV mask. The GM05 mask was obtained by thresholding (>0.5) the averaged gray matter probability images of all subjects and then resampling to 3-mm isotropic voxels. Each subject’s gray matter probability image was obtained from the spatial normalization procedure of the 3D-T1 image. The FOV mask was made with a consideration that the original FOV for ASL and BOLD scanning did not cover the entire brain for a few subjects. After spatial normalization, all images (ASL-rCBF, rGU, mean BOLD) were transformed to binary images. It should be noted that the mean BOLD image was the averaged image over the 230 images. The intersection part of the binary images of all ASL and mean BOLD images with the whole brain mask provided by DPABI was named as the FOV mask. Further, the intersection part of the GM05 mask and FOV mask was named as the intersection mask [22].
Generation of the intersection mask. GM05 mask was obtained by thresholding (> 0.5) the averaged gray matter probability images of all subjects. FOV mask was the brain region covered by the field of view of all ASL and mean BOLD images with the whole brain mask provided by DPABI. Intersection mask was the intersection of GM05 mask and FOV mask. The Z coordinates were from −45 to +65 with a step of 10 mm
Z-Standardization
Each individual image of ASL-rCBF, rGU, fALFF, ReHo, and DC was standardized into a Z-map as follows,
where V is the value of a given metric at a given voxel, and M and SD are the mean and standard deviation, respectively, of a given metric of each subject within the intersection mask.
Statistical Analyses
All statistical analyses were performed in the intersection mask.
One-Sample t Tests of Each Spatial Pattern and Paired t Tests on Each Pair of Metrics
To assess whether a voxel’s value was significantly higher than the mean value (mean z value = 0), one-sample t tests were performed at each voxel for each of the five Z-maps (ASL-rCBF, rGU, fALFF, ReHo, and DC), p < 0.001, uncorrected. Then, paired t tests of each pair of ASL-rCBF, rGU, fALFF, ReHo, and DC were performed to detect the difference between two metrics, p < 0.05, FDR correction.
Correlation Analyses
We performed two kinds of correlation analyses: across-space correlation and across-subject correlation.
Across-space correlation measures the similarity of the spatial patterns in two images in a subject-by-subject manner. To reduce potential partial volume effects (PVE), partial correlation analysis was performed in the individual GM08 mask with gray matter fraction as nuisance covariate. Specifically, the individual GM08 mask was made by thresholding (> 0.8) the gray matter probability image of each subject. We performed two types of across-space correlations. One was voxel level, in which the sample size of n is the total number of voxels in each individual GM08 mask (n = 19,722 ± 866 voxels, varied across participants). The other across-space correlation was region level, in which the sample size of n is the total number of brain regions (n = 90). These 90 brain regions were from the automated anatomical labeling (AAL) template [22]. The mean values of each metric and gray matter fraction were extracted within each region. Partial correlation analysis was performed with gray matter fraction of each region as nuisance covariate. To test whether the across-space correlation was significant, the correlation coefficient r value was transformed into the z value, and one-sample t tests were performed on the z values against zero. Statistical significance level was set at p < 0.001.
It should be noted that the across-space correlation can be performed not only for each pair of images between a single participant (e.g., rGU against ReHo of the same participant A1 or A1, i = 23), but also for the rGU of participant A1 against the ReHo of participant A2 (and vice versa). There were thousands of potential pairwise correlations between different participants (23 participants and 5 image metrics). We thus performed correlations for rGU against ReHo because it was reported that rGU showed higher across-space correlation with ReHo than other RS-fMRI metrics [12, 13]. Only the correlations between rGU of participant Ai against the ReHo of participant Ai + 1(i = 1…22) and between rGU of participant A23 against the ReHo of participant A1 were performed.
Across-Subject Correlation
For each imaging modality and metric, volumes were concatenated across all subjects to reflect the degree to which the different modalities show the linear correlation among subjects. The sample size of n is the total number of subjects in the current study (originally, but one was excluded due to excessive head motion; therefore, n = 23). We performed two types of across-subject correlation. The first was voxel level, in which the partial correlation was performed within the GM05 mask with gray matter fraction of each voxel as nuisance covariate. Another was region level, and the correlation was performed within the GM05 mask with gray matter fraction of each region as nuisance covariate. In the region-level correlation analysis, the mean value of each metric was calculated in each of the 90 AAL regions. Statistical significance level was set at p < 0.01.
Results
Spatial Patterns
As shown in Fig. 2, all metrics (ASL-rCBF, rGU, fALFF, ReHo, and DC) showed overall similar spatial patterns: higher activities in the medial prefrontal cortex, superior temporal gyrus, middle temporal gyrus, posterior cingulate cortex, precuneus, and calcarine fissure.
One-sample t tests on ASL-rCBF, rGU, fALFF, ReHo, and DC (p < 0.001, uncorrected). The Z coordinates were from −45 to +65 with a step of 10 mm. ASL, arterial spin labeling; rCBF, regional cerebral blood flow; rGU, regional glucose metabolism; fALFF, fractional amplitude of low-frequency fluctuations; ReHo, regional homogeneity; DC, degree of centrality
Paired t tests showed significant differences between pairwise metrics (Fig. 3). Only rGU exhibited higher activity in the basal ganglia (e.g., putamen). In the bilateral hippocampus, the signals for rCBF were higher than those of rGU, fALFF, and ReHo. The insula exhibited higher activity for ASL-rCBF and rGU than for other RS-fMRI metrics.
Paired t tests of each pair of ASL-rCBF, rGU, fALFF, ReHo, and DC (p < 0.05, FDR corrected). The Z coordinates were from −45 to +65 with a step of 10 mm. ASL, arterial spin labeling; rCBF, regional cerebral blood flow; rGU, regional glucose metabolism; PET, positron emission tomography; fALFF, fractional amplitude of low-frequency fluctuations; ReHo, regional homogeneity; DC, degree of centrality
Across-Space Correlation
For both voxel-level and region-level across-space correlation analyses, rGU against ReHo had the highest mean correlation coefficients (Fig. 4). rGU had higher correlations with the three RS-fMRI metrics than did ASL-rCBF. Even the correlation coefficients between pairs of different participants were only slightly smaller than that for the same participants (Fig. 4, last column).
Across-Subject Correlation
For the voxel-level across-subject correlation analyses, only a very small portion of the voxels survived the FDR correction for correlation between rGU, ASL-rCBF, and RS-fMRI metrics (Fig. 5), even by a liberal threshold (p < 0.05) without correction (Fig. S1). Table 1 shows voxels with significant correlation between pairwise metrics. Most of the voxels showed significant pairwise across-subject correlations between three RS-fMRI metrics in the brain. The region-level across-subject correlation analyses showed similar results as voxel-level analyses; i.e., no region survived multiple comparison corrections (0.05/900 = 0.00005556) for ASL-rCBF or rGU with other metrics. However, as expected, many regions showed significant region-level across-subject correlation among the three RS-fMRI metrics (Fig. 6).
Across-subject voxel-level correlation analysis of each pair of ASL-rCBF, rGU, fALFF, ReHo, and DC (p < 0.01, FDR corrected). The Z coordinates were from −45 to +65 with a step of 10 mm. ASL, arterial spin labeling; rCBF, regional cerebral blood flow; rGU, regional glucose metabolism; fALFF, fractional amplitude of low-frequency fluctuations; ReHo, regional homogeneity; DC, degree of centrality
Across-subject region-level correlation analysis of each pair of ASL-rCBF, rGU, fALFF, ReHo, and DC (p < 0.05). Abbreviations listed in column are the AAL regions in alphabetical order. ASL, arterial spin labeling; rCBF, regional cerebral blood flow; rGU, regional glucose metabolism; fALFF, fractional amplitude of low-frequency fluctuations; ReHo, regional homogeneity; DC, degree of centrality
Discussion
To the best of our knowledge, this is the first research investigating the relationships among three functional brain imaging modalities, rGU, ASL-CBF, and BOLD fMRI, by simultaneous PET/MR. Our results demonstrated that, in general, each pair of functional metrics showed significant across-space correlation; however, few regions showed significant across-subject correlations for any pair of functional imaging metrics. rGU against ReHo showed the highest mean correlation coefficients. rGU had higher correlations with three RS-fMRI metrics than did ASL-rCBF.
Spatial Patterns Analyses
On visual inspection, the overall spatial distributions are similar among ASL-rCBF, rGU, and BOLD metrics, which is consistent with the findings reported in the literature [11, 23]. Paired t tests between pairwise metrics revealed significant differences in the brain regions such as the putamen, insula, and bilateral hippocampus. We found that the rGU was significantly higher than all other metrics in the putamen. The insula showed higher signals for ASL-rCBF and rGU than for other RS-fMRI metrics. Cha et al. [23] reported that rGU exhibits higher activity in the basal ganglia (e.g., putamen) than CBF. Significant differences among CBF, rGU, and RS-fMRI metrics should be interpreted within the context of the specific physiological processes of each modality. The majority of ASL signal should be from the extracellular space. In contrast, PET captures signal driven by a dynamic balance between different cells [24]. RS-fMRI metrics stem from complex relationships among hemodynamic parameters, including CBF, CBV, and CMRO2 [12]. In addition, RS-fMRI metrics are focused on only a single frequency band of 0.01–0.08 Hz [19]. These factors may be associated with the observed regional differences among CBF, rGU, and RS-fMRI metrics.
Similarity by Across-Space Correlation Analyses
Although the three imaging techniques reflect brain function from different aspects, they are interdependent. Resting-state glucose and oxygen metabolism are closely linked and physiologically related to neural activity. Moreover, both processes are in turn strongly related to resting CBF that delivers O2 and glucose to the tissue. Many groups have done some works about the relationship between these techniques and found that a few functional neuroimaging metrics, including glucose metabolism, rCBF, and voxel-wise metrics of RS-fMRI (e.g., ReHo, fALFF, and DC), showed similar distribution patterns across the brain. For example, these metrics exhibit higher activity in the brain regions of the default mode network [4, 11,12,13, 25, 26]. Such conclusions are supported by each metric’s mean map averaged across all subjects [11], one-sample t test results (Fig. 2, [12]), and voxel-level or region-level across-space correlation analysis (Fig. 4, [11, 12, 27]). Previous PET-fMRI studies have reported that spatial correlations were more prominent among RS-fMRI metrics [11, 12], which is in line with our results.
We found that rGU against ReHo showed the highest mean correlation coefficients. The results are consistent with those reported by Bernier [13], who correlated voxel-wise and regional-wise ReHo, ALFF, and regional global connectivity with cerebral metabolic rate of glucose, finding stronger across-space correlation between glucose metabolism and ReHo than between glucose metabolism and other RS-fMRI metrics. It should be noted that such across-space correlation could be performed between a pair of images (e.g., rGU and ReHo) from not only the same participant, but also two different participants (e.g., rGU of participant 1 with ReHo of participant 2). We thus performed the across-space correlation between rGU of one participant with the ReHo of another participant. The mean correlation coefficients were slightly smaller than those of within-participant comparisons for both voxel and region levels. This suggests that the spatial distribution of different functional modalities was similar either within or across participants.
Similarity and Dissimilarity by Across-Subject Correlation Analyses
Despite the high across-space correlation among functional imaging modalities, the across-subject correlation analyses showed that only the three RS-fMRI metrics showed significant correlation with each other in some brain areas. Neither rGU nor ASL-rCBF showed significant correlation with any other functional imaging modality. Previous studies have reported that a few regions were exhibited significant across-subject correlation [11, 13, 28], which was consistent with our results. We adopted a liberal threshold (p < 0.05) for additional analyses. As shown in Fig. 6, there were significant correlations among the modalities for a few regions. Such discrepancy among modalities was also shown by the results of paired t tests, as we report widespread differences for each pair of comparison.
One reason for the dissimilar results from across-subject correlations may be the different physiological mechanisms. rGU reflects the integrated value or mean value of the glucose metabolism over tens of minutes, ASL-rCBF reflects the mean value of cerebral blood flow, and RS-fMRI reflects the dynamic changes in deoxyhemoglobin. At first, for a given time series, the dynamic change is much different from the mean value from a computational view. As an example, the RS-fMRI metric ALFF is mathematically similar to the standard deviation. We simulated 100 time series and calculated the ALFF and mean value. We then calculated the Pearson linear correlation coefficient between them, and it was not significant (r = 0.05, p = 0.613).
Dissimilarity between mean value and dynamic change may have physiological and pathophysiological relevance. The static and dynamic characteristics of rCBF have been used to investigate the brain activity changes during eyes-open states compared to eyes-closed states using pulsed ASL techniques [29]. The study reported that BOLD-ALFF produced similar results to rCBF-ALFF. However, different computations in the same modality (e.g., rCBF-ALFF and rCBF-mean) demonstrated very different patterns of brain activity changes between eyes-open and eyes-closed states. The discrepancy between mean value and dynamic change may help to understand the pathophysiology of epilepsy. For example, a meta-analysis of rGU confirmed significant hypometabolism in the medial temporal lobe in patients with mesial temporal lobe epilepsy (MTLE) vs. healthy subjects [30], while at least two studies have reported increased BOLD-ALFF in patients with MTLE [31, 32]. The decreased glucose metabolism might be due to a decrease in neurons [33], while the increased ALFF might result from increased spikes of epileptic neurons [32]. These discrepancies have not been reported in simultaneous PET-fMRI studies.
Dissimilarity Between rGU and ASL-rCBF by Across-Subject Correlation Analyses
The aforementioned interpretation of discrepancy between mean value (e.g., rGU and ASL-rCBF) and dynamic change (e.g., RS-fMRI BOLD metrics) may not be applicable for the correlation between rGU and ASL-rCBF in the current study because rGU and ASL-rCBF both measure the mean value over a period of time. A previous study collected rGU and ASL-rCBF separately in a group of participants and performed region-level across-subject correlation [23]. The authors divided the whole brain into 11 larger regions and found significant correlations between rGU and ASL-rCBF in 4 regions (frontal, temporal, putamen, and caudate, with r values 0.52–0.76). In our study, significant correlation between rGU and ASL-rCBF was found in a few regions by a liberal threshold (p < 0.05), without correlation. But there were no regions that survived multiple comparison corrections. Although both rGU and ASL-rCBF measure the mean value over time, we cannot conclude with confidence that rGU has high correlation with ASL-rCBF across subjects. Unlike the interpretation of dissimilarity between mean value of rGU or ASL-rCBF and the dynamic changes of BOLD RS-fMRI metrics, the reasons for the dissimilarity between the mean value of rGU and mean value of ASL-rCBF may be complicated. ASL-rCBF and FDG-PET rGU have different physiological mechanisms and may be affected by different factors. For example, ASL-rCBF is affected by transit time and post-labeling time, while rGU and RS-fMRI metrics are not affected by these factors [23].
A few limitations in the current study should be addressed. First, the sample size is not large, and this may explain why no significant results were found in the across-subject analysis. Second, although we used a gray matter mask and partial correlation analysis to reduce PVE, the RS-fMRI metrics of ReHo and DC must be calculated before masking and hence can be affected by PVC. Third, part of the cerebellum was not covered in some participants. Therefore, the current study is not a true “whole” brain analysis.
Conclusions
We simultaneously recorded rGU, rCBF, and RS-fMRI BOLD data and found significant across-space correlations at both voxel level and region level for all pairs of metrics (rGU, rCBF, ALFF, ReHo, and DC), but no significant across-subject correlations were found between modalities (rGU, rCBF, and BOLD fMRI) except for within RS-fMRI (ALFF, ReHo, and DC). The dissimilarities among functional imaging modalities may reveal different aspects of pathophysiology in brain disorders. These comprehensive findings from hybrid PET/MR might provide complementary information to reveal the underlying mechanisms of the brain activity and basis for further exploring the relationship in pathologic conditions.
Data Availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ogawa S, Lee TM, Kay AR, Tank DW (1990) Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A 87(24):9868–9872. https://doi.org/10.1073/pnas.87.24.9868
Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995) Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34(4):537–541. https://doi.org/10.1002/mrm.1910340409
Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, Liang M, Tian LX, Jiang TZ et al (2007) Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain and Development 29(2):83–91. https://doi.org/10.1016/j.braindev.2006.07.002
Zou QH, Zhu CZ, Yang Y, Zuo XN, Long XY, Cao QJ, Wang YF, Zang YF (2008) An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. J Neurosci Methods 172(1):137–141. https://doi.org/10.1016/j.jneumeth.2008.04.012
Zang Y, Jiang T, Lu Y, He Y, Tian L (2004) Regional homogeneity approach to fMRI data analysis. NeuroImage 22(1):394–400. https://doi.org/10.1016/j.neuroimage.2003.12.030
Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA et al (2009) Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29(6):1860–1873. https://doi.org/10.1523/JNEUROSCI.5062-08.2009
Vanicek T, Hahn A, Traub-Weidinger T, Hilger E, Spies M, Wadsak W, Lanzenberger R, Pataraia E et al (2016) Insights into intrinsic brain networks based on graph theory and PET in right- compared to left-sided temporal lobe epilepsy. Sci Rep 6:28513. https://doi.org/10.1038/srep28513
Chen D, Lu J, Zhou H, Jiang J, Wu P, Guo Q, Ge J, Zhang H et al (2018) Glucose metabolic brain network differences between Chinese patients with Lewy body dementia and healthy control. Behav Neurol 2018:8420658. https://doi.org/10.1155/2018/8420658
Riedl V, Bienkowska K, Strobel C, Tahmasian M, Grimmer T, Forster S, Friston KJ, Sorg C et al (2014) Local activity determines functional connectivity in the resting human brain: a simultaneous FDG-PET/fMRI study. J Neurosci 34(18):6260–6266. https://doi.org/10.1523/JNEUROSCI.0492-14.2014
Soddu A, Gomez F, Heine L, Di Perri C, Bahri MA, Voss HU, Bruno MA, Vanhaudenhuyse A et al (2016) Correlation between resting state fMRI total neuronal activity and PET metabolism in healthy controls and patients with disorders of consciousness. Brain Behav 6(1):e00424. https://doi.org/10.1002/brb3.424
Aiello M, Salvatore E, Cachia A, Pappata S, Cavaliere C, Prinster A, Nicolai E, Salvatore M et al (2015) Relationship between simultaneously acquired resting-state regional cerebral glucose metabolism and functional MRI: a PET/MR hybrid scanner study. NeuroImage 113:111–121. https://doi.org/10.1016/j.neuroimage.2015.03.017
Jiao F, Gao Z, Shi K, Jia X, Wu P, Jiang C, Ge J, Su H et al (2019) Frequency-dependent relationship between resting-state fMRI and glucose metabolism in the elderly. Front Neurol 10:566. https://doi.org/10.3389/fneur.2019.00566
Bernier M, Croteau E, Castellano CA, Cunnane SC, Whittingstall K (2017) Spatial distribution of resting-state BOLD regional homogeneity as a predictor of brain glucose uptake: a study in healthy aging. NeuroImage 150:14–22. https://doi.org/10.1016/j.neuroimage.2017.01.055
Detre JA, Leigh JS, Williams DS, Koretsky AP (1992) Perfusion imaging. Magn Reson Med 23(1):37–45. https://doi.org/10.1002/mrm.1910230106
Zou Q, Wu CW, Stein EA, Zang Y, Yang Y (2009) Static and dynamic characteristics of cerebral blood flow during the resting state. NeuroImage 48(3):515–524. https://doi.org/10.1016/j.neuroimage.2009.07.006
Riederer I, Bohn KP, Preibisch C, Wiedemann E, Zimmer C, Alexopoulos P, Forster S (2018) Alzheimer disease and mild cognitive impairment: integrated pulsed arterial spin-labeling MRI and (18)F-FDG PET. Radiology 288(1):198–206. https://doi.org/10.1148/radiol.2018170575
Yan CG, Wang XD, Zuo XN, Zang YF (2016) DPABI: data processing & analysis for (resting-state) brain imaging. Neuroinformatics 14(3):339–351. https://doi.org/10.1007/s12021-016-9299-4
Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comp Biomed Res An Inte J 29(3):162–173. https://doi.org/10.1006/cbmr.1996.0014
Wang JJ, Shan Y, Dai JD, Cui BX, Shang K, Yang HW, Chen ZW, Shan BC et al (2020) Altered coupling between resting-state glucose metabolism and functional activity in epilepsy. Ann Clin Transl Neurol.In Press. https://doi.org/10.1002/acn3.51168
Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP (2012) Network centrality in the human functional connectome. Cereb Cortex 22(8):1862–1875. https://doi.org/10.1093/cercor/bhr269
Liao XH, Xia MR, Xu T, Dai ZJ, Cao XY, Niu HJ, Zuo XN, Zang YF et al (2013) Functional brain hubs and their test-retest reliability: a multiband resting-state functional MRI study. NeuroImage 83:969–982. https://doi.org/10.1016/j.neuroimage.2013.07.058
Yan CG, Craddock RC, Zuo XN, Zang YF, Milham MP (2013) Standardizing the intrinsic brain: towards robust measurement of inter-individual variation in 1000 functional connectomes. NeuroImage 80:246–262. https://doi.org/10.1016/j.neuroimage.2013.04.081
Cha YH, Jog MA, Kim YC, Chakrapani S, Kraman SM, Wang DJ (2013) Regional correlation between resting state FDG PET and pCASL perfusion MRI. J Cereb Blood Flow Metab: official journal of the International Society of Cerebral Blood Flow and Metabolism 33(12):1909–1914. https://doi.org/10.1038/jcbfm.2013.147
Zimmer L (2017) Contribution of clinical neuroimaging to the understanding of the pharmacology of methylphenidate. Trends Pharmacol Sci 38(7):608–620. https://doi.org/10.1016/j.tips.2017.04.001
Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001) A default mode of brain function. Proc Natl Acad Sci U S A 98(2):676–682. https://doi.org/10.1073/pnas.98.2.676
Nugent AC, Martinez A, D’Alfonso A, Zarate CA, Theodore WH (2015) The relationship between glucose metabolism, resting-state fMRI BOLD signal, and GABAA-binding potential: a preliminary study in healthy subjects and those with temporal lobe epilepsy. J Cereb Blood Flow Metab : official journal of the International Society of Cerebral Blood Flow and Metabolism 35(4):583–591. https://doi.org/10.1038/jcbfm.2014.228
Tomasi D, Wang GJ, Volkow ND (2013) Energetic cost of brain functional connectivity. Proc Natl Acad Sci U S A 110(33):13642–13647. https://doi.org/10.1073/pnas.1303346110
Marchitelli R, Aiello M, Cachia A, Quarantelli M, Cavaliere C, Postiglione A, Tedeschi G, Montella P et al (2018) Simultaneous resting-state FDG-PET/fMRI in Alzheimer disease: relationship between glucose metabolism and intrinsic activity. NeuroImage 176:246–258. https://doi.org/10.1016/j.neuroimage.2018.04.048
Zou Q, Yuan BK, Gu H, Liu D, Wang DJ, Gao JH, Yang Y, Zang YF (2015) Detecting static and dynamic differences between eyes-closed and eyes-open resting states using ASL and BOLD fMRI. PLoS One 10(3):e0121757. https://doi.org/10.1371/journal.pone.0121757
Guedj E, Bonini F, Gavaret M, Trebuchon A, Aubert S, Boucekine M, Boyer L, Carron R et al (2015) 18FDG-PET in different subtypes of temporal lobe epilepsy: SEEG validation and predictive value. Epilepsia 56(3):414–421. https://doi.org/10.1111/epi.12917
Zhang Z, Lu G, Zhong Y, Tan Q, Chen H, Liao W, Tian L, Li Z et al (2010) fMRI study of mesial temporal lobe epilepsy using amplitude of low-frequency fluctuation analysis. Hum Brain Mapp 31(12):1851–1861. https://doi.org/10.1002/hbm.20982
Zhu X, He Z, Luo C, Qiu X, He S, Peng A, Zhang L, Chen L (2018) Altered spontaneous brain activity in MRI-negative refractory temporal lobe epilepsy patients with major depressive disorder: a resting-state fMRI study. J Neurol Sci 386:29–35. https://doi.org/10.1016/j.jns.2018.01.010
Govil-Dalela T, Kumar A, Behen ME, Chugani HT, Juhasz C (2018) Evolution of lobar abnormalities of cerebral glucose metabolism in 41 children with drug-resistant epilepsy. Epilepsia 59(7):1307–1315. https://doi.org/10.1111/epi.14404
Acknowledgements
The authors thank AiMi Academic Services (www.aimieditor.com) for the English language editing and review services. We also acknowledged Xinrong, Wang, GE Healthcare, Beijing for her professional suggestions.
Funding
This work was supported by the National Natural Science Foundation of China (81671662, 81520108016, 81661148045, 31471084); Beijing Municipal Administration of Hospitals Ascent Plan [DFL20180802]; and the Key Project of the Department of Science and Technology of Zhejiang Province (2015C03037).
Author information
Authors and Affiliations
Contributions
Conception and design: JL and YZ; data collection: JW, BC, and HY; data analysis: JW and HS; the first draft of the manuscript was written by JW and YZ; funding acquisition: JL and YZ; review and editing: JW, CD, and YS. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
The study was approved by the ethics committee of Xuanwu Hospital. The procedures used in this study adhere to the tenets of the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Code Availability
DPABI could be freely available from http://rfmri.org/dpabi. SPM12 could be freely available from https://www.fil.ion.ucl.ac.uk/spm. Analysis of Functional NeuroImages (AFNI) could be freely available from https://afni.nimh.nih.gov.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 4963 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Sun, H., Cui, B. et al. The Relationship Among Glucose Metabolism, Cerebral Blood Flow, and Functional Activity: a Hybrid PET/fMRI Study. Mol Neurobiol 58, 2862–2873 (2021). https://doi.org/10.1007/s12035-021-02305-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02305-0