Abstract
Increased amyloid β (Aβ) aggregation is a hallmark feature of Alzheimer’s disease (AD) pathology. The APP/PS1 mouse model of AD exhibits accumulation of Aβ in the retina and demonstrates reduced retinal function and other degenerative changes. The overall molecular effects of AD pathology on the retina remain undetermined. Using a proteomics approach, this study assessed the molecular effects of Aβ accumulation and progression of AD pathology on the retina. Retinal tissues from younger (2.5 months) and older 8-month APP/PS1 mice were analysed for protein expression changes. A multiplexed proteomics approach using chemical isobaric tandem mass tags was applied followed by functional and protein-protein interaction analyses using Ingenuity pathway (IPA) and STRING computational tools. We identified approximately 2000 proteins each in the younger (upregulated 50; downregulated 36) and older set of APP/PS1 (upregulated 85; downregulated 79) mice retinas. Amyloid precursor protein (APP) was consistently upregulated two to threefold in both younger and older retinas (p < 0.0001). Mass spectrometry data further revealed that older APP/PS1 mice retinas had elevated levels of proteolytic enzymes cathepsin D, presenilin 2 and nicastrin that are associated with APP processing. Increased levels of proteasomal proteins Psma5, Psmd3 and Psmb2 were also observed in the older AD retinas. In contrast to the younger animals, significant downregulation of protein synthesis and elongation associated proteins such as Eef1a1, Rpl35a, Mrpl2 and Eef1e1 (p < 0.04) was identified in the older mice retinas. This study reports for the first time that not only old but also young APP/PS1 animals demonstrate increased amyloid protein levels in their retinas. Quantitative proteomics reveals new molecular insights which may represent a cellular response to clear amyloid build-up. Further, downregulation of ribosomal proteins involved in protein biosynthesis was observed which might be considered a toxicity effect.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease makes a substantial contribution to the global disease burden, with an estimated prevalence of 45 million patients worldwide [1]. The disease incidence has been shown to increase from 3 to 32% in the developed countries as the age advances from 65 to 85 years [2]. A hallmark feature of AD pathology is the accumulation of aggregated amyloid β protein and neurofibrillary tangles with hyperphosphorylated tau protein. Amyloid β accumulation and other degenerative changes in the brain have been reported decades prior to the actual clinical diagnosis of the disease [3]. In addition to the amyloid misfolding and its accumulation in the brain, other degenerative changes have been observed which impact cognition, behaviour, olfactory systems, visual pathways, autonomic function and circadian rhythm [4,5,6,7,8].
Amyloid β accumulation and other associated functional and imaging deficits in the retina have previously been reported by us and others [7, 9,10,11]. Retinal pathological changes associated with local inflammation, reduced positive scotopic threshold responses and visual evoked potentials, apoptosis, retinal vascular changes, retinal ganglion cell loss and thinning of retinal nerve fibre layer using optical coherence tomography (OCT) have been identified both in AD animal models and dementia subjects [7, 9, 11,12,13,14,15]. However, it is important to highlight that converging data from various AD, at-risk mildly cognitive impairment cohorts and healthy ageing population indicate that molecular changes in the brain are initiated decades before actual onset of clinically significant cognitive impairment and behavioural changes [16, 17]. The lack of a reliable early disease detection test is primarily attributed to economical and health-related challenges associated with imaging the brain for diagnosis, screening and monitoring the progression especially during prodromal stages of the disease [18]. With mounting evidence suggesting that the eye is affected in AD, retinal changes might demonstrate some early signs and markers that could help diagnose the disease or help to monitor changes in the disease course by disease-modifying therapies [19,20,21]. Most of the biochemical studies have focussed on identifying Aβ reactivity, characterising tau phosphorylation and identifying the inflammatory or glial changes in the eye in AD, but an overall understanding of how various biochemical pathways in the retinas are affected in early stages of the disease is lacking [7, 12, 22, 23]. As a developmental offshoot of the brain, the retina exhibits several biochemical similarities with the brain and delineating retinal changes will help understand molecular mechanisms of the disease onset and its tissue specific effects.
We recently reported proteomics changes in the retinas from human glaucoma and control subjects that highlighted changes in the expression of several proteins that have been implicated in AD pathology [24]. Here, we sought to identify molecular changes in the retinas of APP/PS1 mouse model of AD that demonstrates gradual progression of the disease with age. Cognitive loss including deficits in the Morris water maze and hippocampal long-term potentiation impairments has been reported in these animals at 7 months of age [25, 26]. These mice are also reported to exhibit impaired learning at about 8 months of age [27]. In this study, we investigated molecular changes occurring in the early stages of AD in retina in 2.5-month-old APP/PS1 mice and compared findings with 8 months of age when these animals demonstrate cognitive and behavioural changes and amyloid plaque formation in the brain but without the neuronal loss that is observed at much later ages [28]. An integrated proteomics approach followed by computational analysis of the data has been used to understand molecular neuropathology of early AD in the retina (Fig. 1).
Experimental design and TMT labelling workflow of the study. Retinas from WT and APP/PS1 mice were harvested (n = 5 each). Extracted proteins from these samples were subjected to reduction, alkylation and subsequent digestion with Lys-C and Trypsin. Extracted peptides were quantified and labelled in a 10 plex TMT experiment. Labelled samples were pooled together, fractionated by high pH reversed-phase chromatography, and analysed by LC-ESI-MS/MS on QExactive mass spectrometer. Functional pathway and protein network data analysis were performed using STRING and Ingenuity pathway analysis. Immunostaining and Western blotting were used to validate the analysed data.
Methods
Reagents and Animal Experiments
Anti-β amyloid (Sigma, USA, SIG-39320), anti-Presenilin I (APS18) (Abcam, USA, ab15458), anti-Tau (Tau 46) (Cell Signalling, USA, #4019), anti-phospho-Tau (Ser404) (D2Z4G) (Cell Signalling, USA, #20194), anti-Cathepsin B (S-12) (Santacruz Biotechnology, USA, SC-6493), anti-PSMB2 (Thermofisher Scientific, USA, PA5-55304), anti-MRPL2 (Thermofisher Scientific, USA, PA5-68062), anti-EEF1E1 (Thermofisher Scientific, USA, PA5-28283), anti-αB crystallin (FL-175) (Santacruz Biotechnology, USA, SC22744), anti-βB2 crystallin (D-1) (Santacruz Biotechnology, USA, SC376856), anti-βB3 crystallin (I-20) (Santacruz Biotechnology, USA, SC22413), NeuN (Ab104225 abcam; Mab 377 Merck) and anti-nicastrin (Abcam, USA, ab45425) antibodies were used for probing Western blots and immunohistochemistry of retinal sections. β-Actin (Sigma, USA, AC-40) antibody was used for Western blotting. Alexa Fluor 488 affinipure donkey anti-mouse secondary antibody (Green), Cy3 affinipure donkey anti-rabbit secondary antibody (Red) and Fluorescein (FITC) affinipure donkey anti-goat secondary antibody (Green) were from Jackson Immuno research laboratories Inc., USA for the immunohistochemistry of retinal sections. All other analytical grade reagents were from Sigma, USA. APP/PS1 mice (2.5 and 8 months) were used in the study and were maintained in an air-conditioned room with controlled temperature (21 ± 2 °C) and ambient light at 12-h light/dark cycles. Animal experiments were performed according to the Australian code of practice for the care and use of animals for scientific purposes and as per the ARVO guidelines for the use of animals in ophthalmic and vision research and approved by the Macquarie University, Animal Ethics Committee. The number of samples (n) represents tissues from different animals.
Preparation of Protein Samples
The retinas were carefully extracted from the mice eyes using winkling technique employing curved forceps. The procedure was carried out under a surgical microscope (Carl Zeiss, Oberkochen, Germany) and tissues flash frozen in liquid nitrogen [29, 30]. Retinal tissues were lysed in lysis buffer (20 mM HEPES, pH 7.4, 1% Triton X-100, 1 mM EDTA) containing protease and phosphatase inhibitor cocktails and sonicated using a probe sonicator (3 pulses/15 s) at 50 Hz frequency on ice. Insoluble materials were removed by centrifugation at 15,000g for 10 min at 4 °C. Extracted proteins were reduced with 5 mM of dithiothreitol (DTT) for 15 min at room temperature, and then alkylated with 10 mM of iodoacetamide for 30 min in the dark at 4 °C. The alkylation reaction was then quenched with the addition of 5 mM DTT for 15 min in the dark. The proteins were precipitated using chloroform methanol precipitation procedure [24] and the pellet resuspended in 100 μl of 8 M urea in 50 mM Tris (pH 8.8) buffer. Protein concentration was determined by BCA assay kit (Pierce, Rockford, USA) using bovine serum albumin (BSA) as a standard. Dual digestion was carried out on 100 μg of protein, initially with Lys-C (Wako, Japan) at a 1:100 enzyme:protein ratio overnight at room temperature, followed by trypsin (Promega, Madison, WI) at a 1:100 enzyme:protein ratio for at least 4 h at 37 °C. Samples were then acidified with TFA to a final concentration of 1% (pH 2 to 3) and desalted using SDB-RPS (3M-Empore) stage Tip columns.
TMT Labelling
To accommodate the 20 biological samples (5 WT and 5 APP/PS1 2.5 months; 5 WT and 5 APP/PS1 8 months), 2 separate 10 plex TMT experiments were carried out (TMT1 for 2.5 months and TMT2 for 8-month group). Briefly, dried peptides from each sample were resuspended in 100 mM HEPES (pH 8.2) buffer and peptide concentration measured using the MicroBCA protein assay kit (Thermo Scientific- Rockford, IL). Fifty micrograms of peptides from each sample was subjected to TMT labelling with 0.8 mg of reagent per tube. Labelling was carried out at room temperature for 1 h with continuous vortexing. To quench any remaining TMT reagent and reverse the tyrosine labelling, 8 μl of 5% hydroxylamine was added to each tube, followed by vortexing and incubation for 15 min at room temperature. For each of respective ten plex experiments, the ten labelled samples were combined, and then dried down by vacuum centrifugation. Prior to BpRP fractionation, the digested and TMT-labelled peptide samples were cleaned using a reverse-phase C18 clean-up column (Sep-pak, Waters) and dried in vacuum centrifuge. The peptide mixture was resuspended in loading buffer (5 mM ammonia solution (pH 10.5), separated into a total of 96 fractions using an Agilent 1260 HPLC system equipped with quaternary pump, a degasser and a Multi-Wavelength Detector (MWD) (set at 210-, 214- and 280-nm wavelength). Peptides were separated on a 55-min linear gradient from 3 to 30% acetonitrile in 5 mM ammonia solution pH 10.5 at a flow rate of 0.3 mL/min on an Agilent 300 Extend C18 column (3.5 μm particles, 2.1 mm ID and 150 mm in length). The 96 fractions were finally consolidated into 17 fractions. Each peptide fraction was dried by vacuum centrifugation, resuspended in 1% formic acid and desalted again using SDB-RPS (3M-Empore) stage tips [24].
Nanoflow Liquid Chromatography Electrospray Ionisation Tandem Mass Spectrometry (nano LC-ESI-MS/MS)
Fractionated and dried peptides were reconstituted in 40 μL of 0.1% formic acid and analysed on a Q Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) coupled to an EASY-nLC1000 nanoflow HPLC system (Thermo Scientific, San Jose, CA). Reversed-phase chromatographic separation was performed on an in-house packed reverse-phase column (75 μm × 10 cm Halo® 2.7 μm 160 Å ES-C18, Advanced Materials Technology). Labelled peptides were separated for 2 h using a gradient of 1–30% solvent B (99.9% acetonitrile/0.1% formic acid) and Solvent A (97.9% water/2% acetonitrile/0.1% formic acid). The Q Exactive mass spectrometer was operated in the data-dependent acquisition (DDA) mode to automatically switch between full MS and MS/MS acquisition. Following the Full MS scan from m/z 350–1850, MS/MS spectra were acquired at resolution of 70,000 at m/z 400 and an automatic gain control target value of 1 × 106 ions. The top 10 most abundant ions were selected with precursor isolation width of 0.7 m/z for higher-energy collisional dissociation (HCD) fragmentation. HCD-normalised collision energy was set to 35% and fragmentation ions were detected in the Orbitrap at a resolution of 70,000. Target ions that had been selected for MS/MS were dynamically excluded for 90 s.
Database Searching, Peptide Quantification and Statistical Analysis
Raw data files were processed in Proteome Discoverer V2.1 (Thermo Scientific, San Jose, CA) using Mascot (Matrix Science, UK). Data were matched against the reviewed SwissProt Mus musculus protein database (16,953 sequences, Feb 2018). The MS1 tolerance was set to ± 10 ppm and the MS/MS tolerance to 0.02 Da. Carbamidomethyl (C) was set as static modification, while TMT 10-plex (N-term, K), Oxidation (M), Deamidation (N, Q), Glu->pyro-Glu (N-term E), Gln->pyro-Glu (N-term Q) and Acetylation (Protein N-Terminus) were set as dynamic modifications. The percolator algorithm was used to discriminate correct from incorrect peptide-spectrum matches, and calculate statistics including q value (FDR) and posterior error probabilities (PEPs). Search results were further filtered to retain protein with FDR of < 1% and only master proteins assigned via protein grouping algorithm were retained. Relative quantitation of proteins was achieved by pairwise comparison of TMT reporter ion signal to noise (S/N) ratios, for example, the ratio S/N of the labels for each of the treatment replicates (numerator) versus the labels of their corresponding control replicates (denominator). Proteins were further analysed using the TMTPrepPro analysis pipeline [31]. The TMTPrepPro scripts are implemented in the R programming language and are available as an R package, which was accessed through a graphical user interface provided via a local GenePattern server. All protein ratios with respect to the reference (label-131) were extracted. In pairwise comparison tests, relative quantitation of protein abundance was derived from the ratio of the TMT label S/N detected in each condition (APP/PS1 and WT), and differentially expressed proteins were identified based on Student t tests between APP/PS1 and WT ratios (log-transformed). The overall fold changes were calculated as geometric means of the respective ratios. Differential expression required the proteins to meet both a ratio fold change (> 1.10 (up-regulated) or < 0.85 (down-regulated)) and a p value cut-off (t test p value < 0.05) [32, 33].
Functional Network and Computational Analysis
The identified differentially expressed proteins through the pairwise comparisons were subjected to pathway enrichment analysis using Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com/index.html). Identified proteins were mapped to genes using Ingenuity Pathway Knowledge Base. Significant interaction networks (p < 0.05) and molecular and cellular functions were identified based on known protein–protein interactions in the Ingenuity knowledgebase. Network-naming was based on the most common functional group(s) present. Canonical pathway analysis is used to identify the function-specific genes amongst the up- and down-regulated proteins. Further, as a separate orthogonal approach, the differentially expressed proteins identified with ANOVA analysis (p value < 0.05, maximum absolute value of fold change ≥ 1.1) were classified according to KEGG pathways and biological processes using the Cytoscape stringApp plugin (http://apps.cytoscape.org/apps/stringapp). Significantly changed proteins were loaded into Cystoscape, and the Mus Musculus database in the StringDB was selected to reveal the protein interactions in the context of enriched pathways.
Western Blotting
Western blot analyses were carried out on APP, PS1, Tau, pTau (Ser 404), Cathepsin B, PSMB2, MRPL2, EEF1E1 and crystallins to establish the protein changes underlying APP/PS1 mice retinas at indicated time points. Twenty micrograms of protein from each sample was separated by 10% SDS-PAGE and transferred to PVDF membrane. Membranes were blocked by Tris-buffered saline containing 5% skimmed milk buffer, and primary antibodies added. After incubation with the primary antibody overnight at 4 °C, blots were thoroughly washed with Tris-buffered saline and incubated with anti-IgG antibody linked to horseradish peroxidase for 2 h. Chemiluminescent signals were detected using the enhanced chemiluminescent substrate (BioRad) and Western blot band intensities quantified within the linear range of detection [34,35,36].
Tissue Sectioning and Immunofluorescence Analysis
The animals were subjected to transcardial perfusion with paraformaldehyde (PFA, 4%), the eyes were removed and subjected to 4% PFA treatment for 1 h. The tissues were washed with PBS and incubated overnight in 30% sucrose solution. The tissues were embedded in OCT media and 15-μm thick sections were prepared and treated with cold EtOH for permeabilization. The sections were subjected to incubation with primary antibodies (1:300) overnight at 4 °C. Subsequently, the tissues were incubated with indicated secondary antibodies (1:400 in Tris phosphate saline buffer) for 1 h in the dark and mounted on slides using antifade mounting media with Prolong DAPI. The pictures were captured using a microscope (Carl Zeiss) as described previously [37, 38].
Results
Differential Regulation of Protein Expression in APP/PS1 Mice Retinas
A non-redundant total of 2079 and 2152 proteins were identified and quantified from the younger (2.5-months; TMT1) and older (8-months; TMT2) animal retinas, respectively. The data was analysed using a series of established tests to assess the data quality. The distribution pattern of proteins was assessed through heat maps and volcano plots with log transformed protein abundance ratios (Fig. 2). No major asymmetric bias was observed in the data reflecting overall similar protein abundance distributions of the individual samples. The pairwise analysis of APP/PS1 vs. WT retinas identified 50 upregulated and 36 downregulated proteins (p value ≤ 0.05 and ≥ 1.15-fold difference) for 2.5-month-old animals. On the other hand, we identified 85 upregulated and 79 downregulated proteins in the retinas of 8-month-old mice (p value ≤ 0.05 and ≥ 0.85-fold difference) as shown in Fig. 2C, D. Hierarchical clustering maps of proteins separated the WT and APP/PS1 retinal samples well at both the ages (Fig. 2A, B). The two proteomes were qualitatively similar with no remarkable outliers. The complete TMT datasets for both including protein identifications and differentially expressed protein for each comparison tests can be viewed in the supplementary tables S1 and 2.
Results of proteomics analysis and quality control measurements. A, B Heatmaps (hierarchical clustering) of the log-transformed ratios of differentially expressed proteins (WT vs. APP/PS1) in the retinas of young and ageing animals. Column colours indicate treatment type. Red and blue colour-coding indicate relative increase or decrease in protein abundance, respectively. C, D Volcano plots demonstrating the dual thresholds for differentially regulated proteins. Each data point represents a single quantified protein. The x-axis represents log2 fold change in abundance (APP/PS1/ WT mice). Vertical dotted lines indicate 1.1 and 0.9 ratios. The −log10 (p value) is plotted on the y-axis. Proteins above the horizontal dotted line indicate significance ≤ 0.05. Proteins within the upper and outer quadrants meet both the fold change and p value cut-off and are therefore considered as differentially regulated.
The top 10 differentially regulated proteins in both the 2.5-month and 8-month-old APP/PS1 mice compared to the respective WT controls are shown in Fig. 3A, B. Interestingly, the expression of APP was highly elevated in both the young and older mice (two to threefold upregulation). CLSTN1 and AP1S2 were the other proteins that demonstrated a twofold increase in the younger retinal tissues. For the older mice, CSTF2 was observed to be increased twofold compared to the WT tissues. The crystallin family of proteins (fivefold downregulation) along with elongation factors were downregulated in the older mice retinas while the younger mice tissues exhibited a modest downregulation of transcription regulatory proteins and Cystatin B (1.3–1.6-fold downregulation). Eleven of the differentially expressed proteins were common between the 2.5- and 8-month-old APP/PS1 mice groups. Out of these, APP, Prdx1 and Atp6v1e1 were consistently upregulated across both of these age groups, while Galnt13, Crx and Mad2l1 were downregulated in both (Fig. 3C). Interestingly, some of the proteins such as Stag2, Pcmtd1, Puf60 and Mien1 were downregulated in the older retinas although they were found to be initially upregulated in the 2.5-month-old eyes. These specific protein differences are attributed to their differential regulation in APP/PS1 versus WT genotypes and the potential effects of AD pathology on the retina with respect to age.
A, B List of 10 most upregulated and downregulated proteins in the young and older APP/PS1 mice retinas, respectively. C Venn diagram representing the overlap between the differentially regulated proteins quantified between the young (2.5 months) and older (8 months) APP/PS1 mice retinas (p value ≤ 0.05, ≥ 1.1-fold or ≤ 0.9-fold). The common differentially affected proteins in the younger (Y) and older (O) cohorts are listed (red, up and green, down-regulated)
Protein Network and Functional Pathway Analysis Identifies Functionally Coordinated Protein Abundance Changes in APP/PS1 Mice Retinas
The differentially expressed proteins in the younger (86 proteins) and older (164 proteins) APP/PS1 mice were subjected to cellular pathway enrichment and functional protein network analyses to understand the biochemical pathways and physiological process that are affected in the retina with the advancement of AD pathology.
The functional connectivity of the differentially expressed proteins was plotted and assessed using the STRING Cytoscape analysis tool. The APP processing, proteasomal pathways and ribosomal machinery protein networks were differentially affected and prominently highlighted in the older mice tissues (Fig. 4A and S1). While the APP processing module was distinct in both age groups, in the younger mice, we observed the modulation of prdx1, pdia3, GSR and GPX1 proteins, which are mainly involved in ER stress and oxidative stress pathways. The greater interconnectivity and protein clusters in the older mice correlated with higher number of differentially expressed proteins identified in the mice tissues at this age. This computational analysis was followed by specific investigations into several representative proteins and pathways to understand the molecular effects of AD development on the retina and also for the validation of the proteomics data.
Results of functional protein interaction network and pathway analysis. A Functional interaction networks of differentially expressed proteins (older retina: APP/PS1 vs WT) analysed using Cytoscape tool with the String DB plugin. The enriched networks are: proteasome, ribosomal machinery and APP processing proteins. B A table of proteins included in the networks along with their fold changes (red, up; green, down). C, D Comparison of the top canonical pathways, disease and biological pathways enriched from IPA analysis of differentially regulated proteins from younger and older retinas of APP/PS1 animals compared to the retina of WT animals (based on the −log (p value)
Analysis of the differentially regulated proteins using IPA based on p value ≤ 0.05 indicated that biochemical canonical pathways associated with phagosome maturation, protein ubiquitination, mitochondrial dysfunction, amyloid processing, retinoic acid receptor activation and oxidative stress response were over-represented in both the younger and older APP/PS1 mice retinas compared to the WT counterparts (Fig. 4C). Disease and biological functions associated with cytoskeletal and microtubule organisation, neuritogenesis, apoptosis, neuronal development and differentiation were prominently affected (Fig. 4D). Overall, our data suggested that several networks linked with AD pathology and neurological disorders such as amyloid processing, neuronal function and development were affected and the changes were more prominent in the retinas of older animals (Fig. 4D). Combined analysis using STRING and IPA revealed the most significantly affected cellular pathways in the retinas of both young and older APP/PS1 and control animals.
Altered Processing of Amyloid Precursor Protein and Its Processing Enzymes
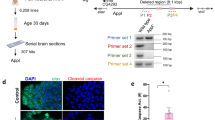
Following the quantitative mass spectrometric analysis of the APP and related enzymes, we performed Western blot and immunohistochemistry analysis to corroborate the expression patterns of these proteins. Mass spectrometry identified upregulation of APP in retinas of both younger (threefold) and older (2.3-fold) APP/PS1 mice compared to WT animals (Fig. 3A, B). Subsequently, Western blotting analysis confirmed significantly increased expression of amyloid precursor protein band intensity in 2.5-month-old mice compared to the wild-type controls (p < 0.0001). Band corresponding to the amyloid β monomer peptide was also elevated in the APP/PS1 mice retinas (p < 0.001) (Fig. 5A). We evaluated APP expression changes in the retinas of 8-month-old APP/PS1 mice and interestingly, at this age we observed that immunoreactivity of the APP parent protein was significantly reduced compared to the WT counterparts (p < 0.05). However, the levels of soluble Aβ monomeric peptide band (p < 0.0001) and its oligomeric forms (p < 0.0002) were significantly increased (Fig. 5B). Actin was used as an internal control for data normalisation in each case. APP immunostaining analysis of sections from the retinas of 8-month-old APP/PS1 and WT mice sections also revealed increased reactivity in the APP/PS1 mice tissues indicating altered APP processing in the retinas of AD mouse model in early stages of the disease (Fig. 5C, S3).
A Western blots of retinal lysates from young WT and APP/PS1 mice were probed with amyloid β and used actin as a loading control. The relative intensities of WB bands were quantified and plotted (n = 4). Both APP and monomeric forms of amyloid β expressions were significantly increased in young APP/PS1 retinal lysates compared to the WT retinal lysates [1, 2]. B Western blots of retinal lysates from 8-month-old WT and APP/PS1 mice were probed with amyloid β with actin as a loading control. The relative intensities of WB bands were quantified and plotted (n = 3; **p < 0.05). Both monomeric and oligomeric amyloid β species were significantly increased when compared to the WT retinal lysates [2, 3]. C Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating amyloid (green, Alexa Fluor 488) and NeuN (red, Cy3, Ab104225) expression (blue, DAPI) (n = 3). Expression of the amyloid β in the retinal ganglion cell layer of APP/PS1 retinal sections is indicated by the white arrowheads. Scale = 50 μm
We also evaluated the presenilin 1 (PS1) expression in the mice retinas at both of these ages. Western blot analysis revealed that PS1 expression remained largely unchanged in early life, however the expression was significantly increased in the older APP/PS1 group compared to WT mice (p < 0.05). Actin was used as loading control and the band intensities quantified within the linear range of detection (Fig. 6A). Immunofluorescence analysis of the retinal sections of 8-month-old mice also revealed increased reactivity of the PS1 protein, with most of the reactivity predominantly localised within the inner retinal layers (Fig. 6B, S3).
A Western blots of retinal lysates from young (2.5 months) and older (8 months), WT and APP/PS1 mice were probed with PS1 with actin as a loading control. The relative intensities of WB bands were quantified and plotted (n = 3). Older APP/PS1 retinal lysates indicate significant overexpression of PS1 (p < 0.05). B Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating PS1 (green, Alexa Fluor 488), NeuN (red-Cy3, Ab104225) expression (blue, DAPI) (n = 3). Expression of the PS1 on the ganglion cell layer of APP/PS1 retinal sections was indicated by the white arrowheads. C Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating Tau (green, Alexa Fluor 488), NeuN (red, Cy3, Ab104225) expression (blue, DAPI) (n = 3). APP/PS1 retinal sections did not indicate any overexpression of the Tau proteins on retinal ganglion cell layer. D Expression of the pTau (red, Cy3), NeuN (green, Alexa Flour 488, Mab 377) on the ganglion cell layer of APP/PS1 retinal sections was increased compared to the WT sections. Overexpressed pTau (Ser404) indicated by the white arrowheads. Scale = 50 μm
Tau is another important protein that has been consistently reported to undergo increased phosphorylation and form intracellular tangles in the cells in AD brain. Our data concerning the older group of APP/PS1 mice revealed that the tau protein staining or localisation was not altered noticeably in the retinas (Fig. 6C). Interestingly, pTau (Ser404) immunoreactivity was much increased in the APP/PS1 mice retinas compared to the WT mice (Fig. 6D, S3). Together, these studies demonstrate that levels of APP, PS1 and pTau, which are hallmark proteins associated with AD pathology, were significantly perturbed in the mice retinas in AD pathology.
Activation of Proteasome and Proteolytic Enzymes in the APP/PS1 Mice Retinas with Age
Our data showed increased abundance of several proteolytic enzymes and the proteasome associated proteins in the older mice retinas. To further confirm the expression changes of selected proteolytic enzymes and proteasomal proteins, we performed Western blotting and immunostaining experiments. Western blot analysis for cathepsin B did not reveal any significant changes in the expression of the protein. Immunofluorescence analysis, nevertheless, showed that the Cathepsin B expression was elevated in the ganglion cell layer (GCL) region of the APP/PS1 mice (Fig. 7A, B, S3). Immunoblot analysis of PSMB2 also reflected no noticeable expression changes in the young or older APP/PS1 mice retinas compared to the control eyes. However, as proteomics data suggested an increased level of PSMB2 in older mice retinas, we performed immunostaining of the retinal sections of older mice, revealing a locally increased expression of the protein in the GCL region (Fig. 7C, D, S3). Similarly, Nicastrin expression was more prominent in the inner retinal layers of the older APP/PS1 mice tissues parallel to the changes that were identified in the mass spectrometry data (Fig. S2).
A Western blots of retinal lysates from younger and older, WT and APP/PS1 mice were probed with Cathepsin B. The relative intensities of WB bands were quantified and plotted (n = 3). B Immunostaining of the old retinal sections from APP/PS1 mice illustrating overexpression of cathepsin B (green, FITC), NeuN (red, Cy3, Ab104225) on the retinal ganglion cells (blue, DAPI) (n = 3). Scale = 50 μm. C Western blots of retinal lysates from younger and older, WT and APP/PS1 mice were probed with PSMB2 with actin as a loading control. The relative intensities of WB bands were quantified and plotted (n = 3). D Over-expression of the PSMB2 (red, Cy3) with RGC marker NeuN (green, Alexa Flour 488, Mab377) in the ganglion cell layer of the older APP/PS1 retina compared to WT retinal sections (blue, DAPI) (n = 3)
Down-regulation of the Protein Synthesis and Elongation-Associated Proteins
In our data, we also identified that a subset of proteins involved in protein elongation and synthesis were decreased in abundance in the 8-month-old mice retinas compared to WT animals. We sought to establish and validate these changes using a combination of Western blotting and immunofluorescence analysis of the retinal tissue lysates. MRPL2 immunoblotting did not reveal significant expression changes in the younger or older mice tissues. Therefore, we further analysed the older mice retinal sections using IF analysis, which interestingly revealed reduced immunoreactivity in the GCL region of the retina in ageing animals (Fig. 8A, B, S3). The expression changes for elongation factor EEF1E1 were also analysed for the younger and older mice tissues. In this case also, no significant protein expression changes were observed in the APP/PS1 mice retinas compared to the WT counterparts. Immunofluorescence analysis of the older mice retinas revealed much higher expression of the protein in the inner retinal layers of age matched WT mice compared to the APP/ PS1 mice tissues (Fig. 8C, D, S3).
A Western blots of retinal lysates from young (2.5 months) and older (8 months), WT and APP/PS1 mice were probed with MRPL2. The relative intensities of WB bands were quantified and plotted (n = 3). B Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating MRPL2 (red, Cy3), NeuN (green, Alexa Flour 488, Mab377) expression (blue, DAPI) (n = 3). C Western blots of retinal lysates from young and older, WT and APP/PS1 mice were probed with EEF1E1 (p < 0.05). Scale = 50 μm. The relative intensities of WB bands were quantified and plotted (n = 3). D Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating EEF1E1 (red, Cy3), NeuN (green, Alexa Flour 488, Mab377) expression (blue, DAPI) (n = 3)
Downregulation of Crystallins in the Ageing APP/PS1 Mice Retinas
The most significant differentially downregulated proteins in our proteomics data belonged to the crystallin family of proteins in the 8-month-old APP/PS1 mice retinas compared to the WT mice tissues (Fig. 3A, B). We further evaluated crystallin changes in older mice retinas using specific antibodies for αβ-crystallin, βB2-crystallin and βB3 crystallins. Western blotting and immunohistochemical evidence established that there was reduced expression of the αβ-crystallin mainly in the GCL region (Fig. 9A, B, S3). A profound loss of βB2-crystallin expression was evident throughout various layers of the retina in APP/PS1 mice and interestingly this isoform was the most differentially regulated protein in the whole data set (Fig. 9C, D, S3). βB3 crystallin Western blotting and immunostaining data also indicated reduced levels of the protein in the APP/PS1 mice tissue (Fig. 9E, F, S3). Collectively, these results suggest that several crystallin family proteins were significantly downregulated in the retinal tissues in AD pathology.
A Western blots of older retinal lysates indicate significantly decreased αB Crystallin levels in APP/PS1 mice. B Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating αB Crystallin (red, Cy3), NeuN (green, Alexa Flour 488, Mab377) expression (blue, DAPI). C Significantly decreased levels of βB2-Crystallin observed in older APP/PS1 retinas compared to the WT retinal lysates. D Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating βB2-Crystallin (green, Alexa Fluor 488), NeuN (red, Cy3, Ab104225) expression (blue, DAPI). Scale = 50 μm. E Western blot analysis indicates decreased βB3-Crystallin levels in older APP/PS1 mice retinas compared to the WT older retinal lysates. F Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating βB3-Crystallin (green, FITC), NeuN (red, Cy3, Ab104225) expression (blue, DAPI). (n = 3 each; p < 0.05)
Discussion
This study focussed on protein expression changes in the retinas of 2.5-month-old APP/PS1 mice and compared the findings with 8-month-old animals. These ages were selected because in these mice, amyloid deposition has been identified starting from 6 weeks in brain cortex and deposition in the hippocampus has been observed starting from about 3 to 4 months, and then gradually, deposits are observed in other brain regions such as thalamus, striatum and brainstem regions at 4 to 5 months. At 8 months, the animals demonstrate significant cognitive and behavioural deficits but without the neuronal loss, which can mask the accurate protein expression changes [26,27,28].
Several studies have been performed on the proteomics changes in post-mortem tissues of AD subjects but there are a limited number of studies involving investigation of proteome alterations in the brains of APP/PS1 mice model [39,40,41]. While an increasing number of studies implicate that the retina is affected in AD pathology, there is limited evidence of the molecular pathways that underlie functional imaging and histological observations [7, 9]. Further, several studies highlight increased amyloid β accumulation and tau immunostaining in the tissues of AD mice models [7, 22, 42], but to the best of our knowledge, a comprehensive investigation of which biochemical pathways are affected and the protein alterations that occur in the retina and their quantification has not been performed previously. Here, we analysed the retinal proteome of APP/PS1 mice and identified and quantified over 2000 non-redundant proteins, providing the first evidence of retinal molecular changes that occur in early stages of the disease. We compared the findings with protein changes in retinas of older (8 months) animals, at which age, these mice manifest clinical symptoms of AD but without the neuronal loss that ensues in the later stages of the disease. We report differential expression of 86 proteins in the younger and 164 proteins in the older APP/PS1 mice retinas that signifies an altered biochemical environment in the retina in AD pathology. These molecular changes in the retina provide a valuable resource to help understand the disease mechanisms and may highlight protein targets for drug development and diagnostic and prognostic biomarker candidates. A comparison with brain proteomics alterations in AD will further unravel the molecular overlap between the two CNS regions and delineate the spatial and temporal dimensions of AD pathology in the two tissues.
Functional network analysis of the proteomics data revealed that APP processing was perturbed in both the younger and older mice and here we report for the first time, approximately a threefold increase in the APP protein expression in the retina. This order of increased human APP transgene expression in these animals compared to the endogenous APP in the brain has previously been reported [43]. Retinas from older animals revealed specific upregulation of proteasomal proteins and proteolytic enzymes and indicated that proteins associated with ribosomal machinery such as protein synthesis and elongation were significantly downregulated.
This study established an altered processing of APP and its processing enzymes in the retina even in the younger animals suggesting that the retina is affected much earlier in the disease pathology than previously thought which highlights the importance of monitoring the retinal tissues in AD. It also raises the potential that retinal monitoring could be used to follow the disease progression, if molecular changes can be imaged in vivo. Immunoblotting studies demonstrated that APP expression was significantly elevated with slightly elevated levels of Aβ peptide. In the older mice, however, the parent APP levels were significantly diminished, but a concomitantly increased expression of Aβ and other APP bands was observed illustrating altered protein processing and Aβ oligomerisation. These findings are consistent with increased Aβ plaque formation observed in the brain neocortex region starting from 6 weeks of age which then extends to the hippocampus and other brain regions between 3 and 6 months of age [27]. Immunostaining analysis of the retinas of 8-month-old mice, however, did not reveal any deposits or plaque type structures in the retina. The APP staining was observed to be increased and was predominantly localised to the inner retinal layers. This is in agreement with previous reports that APP/PS1 mice demonstrate inner retinal disturbances and Aβ accumulation in the aged animals [7].
In the older animals, there was also increased presenilin expression which was interestingly not affected in the younger mice retinas suggesting a chronology of expression changes in the retina as the disease progressed. Phosphorylated tau immunoreactivity has been observed in the vicinity of amyloid deposits in the brain [44]. Tau hyperphosphorylation has also been reported in the retinas of animal models of AD and glaucoma [45, 46]. Accordingly, we observed increased tau phosphorylation in the older mice retinas using immunofluorescence staining although no changes in total tau protein expression were observed. Increased tau expression and its phosphorylation have previously been reported in 3-month and 6-month-old APP/PS1 animals and the expression was observed to be significantly increased in CSF of 6-month-old mice [47, 48].
Activation of proteolytic enzymes in response to amyloid β treatment or in the APP/PS1 mice brain tissues has been extensively reported. For instance, plasmin was shown to undergo activation upon stimulation with Aβ [49] which in turn may play a role in inducing Aβ degradation [50]. Cathepsin B has similarly been shown to play a role in Aβ modulation. While inhibition of cathepsin B protease was shown to play a role in reducing amyloid β production and also improve memory [51, 52], there are studies that also demarcate the role of cathepsin B activation in lowering of Aβ levels [53]. Our study revealed that cathepsin B was locally increased in the GCL region of the retina in older mice tissues. Similarly, nicastrin which is a member of the γ secretase complex that is involved in generation of amyloid β peptide [54] has been shown to reduce amyloid deposition in the 5xFAD mouse model of AD [55]. Its increased expression has been reported in the astrocytes and microglia following brain injury [56]. The inner retinal layers of older APP/PS1 mice following retinal immunofluorescence analysis revealed increased expression of nicastrin which validated the mass spectrometry results. The proteasome plays a key role in digesting the proteins and regulating the protein quality control and is associated with proteolytic processing of both amyloid β and tau proteins [57]. There are reports that proteasome activity and ubiquitination processes are diminished in brains of AD subjects [58]. There are also reports that indicate that Aβ accumulation may induce dysfunctional proteasomal activation in cells and mice models of AD [59, 60]. These varied effects can possibly be correlated with stages of disease pathogenesis or tissue or model specific effects. We observed striking activation of various proteasome components in older mice retinas in our proteomics data. Increased expression of PSMB2 proteasome component was locally observed in the retinal layers in immunofluorescence analysis. Follow-up analysis of the aged mice retinas will reveal how the proteasomal activity is affected with the advancement of AD pathology.
Ribosomal dysfunction and altered protein synthesis have been observed in both early and advanced stages of AD pathogenesis [61,62,63]. Our proteomics data revealed several proteins that are involved in protein synthesis, transcription and elongation as negatively modulated in the retinas of older animals. We previously reported dysregulation of seven different mitochondrial ribosomal proteins in the eyes of glaucoma subjects [24], suggesting a possible molecular overlap between the neurodegenerative disorders. Also, recent studies have highlighted the role of tau pathology in promoting ribosomal dysfunction and suppressing protein synthesis which might impair the synaptic function and plasticity of neurons [64, 65]. Wu et al. (2016) recently reported that APP over-expressing cells negatively affected various aspects of translation including protein initiation and elongation [66]. A specific downregulation of the MRPL2 and EEF1E1 in the inner retina was identified in older mice suggesting a tissue response as a consequence of developing disease pathology. Localisation of proteasomal activation and ribosomal suppression to the inner retinal layers further suggested this region to be a major or early site of AD induced pathogenesis. This corresponded with our previous studies that showed decreased positive scotopic threshold response (pSTR) and reduced GCL (both inner retinal) as well as reduced optic nerve axonal density in ageing APP/PS1 mice retinal tissues.
The crystallin family of proteins was the most differentially regulated group of proteins identified in the older mice retinal tissues. Crystallins are the heat-shock and adapter protein molecules that have been shown to play a role in neuro-inflammation in glial cells and are associated with neurofibril formation in neurons [67, 68]. They inhibit apoptosis-induced cell death and have been consistently shown to be upregulated in response to a multitude of stresses in the retinal neurons including ischemia injury and also play a key role in axonal regeneration and molecular defence against dysregulated proteins [69, 70]. Interestingly, recent studies have indicated that intravitreal administration of β crystallin B2 is neuroprotective for the retinal ganglion cells in an animal model of glaucoma [71]. We previously reported downregulation of eleven crystallin family proteins in the eyes of glaucoma subjects [24]. Downregulation of crystallins in both glaucoma and AD conditions suggests molecular overlap of the mechanisms that underlie these two neurodegenerative conditions. Immunofluorescence analysis of the APP/PS1 mice sections revealed that αβ, βB2 and βB3 crystallins were downregulated in various layers of the retina in the older mice tissues. These findings suggest that neuroprotective pathways mediated through the crystallins were compromised in the older APP/PS1 mice retinas. αB crystallin has previously been shown to colocalise with Aβ and the chaperone protein was shown to inhibit Aβ fibril formation in vitro [72]. αB crystallin was reported to be a component of neurofibrillary tangles and its overexpression correlated with reduced tau phosphorylation in N2a neuroblastoma cells [73], suggesting that reduced crystallin levels might further promote Aβ oligomerisation and tau phosphorylation and exacerbate the AD effects on the retinal tissue. AAV mediated overexpression of crystallin isoforms in the retinas of APP/PS1 mice will highlight their specific roles in potentially protecting against the AD-induced degenerative changes. Conversely, crystallin silencing in the retinas of normal healthy mice will elucidate the biochemical pathways that are affected in response to crystalline downregulation for future therapeutic development.
The proteomics part of this study involved analysing whole retinal tissue, and thus molecular changes in each individual retinal layer could not be distinguished. Use of techniques such as laser capture dissection microscopy followed by analysing the tissues using highly sensitive mass spectrometry approaches will help establish proteome alterations in various populations of retinal cells. Nonetheless, we performed immunofluorescence analysis of the APP/PS1 and WT mice sections with specific markers to elucidate protein expression and localisation changes in different layers of the retina in these animals. Further, we provide quantitative comparison of the protein expression changes in young and older APP/PS1 mice with respect to the age-matched WT-type animals. Independent nature of the 10 plex TMT runs in the experiment however meant that we were not able to directly compare and quantify protein changes specifically within the WT or APP/PS1 mice groups. Future studies will highlight the trajectory of protein expression changes that occur within each of these genotypes as the animals’ age. APP/PS1 mice show high APP expression as a result of how these transgenic animals have been programmed and a possible limitation is that APP/PS1-based studies inadvertently presume amyloid aggregation as a basic feature of the disease pathogenesis. This may confound the proteomics analysis as AD is a complex neurodegenerative disorder and its dimensions likely extend well beyond Aβ. Future retinal studies from post-mortem eyes of prodromal, established and advanced AD subjects will help validate these findings. Further, it is possible that some of the proteomics changes may be caused by altered axoplasmic flow through the RGC axons, and though still valuable data, it may not reflect the bonafide retinal cellular changes in APP/PS1 mice. Aβ aggregation and its reduced solubility may also have some effects on protein quantification levels. Nonetheless, this study reports for the first time, biochemical pathways that are affected in early stages of disease development in APP/PS1 mice retinas. Identification of the molecular networks and differentially affected proteins could help to develop novel drug targets to modulate these pathways in early stages of the disease. A comparison of retinal findings in AD with alterations in different regions of the brain will provide mechanistic insights into tissue specific effects of disease. Similarly, contrasting the retinal proteomics changes in AD with other retinal neurodegenerative disorders such as glaucoma will help understand the specific molecular effects of AD pathology.
References
Group, G. B. D. N. D. C (2017) Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol 16:877–897. https://doi.org/10.1016/S1474-4422(17)30299-5
Hebert LE, Weuve J, Scherr PA, Evans DA (2013) Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology 80:1778–1783. https://doi.org/10.1212/WNL.0b013e31828726f5
Bilgel M et al (2016) Individual estimates of age at detectable amyloid onset for risk factor assessment. Alzheimers Dement 12:373–379. https://doi.org/10.1016/j.jalz.2015.08.166
Villemagne VL et al (2013) Amyloid beta deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: a prospective cohort study. Lancet Neurol 12:357–367. https://doi.org/10.1016/S1474-4422(13)70044-9
Chi S, Yu JT, Tan MS, Tan L (2014) Depression in Alzheimer's disease: epidemiology, mechanisms, and management. J Alzheimers Dis 42:739–755. https://doi.org/10.3233/JAD-140324
Wu N, Rao X, Gao Y, Wang J, Xu F (2013) Amyloid-beta deposition and olfactory dysfunction in an Alzheimer's disease model. J Alzheimers Dis 37:699–712. https://doi.org/10.3233/JAD-122443
Gupta VK et al (2016) Amyloid beta accumulation and inner retinal degenerative changes in Alzheimer's disease transgenic mouse. Neurosci Lett 623:52–56. https://doi.org/10.1016/j.neulet.2016.04.059
Holth J, Patel T, Holtzman DM (2017) Sleep in Alzheimer's disease - beyond amyloid. Neurobiol Sleep Circadian Rhythms 2:4–14. https://doi.org/10.1016/j.nbscr.2016.08.002
Golzan SM et al (2017) Retinal vascular and structural changes are associated with amyloid burden in the elderly: ophthalmic biomarkers of preclinical Alzheimer's disease. Alzheimers Res Ther 9:13. https://doi.org/10.1186/s13195-017-0239-9
Criscuolo C et al (2018) The retina as a window to early dysfunctions of Alzheimer's disease following studies with a 5xFAD mouse model. Neurobiol Aging 67:181–188. https://doi.org/10.1016/j.neurobiolaging.2018.03.017
Perez SE, Lumayag S, Kovacs B, Mufson EJ, Xu S (2009) Beta-amyloid deposition and functional impairment in the retina of the APPswe/PS1DeltaE9 transgenic mouse model of Alzheimer's disease. Invest Ophthalmol Vis Sci 50:793–800. https://doi.org/10.1167/iovs.08-2384
Liu B et al (2009) Amyloid-peptide vaccinations reduce {beta}-amyloid plaques but exacerbate vascular deposition and inflammation in the retina of Alzheimer's transgenic mice. Am J Pathol 175:2099–2110. https://doi.org/10.2353/ajpath.2009.090159
Berisha F, Feke GT, Trempe CL, McMeel JW, Schepens CL (2007) Retinal abnormalities in early Alzheimer's disease. Invest Ophthalmol Vis Sci 48:2285–2289. https://doi.org/10.1167/iovs.06-1029
Sivak JM (2013) The aging eye: common degenerative mechanisms between the Alzheimer's brain and retinal disease. Invest Ophthalmol Vis Sci 54:871–880. https://doi.org/10.1167/iovs.12-10827
M Mirzaei VBG, Gupta VK (2018) Retinal changes in Alzheimer's disease: disease mechanisms to evaluation perspectives. J Neurol Neuromed 3:11–13
Sperling RA et al (2011) Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 7:280–292. https://doi.org/10.1016/j.jalz.2011.03.003
Buchhave P et al (2012) Cerebrospinal fluid levels of beta-amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry 69:98–106. https://doi.org/10.1001/archgenpsychiatry.2011.155
Johnson KA et al (2013) Appropriate use criteria for amyloid PET: a report of the Amyloid Imaging Task Force, the Society of Nuclear Medicine and Molecular Imaging, and the Alzheimer's Association. Alzheimers Dement 9:e-1–e16. https://doi.org/10.1016/j.jalz.2013.01.002
Hart NJ, Koronyo Y, Black KL, Koronyo-Hamaoui M (2016) Ocular indicators of Alzheimer's: exploring disease in the retina. Acta Neuropathol 132:767–787. https://doi.org/10.1007/s00401-016-1613-6
Chiu K et al (2012) Neurodegeneration of the retina in mouse models of Alzheimer's disease: what can we learn from the retina? Age (Dordr) 34:633–649. https://doi.org/10.1007/s11357-011-9260-2
Gupta V et al (2016) One protein, multiple pathologies: multifaceted involvement of amyloid beta in neurodegenerative disorders of the brain and retina. Cell Mol Life Sci 73:4279–4297. https://doi.org/10.1007/s00018-016-2295-x
Gasparini L et al (2011) Tau inclusions in retinal ganglion cells of human P301S tau transgenic mice: effects on axonal viability. Neurobiol Aging 32:419–433. https://doi.org/10.1016/j.neurobiolaging.2009.03.002
Ning A, Cui J, To E, Ashe KH, Matsubara J (2008) Amyloid-beta deposits lead to retinal degeneration in a mouse model of Alzheimer disease. Invest Ophthalmol Vis Sci 49:5136–5143. https://doi.org/10.1167/iovs.08-1849
Mirzaei M et al (2017) Age-related neurodegenerative disease associated pathways identified in retinal and vitreous proteome from human glaucoma eyes. Sci Rep 7:12685. https://doi.org/10.1038/s41598-017-12858-7
Serneels L et al (2009) Gamma-Secretase heterogeneity in the Aph1 subunit: relevance for Alzheimer's disease. Science 324:639–642. https://doi.org/10.1126/science.1171176
Gengler S, Hamilton A, Holscher C (2010) Synaptic plasticity in the hippocampus of a APP/PS1 mouse model of Alzheimer's disease is impaired in old but not young mice. PLoS One 5:e9764. https://doi.org/10.1371/journal.pone.0009764
Radde R et al (2006) Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep 7:940–946. https://doi.org/10.1038/sj.embor.7400784
Rupp NJ, Wegenast-Braun BM, Radde R, Calhoun ME, Jucker M (2011) Early onset amyloid lesions lead to severe neuritic abnormalities and local, but not global neuron loss in APPPS1 transgenic mice. Neurobiol Aging 32(2324):e2321–e2326. https://doi.org/10.1016/j.neurobiolaging.2010.08.014
Winkler BS (1972) The electroretinogram of the isolated rat retina. Vision Res 12:1183–1198
Gupta V et al (2017) Glaucoma is associated with plasmin proteolytic activation mediated through oxidative inactivation of neuroserpin. Sci Rep 7:8412. https://doi.org/10.1038/s41598-017-08688-2
Mirzaei M et al (2017) in Proteome bioinformatics (eds Shivakumar Keerthikumar & Suresh Mathivanan) 45-66 (Springer New York)
Margolin AA et al (2009) Empirical Bayes analysis of quantitative proteomics experiments. PloS one 4:e7454. https://doi.org/10.1371/journal.pone.0007454
Kammers K, Cole RN, Tiengwe C, Ruczinski I (2015) Detecting significant changes in protein abundance. EuPA open Proteomics 7:11–19. https://doi.org/10.1016/j.euprot.2015.02.002
Gupta VK, Rajala A, Daly RJ, Rajala RV (2010) Growth factor receptor-bound protein 14: a new modulator of photoreceptor-specific cyclic-nucleotide-gated channel. EMBO Rep 11:861–867. https://doi.org/10.1038/embor.2010.142
Gupta VK, Rajala A, Rajala RV (2012) Insulin receptor regulates photoreceptor CNG channel activity. Am J Physiol Endocrinol Metab 303:E1363–E1372. https://doi.org/10.1152/ajpendo.00199.2012
Chitranshi N et al (2017) Ptpn11 induces endoplasmic stress and apoptosis in Sh-Sy5y cells. Neuroscience 364:175–189. https://doi.org/10.1016/j.neuroscience.2017.09.028
Rajala A, Gupta VK, Anderson RE, Rajala RV (2013) Light activation of the insulin receptor regulates mitochondrial hexokinase. A possible mechanism of retinal neuroprotection. Mitochondrion 13:566–576. https://doi.org/10.1016/j.mito.2013.08.005
Chitranshi N et al (2018) Loss of Shp2 rescues BDNF/TrkB signaling and contributes to improved retinal ganglion cell neuroprotection. Mol Ther. https://doi.org/10.1016/j.ymthe.2018.09.019
Sultana R et al (2006) Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis 22:76–87. https://doi.org/10.1016/j.nbd.2005.10.004
Yu L et al (2018) Targeted brain proteomics uncover multiple pathways to Alzheimer's dementia. Ann Neurol 84:78–88. https://doi.org/10.1002/ana.25266
Kempf SJ et al (2016) An integrated proteomics approach shows synaptic plasticity changes in an APP/PS1 Alzheimer's mouse model. Oncotarget 7:33627–33648. https://doi.org/10.18632/oncotarget.9092
Koronyo Y et al (2017) Retinal amyloid pathology and proof-of-concept imaging trial in Alzheimer's disease. JCI Insight 2. https://doi.org/10.1172/jci.insight.93621
Johnson-Wood K et al (1997) Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci U S A 94:1550–1555
Oliveira JM, Henriques AG, Martins F, Rebelo S, da Cruz e Silva OA (2015) Amyloid-beta modulates both AbetaPP and Tau phosphorylation. J Alzheimers Dis 45:495–507. https://doi.org/10.3233/JAD-142664
Chiasseu M et al (2016) Tau Accumulation, altered phosphorylation, and missorting promote neurodegeneration in glaucoma. J Neurosci 36:5785–5798. https://doi.org/10.1523/JNEUROSCI.3986-15.2016
Zhao H et al (2013) Hyperphosphorylation of tau protein by calpain regulation in retina of Alzheimer's disease transgenic mouse. Neurosci Lett 551:12–16. https://doi.org/10.1016/j.neulet.2013.06.026
Pedros I et al (2014) Early alterations in energy metabolism in the hippocampus of APPswe/PS1dE9 mouse model of Alzheimer's disease. Biochim Biophys Acta 1842:1556–1566. https://doi.org/10.1016/j.bbadis.2014.05.025
Bennett RE et al (2017) Enhanced Tau aggregation in the presence of amyloid beta. Am J Pathol 187:1601–1612. https://doi.org/10.1016/j.ajpath.2017.03.011
Kranenburg O et al (2005) Amyloid-beta-stimulated plasminogen activation by tissue-type plasminogen activator results in processing of neuroendocrine factors. Neuroscience 131:877–886. https://doi.org/10.1016/j.neuroscience.2004.11.044
Tucker HM et al (2000) The plasmin system is induced by and degrades amyloid-beta aggregates. J Neurosci 20:3937–3946
Hook V et al (2005) Inhibition of cathepsin B reduces beta-amyloid production in regulated secretory vesicles of neuronal chromaffin cells: evidence for cathepsin B as a candidate beta-secretase of Alzheimer's disease. Biol Chem 386:931–940. https://doi.org/10.1515/BC.2005.108
Hook VY, Kindy M, Hook G (2008) Inhibitors of cathepsin B improve memory and reduce beta-amyloid in transgenic Alzheimer disease mice expressing the wild-type, but not the Swedish mutant, beta-secretase site of the amyloid precursor protein. J Biol Chem 283:7745–7753. https://doi.org/10.1074/jbc.M708362200
Embury CM et al (2017) Cathepsin B improves ss-amyloidosis and learning and memory in models of Alzheimer's disease. J Neuroimmune Pharmacol 12:340–352. https://doi.org/10.1007/s11481-016-9721-6
Pamren A et al (2011) Mutations in nicastrin protein differentially affect amyloid beta-peptide production and Notch protein processing. J Biol Chem 286:31153–31158. https://doi.org/10.1074/jbc.C111.235267
Sesele K et al (2013) Conditional inactivation of nicastrin restricts amyloid deposition in an Alzheimer's disease mouse model. Aging Cell 12:1032–1040. https://doi.org/10.1111/acel.12131
Nadler Y et al (2008) Increased expression of the gamma-secretase components presenilin-1 and nicastrin in activated astrocytes and microglia following traumatic brain injury. Glia 56:552–567. https://doi.org/10.1002/glia.20638
Ciechanover A, Kwon YT (2015) Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med 47:e147. https://doi.org/10.1038/emm.2014.117
Keck S, Nitsch R, Grune T, Ullrich O (2003) Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J Neurochem 85:115–122
Morozov AV et al (2016) Amyloid-beta increases activity of proteasomes capped with 19S and 11S regulators. J Alzheimers Dis 54:763–776. https://doi.org/10.3233/JAD-160491
Aso E et al (2012) Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer's disease. Brain Pathol 22:636–653. https://doi.org/10.1111/j.1750-3639.2011.00560.x
Ding Q, Markesbery WR, Chen Q, Li F, Keller JN (2005) Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci Off J Soc Neurosci 25:9171–9175. https://doi.org/10.1523/JNEUROSCI.3040-05.2005
Hernandez-Ortega K, Garcia-Esparcia P, Gil L, Lucas JJ, Ferrer I (2016) Altered machinery of protein synthesis in Alzheimer's: from the nucleolus to the ribosome. Brain Pathol 26:593–605. https://doi.org/10.1111/bpa.12335
Garcia-Esparcia P et al (2017) Altered mechanisms of protein synthesis in frontal cortex in Alzheimer disease and a mouse model. Am J Neurodegener Dis 6:15–25
Meier S et al (2016) Pathological Tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci Off J Soc Neurosci 36:1001–1007. https://doi.org/10.1523/JNEUROSCI.3029-15.2016
Bou Samra E et al (2017) A role for Tau protein in maintaining ribosomal DNA stability and cytidine deaminase-deficient cell survival. Nat Commun 8:693. https://doi.org/10.1038/s41467-017-00633-1
Wu Y et al (2016) Regulation of global gene expression and cell proliferation by APP. Sci Rep 6:22460. https://doi.org/10.1038/srep22460
Mao JJ et al (2001) The relationship between alphaB-crystallin and neurofibrillary tangles in Alzheimer's disease. Neuropathol Appl Neurobiol 27:180–188
Dulle JE, Fort PE (2016) Crystallins and neuroinflammation: the glial side of the story. Biochim Biophys Acta 1860:278–286. https://doi.org/10.1016/j.bbagen.2015.05.023
Piri N, Kwong JM, Caprioli J (2013) Crystallins in retinal ganglion cell survival and regeneration. Mol Neurobiol 48:819–828. https://doi.org/10.1007/s12035-013-8470-2
Liedtke T, Schwamborn JC, Schroer U, Thanos S (2007) Elongation of axons during regeneration involves retinal crystallin beta b2 (crybb2). Mol Cell Proteomics 6:895–907. https://doi.org/10.1074/mcp.M600245-MCP200
Anders F et al (2017) Intravitreal injection of beta-crystallin B2 improves retinal ganglion cell survival in an experimental animal model of glaucoma. PloS one 12:e0175451. https://doi.org/10.1371/journal.pone.0175451
Shammas SL et al (2011) Binding of the molecular chaperone alphaB-crystallin to Abeta amyloid fibrils inhibits fibril elongation. Biophys J 101:1681–1689. https://doi.org/10.1016/j.bpj.2011.07.056
Bjorkdahl C et al (2008) Small heat shock proteins Hsp27 or alphaB-crystallin and the protein components of neurofibrillary tangles: tau and neurofilaments. J Neurosci Res 86:1343–1352. https://doi.org/10.1002/jnr.21589
Acknowledgements
We acknowledge the support from the Ophthalmic Research Institute of Australia, National Health and Medical Research Council (NHMRC) and Hillcrest Foundation. The mass spectrometry analysis in this study was conducted at the Australian Proteome Analysis Facility supported by the Australian Government’s National Collaborative Research Infrastructure Scheme (NCRIS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Supplementary Figure 1:
Protein interaction and functional analysis. A, B. both young and older APP/PS1 mice respectively used to understand biochemical and physiological retinal anomalies cause due to AD progression with age. (PNG 395 kb)
Supplementary Figure 2:
Immunostaining of the old retinal sections from WT and APP/PS1 mice illustrating nicastrin (Red–Cy3) expression (blue-DAPI) (n=3). Overexpressed locations were indicated by the white arrowheads. Scale=50 μm. (PNG 261 kb)
Supplementary Figure 3:
Relative fluorescence intensity of the GCL region of immunostained sections as labelled above each graph. The sections were compared between WT and APP retinal staining. Aβ (Fig. 5C); Presenilin 1, Tau, pTau Ser404 (Fig. 6 B,C,D), Cathepsin B (Fig. 7B), PSMB2 (Fig. 7D), MRPL2 (Fig. 8B), EEF1E1 (Fig. 8D), αB Crystallin (Fig. 9B), βB2 Crystallin (Fig. 9D) and βB3 Crystallin (Fig. 9F), (n=3 each) (****p<0.0001, ***p<0.001, *p<0.01). (PNG 303 kb)
Supplementary Table 1:
The list of proteins identified in the young retinal samples, including differentially expressed proteins identified from pairwise comparison test of APP/Ps1 vs. WT. The table 1 contains 4 independent sheets; Sheet 1- list of quantified proteins in the TMT experiments number 1 (younger retina), Sheet 2- list of differentially expressed proteins (APP vs WT comparison student t-test, p-value ≤ 0.05 and ≥ 1.15-fold difference), Sheet 3 and Sheet 4- list of down-regulated protein and list of up-regulated proteins respectively. Column A represents: UniProt ID; Column B: Peptide counts; Column C-G: fold changes ratios obtained from the TMT reporter ion intensities of APP replicates (129N, 129C, 130 N, 130 C and 131) versus the reference replicate 126 (one of the WT replicates); Column H-K: fold change ratios obtained from the TMT reporter ion intensities of WT replicates (127N, 127C, 128N, 128C) versus the reference replicate 126 (one of the WT replicates); Column L: p-values generated from APP vs WT t-test comparison test (p-value ≤ 0.05 highlighted as yellow); Column M: Max fold change of mean ration of APP vs WT; Column N: mean value of the fold change ratio of APP replicates vs reference replicate (126), purple: up-regulated, green: down-regulated; Column O: mean value of the fold change ratio of WT replicates vs reference replicate (126), purple: up-regulated, green: down-regulated ; Column P: Protein name/description and Column Q: Gene ontology information of every protein. (XLSX 840 kb)
Supplementary Table 2:
The list of proteins identified in the old retina samples, including differentially expressed proteins from pairwise comparison test of APP/Ps1 vs. WT. The table 2 contains 4 independent sheets; Sheet 1-list of quantified proteins in the TMT experiments number 1 (older retina), Sheet 2- list of differentially expressed proteins (APP vs WT comparison student t-test, p-value ≤ 0.05 and ≥ 1.15-fold difference), Sheet 3 and 4- list of down-regulated protein and list of up-regulated proteins respectively. Column A represents : UniProt ID; Column B: Peptide counts; Column C-G: fold changes ratios obtained from the TMT reporter ion intensities of APP replicates (129N, 129C, 130 N, 130 C and 131) versus the reference replicate 126 (one of the WT replicates); Column H-K: fold change ratios obtained from the TMT reporter ion intensities of WT replicates (127N, 127C, 128N, 128C) versus the reference replicate 126 (one of the WT replicates); Column L: P-values generated from APP vs WT t-test comparison test (p-value ≤ 0.05 highlighted as yellow); Column M: Max fold change of mean ration of APP vs WT; Column N: mean value of the fold change ratio of APP replicates vs reference replicate (126) - purple: up-regulated, green: down-regulated; Column O: mean value of the fold change ratio of WT replicates vs reference replicate (126), purple: up-regulated, green: down-regulated; Column P: Protein name/description; Column Q: Gene ontology information of every protein (XLSX 884 kb)
Rights and permissions
About this article
Cite this article
Mirzaei, M., Pushpitha, K., Deng, L. et al. Upregulation of Proteolytic Pathways and Altered Protein Biosynthesis Underlie Retinal Pathology in a Mouse Model of Alzheimer’s Disease. Mol Neurobiol 56, 6017–6034 (2019). https://doi.org/10.1007/s12035-019-1479-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1479-4