Abstract
Autism is a neurodevelopmental disorder characterized by stereotypic repetitive behaviors, impaired social interactions, and communication deficits. Numerous immune system abnormalities have been described in individuals with autism including abnormalities in the ratio of Th1/Th2/Th17 cells; however, the expression of the transcription factors responsible for the regulation and differentiation of Th1/Th2/Th17/Treg cells has not previously been evaluated. Peripheral blood mononuclear cells (PBMCs) from children with autism (AU) or typically developing (TD) control children were stimulated with phorbol-12-myristate 13-acetate (PMA) and ionomycin in the presence of brefeldin A. The expressions of Foxp3, RORγt, STAT-3, T-bet, and GATA-3 mRNAs and proteins were then assessed. Our study shows that children with AU displayed altered immune profiles and function, characterized by a systemic deficit of Foxp3+ T regulatory (Treg) cells and increased RORγt+, T-bet+, GATA-3+, and production by CD4+ T cells as compared to TD. This was confirmed by real-time PCR (RT-PCR) and western blot analyses. Our results suggest that autism impacts transcription factor signaling, which results in an immunological imbalance. Therefore, the restoration of transcription factor signaling may have a great therapeutic potential in the treatment of autistic disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism is a neurodevelopmental disorder characterized by social shortfalls and limited or stereotypic behavior patterns that is diagnosed in children at around 3 years of age [1, 2]. While the biological basis of autism is unclear, this disease likely involves a relationship between genetic liability and environmental exposures [3]. Moreover, autoimmunity against the central nervous system may have a role in the pathogenesis of autism [4, 5]. There are no reliable biological markers for autism, and diagnosis is grounded on behavioral traits and developmental history [6]. Moreover, the most reliable treatment for autism is behavioral therapy, which is most effective when implemented early in life [7]. Immune system abnormalities have been implicated in the pathogenesis of autism. In fact, children with a diagnosis of autism often present with immune dysfunction at a young age [8]. This immunological dysregulation is complex and may be connected to modifications in the prenatal immune environment, which may result in increased risk of autism [9]. However, it is unexplored whether immunological changes arise in children due to an imbalance in the transcription factor signaling that regulates Th1/Th2/Th17/Treg cell development and function.
Transcription factors play important roles in many immune disorders. We focus on the role of key transcription factors such as forkhead box P3 (Foxp3), signal transducer and activator of transcription 3 (STAT3), retinoid-acid receptor-related orphan receptor gamma t (RORγt), T-box transcription factor (T-bet), and GATA binding protein 3 (GATA3), which play important roles in differentiation of Treg, Th17, Th1, and Th2 cells, respectively [10–12]. Given that Tregs are critical for limiting immune activation and preventing self-reactivity, their deficiency may result in activation of inflammatory pathways as observed in autism and autoimmune diseases [13]. The balance of Th1/Th2 cells is tightly controlled and there is an overactivity of either of these cell types in immune disease; there is evidence of a shift from Th1 and Th2 cells in autism [14]. IL-17 is a cytokine produced by Th17 CD4+ T cells, which are known to secrete IL-17, IL-17 F, IL-21, and IL-22 [15]. Previous reports indicated increased serum levels of IL-17 in individuals with autism spectrum disorders [16]. Alterations in transcription factor signaling may lead to imbalance in the immune systems of autistic children. For example, decreased levels of regulatory cytokines and increased levels of proinflammatory cytokines including IL-17 has been shown in children with autism [16–19]. The GATA-3 pathway has been involved and has the ability to enhance IL-4 and IL-5 and to inhibit IFN-γ cytokine production in differentiated T cells [20]. Indeed, during neurodevelopment, IL-6 activates the STAT3 pathway to maintain homeostasis between neurogenesis and gliogenesis [21]. STAT3 signaling has been proposed as a neurobiological mechanism to mediate the effects of the maternal immune system (MIA) on the developing fetal brain [22]. In addition, STAT3 is the major transcription factor involved in the differentiation of Th17 cells [23].
Based on the abnormal immune responses observed in children with autism, we assessed the expression of transcription factors associated with Th1, Th2, Th17, and Treg development and function. To our best knowledge, this is the first study to show an increase in Th1/Th2/Th17-related transcription factors and a decrease in Treg-related transcription factors in autistic children. Based on our findings, we hypothesize that alterations in transcription factor signaling may be responsible for the dysregulated immune system in children with autism.
Materials and Methods
Participants
This cross-sectional study was conducted on 40 children, who had classic-onset autism, over a period of 6 months from the beginning of June 2015 to the end of November 2015. The autistic group comprised 40 children (30 males and 10 females) recruited from the Autism Research and Treatment Center, Faculty of Medicine, King Saud University, Riyadh, Saudi Arabia. Subjects were fulfilling the criteria for the diagnosis of autism according to the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders [2]. Their ages ranged between 3 and 11 years (mean ± SD = 7.69 ± 2.26 years). Subjects included in this study had no associated neurological diseases (such as cerebral palsy, tuberous sclerosis) or metabolic disorders (e.g., phenylketonuria) because these associated comorbidities with autism may influence the results. Also, the included subjects were not receiving any medications.
The control group comprised 32 age- and sex-matched healthy children (24 males and 8 females). They were the healthy older siblings of the healthy infants who attend the Well Baby Clinic, King Khalid University Hospital, Faculty of Medicine, King Saud University, Riyadh, Saudi Arabia for routine following up of their growth parameters. The control children were not related to the children with autism and demonstrated no clinical findings suggestive of immunological or neuropsychiatric disorders. Their ages ranged between 3 and 11 years (mean ± SD = 7.76 ± 2.45 years). The local Ethical Committee of the Faculty of Medicine, King Saud University, Riyadh, Saudi Arabia approved this study. In addition, an informed written consent of participation in the study was signed by the parents or the legal guardians of the studied subjects.
Study Measurements
Clinical evaluation of autistic subjects: This was based on clinical history taking from caregivers, clinical examination, and neuropsychiatric assessment. In addition, the disease severity was assessed by using the Childhood Autism Rating Scale (CARS) [24] which rates the child on a scale from one to four in each of 15 areas (relating to people; emotional response; imitation; body use; object use; listening response; fear or nervousness; verbal communication; non-verbal communication; activity level; level and consistency of intellectual response; adaptation to change; visual response; taste, smell, and touch response and general impressions).
Chemicals and Antibodies
The chemicals and antibodies used in this study were purchased from the following companies. Primary antibodies [RORγ (no. sc-28559), Foxp3 (no. sc-130666), T-bet (no. sc-21763), GATA-3 (no. sc-268), STAT3 (no. sc-8019)], secondary antibodies [anti-mouse, anti-rabbit and anti-goat horseradish peroxidase-conjugated antibodies (no. sc-2005, no. sc-2004 and no. sc-2020 respectively)], and broad range markers (no. sc-2361) were purchased from Santa Cruz Biotechnology, Inc, USA. Histopaque-1077 (no. 10771), heparin (no. H3393), phorbol-12-myristate 13-acetate (no. P8139) and ionomycin (no. 9657) were purchased from Sigma-Aldrich, USA. Fluoro isothiocyanate-labeled CD4 (no. 357406) anti-human monoclonal antibody, and phycoerythrin labeled Foxp3 (no. 320008), RORγt (no. 563081), T-bet (no. 644810), and GATA-3 (no. 653804) anti-human monoclonal antibodies, GolgiStop (no. 554724), FcR blocking reagent (no. 130-059-901), fixation (no. 420801) and permeabilizing buffers (no. 421002) were obtained from (Miltenyi Biotech, Germany; BioLegend and BD Biosciences, USA). Primers used to assess gene expressions were purchased from Applied Bio Systems (Paisley, UK) and Genscript (Piscataway, USA); TRIzol reagent (15596018) was purchased from Life Technologies (Grand Island, USA); High Capacity cDNA Reverse Transcription kit (no. 4368814) and SYBR® Green PCR Master Mix (1509503) were purchased from Applied Bio systems (Paisley, UK). Luminata forte Western HRP substrates (no. WBLUF0100) were purchased from Merck Millipore UK; nitrocellulose membranes (LC2001) were purchased from Life technologies, USA.
Isolation of PBMCs
Peripheral blood was collected in an acid-citrate-dextrose Vacutainer tube (BD Biosciences; USA) and processed for flow cytometry and gene/protein expression analyses. Peripheral blood was mixed 1:1 with Hanks balanced salt solution (HBSS; Gibco, Gaithersburg, MD, USA), and the diluted blood was then carefully layered over a Ficoll-Paque gradient (Sigma-Aldrich, USA) and centrifuged at 400×g for 40 min at room temperature. This centrifugation separated lymphocytes, monocytes, and plasma. The peripheral blood mononuclear cell (PBMC) layers were carefully removed from the tube and transferred to a new conical tube followed by two washes with HBSS [25]. After centrifugation, the PBMCs were resuspended in the appropriate volume of 1× PBS and were stored at −80 °C.
Intracellular Staining and Flow Cytometric Analysis
Transcription factors were assessed by flow cytometric analysis of PBMCs stimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (10 μg/ml, Sigma, USA) in the presence of 5 μg/ml brefeldin A (Bio legend, USA) for 4 h, using methods previously described for intracellular staining [26, 27]. Briefly, PBMCs were stained for the CD4 T cell surface marker (Bio legend, USA), followed by incubation at room temperature in the dark for 10 min. The freshly prepared fixation/permeabilization solution (Miltenyi Biotech, Germany; Biolegend and BD Biosciences, USA) was added to the pellet. Cells were permeabilized and incubated for 10 min at room temperature followed by adding, human Fc receptor blocking solution (Miltenyi Biotech, Germany). After washing with wash buffer, anti-human Foxp3, RORγt, T-bet, and GATA-3 (Biolegend and BD Biosciences, USA) were added to the cells, followed by incubation for 30 min at room temperature in the dark. All cells were analyzed for the expression of phenotypic markers and transcription factors on FC500 flow cytometer (Beckman Coulter, USA) using CXP software, and 10,000 events were counted. To analyze the staining of the CD4, Foxp3, RORγt, T-bet, and GATA-3, the lymphocytes were first gated by their physical properties (forward and side scatter). Then, a second gate was drawn based on the immunofluorescence characteristics of the gated cells. The data are presented as dot plots as previously described [28].
Isolation of Total RNA and RT-PCR
Total RNA was extracted using Trizol (Invitrogen; Life Technologies, USA) according to method described earlier [29, 30]. cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems, USA) according to the manufacturer’s instructions. The primers used in these assays were selected from PubMed and other databases, which are listed in Table 1. Assay controls were incorporated into the same plate, which consisted of no-template controls to test for contaminations of any of the assay reagents. The real-time PCR data were analyzed using the relative gene expression (i.e., ΔΔCT) method, as described in Applied Biosystems User Bulletin No. 2. Briefly, the data are presented as the fold change in gene expression normalized to an endogenous reference gene (GAPDH) and relative to a calibrator.
Protein Extraction and Western Blot Analysis
The total cellular proteins were extracted from PBMCs by a method as previously described [31]. Protein concentrations were measured by the Lowry method [32]. Western blot analysis was performed using a previously described method [32]. Briefly, 25–50 μg of protein from each group was separated by 10 % SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred into nitrocellulose membranes (Bio-Rad, USA). Protein blots were blocked overnight at 4 °C, followed by incubations with primary antibodies against Foxp3, RORγ, T-bet, GATA-3, and STAT3 (Santa Cruz Biotechnology, Inc, USA), followed by incubation for 2 h with peroxidase-conjugated secondary antibodies at room temperature. The Foxp3, RORγ, STAT3, T-bet, and GATA-3 bands were visualized using the enhanced chemiluminescence method (GE Health care, Mississauga, Canada) and quantified relative to β-actin bands using the ImageJ® image processing program (National Institutes of Health, Bethesda, USA). Images were taken on C-Digit chemiluminescent Western blot scanner (LI-COR, Lincoln, USA).
Statistical Analysis
The results were analyzed using a commercially available software package (GraphPad InStat Software, La Jolla USA). The data were expressed as mean ± SEM. The results were analyzed by Student’s t test. The level of statistical significance was set at p < 0.05 for differences between the groups.
Results
Transcription Factor Expression Correlates with Immunological Imbalance in Autism
The importance of transcription factors in the development of several neurodevelopmental disorders was shown in earlier studies [33]. As seen in Fig. 1a, children with AU display decreased Foxp3+ cells in the peripheral blood as compared to TD. This deficit is also revealed when looking specifically at CD4+Foxp3+ T cells (Fig. 1a). Further, the mRNA and protein expression of Foxp3 were significantly reduced in the PBMCs from children with AU as compared to TD (Fig. 1b, c). Taken together, these findings indicate a systemic deficit in the abundance of Tregs in children with autism.
a Flow cytometric analysis of Foxp3+ transcription factor production by all cells and specifically by CD4+ T cells in peripheral blood mononuclear cells (PBMCs). b Quantitative RT-PCR of Foxp3 mRNA expression in PBMCs. c Western blot analysis of Foxp3 protein expression in lysates generated from PBMCs. Peripheral blood was incubated with PMA/ionomycin (10 μg/ml) for 4 h. d Dot plots represent events from PBMCs taken from one typically developing (TD) control child and one autistic (AU) child. Statistical analyses were performed using Student’s t test. The level of significance was set at *p < 0.05 compared with the TD group
RORγt-deficient mice have attenuated autoimmune disease and lack tissue-infiltrating Th17 cells [34]. In addition, a number of reports show increased production of IL-17 in children with autism [16, 19]. Therefore, we assessed the expression of RORγt in children with autism and found it to be considerably increased in both CD4+ and CD4- T cells in AU children as compared to TD controls (Fig. 2a). Further, RT-PCR and western blotting analysis confirmed the strong induction of RORγt mRNA and RORγ protein expression in PBMCs from AU children as compared to TD children (Fig. 2b, c). These results provide evidence that RORγt, a key regulator of Th17 cell differentiation, is upregulated in autistic children.
a Flow cytometric analysis of RORγt+ transcription factor production by all cells and specifically by CD4+ T cells in peripheral blood mononuclear cells (PBMCs). b Quantitative RT-PCR of RORγt mRNA expression in PBMCs. c Western blot analysis of RORγ protein expression in lysates generated from PBMCs. Peripheral blood was incubated with PMA/ionomycin (10 μg/ml) for 4 h. d Dot plots represent events from PBMCs taken from one typically developing (TD) control child and one autistic (AU) child. Statistical analyses were performed using Student’s t test. The level of significance was set at *p < 0.05 compared with the TD group
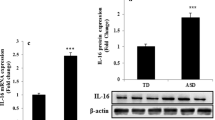
Given the increasing indications of the involvement of STAT3 in neurological disorders, and its association with the development of Th17 cells, we wanted to extend our results to determine whether STAT3 mRNA expression levels are affected in AU children [35]. Thus, we assessed STAT3 mRNA expression in the PBMCs of children with autism and found it to be upregulated compared to TD controls (Fig. 3a). Next, we examined STAT3 protein expression by western blot analysis and found a significantly higher level of STAT3 expression in samples from children with autism than in TD controls (Fig. 3b). These observations suggest increased STAT3/RORγt signaling in autistic children.
a Quantitative RT-PCR of the mRNA expression of STAT3 in peripheral blood mononuclear cells (PBMCs). b Western blot analysis of STAT3 protein expression in lysates generated from PBMCs. Statistical analyses were performed using Student’s t test. The level of significance was set at *p < 0.05 compared with the TD group.
T-bet regulates Th1 responses by mediating essential effects on GATA-3 function [36]. In children with autism, there was a significant increase in the proportion of T-bet+ cells and in the number of CD4+ T cells expressing T-bet in peripheral blood as compared to the TD controls (Fig. 4a). Furthermore, children with autism showed a significant increase in both the mRNA and protein expression of T-bet as compared to TD controls (Fig. 4b, c). These data suggest that T-bet may also play a significant role in the development of autism.
a Flow cytometric analysis of T-bet+ transcription factor production by all cells and specifically by CD4+ T cells in peripheral blood mononuclear cells (PBMCs). b Quantitative RT-PCR of T-bet mRNA expression in PBMCs. c Western blot analysis of T-bet protein expression in lysates generated from PBMCs. Peripheral blood was incubated with PMA/ionomycin (10 μg/ml) for 4 h. d Dot plots represent events from PBMCs taken from one typically developing (TD) control child and one autistic (AU) child. Statistical analyses were performed using Student’s t test. The level of significance was set at *p < 0.05 compared with the TD group.
GATA-3 is involved in the brain development and regulates many processes in the body including cell differentiation, immune responses, and the synthesis of neurotransmitters and hormones [37]. In PBMCs from AU children, we observed an upregulation in the percentage of GATA-3+ producing cells compared to TD controls (Fig. 4a). A similar trend was noted when looking specifically at CD4+GATA-3+ T cells (Fig. 5a). Similarly, GATA-3 mRNA and protein were highly expressed in autistic children compared to TD controls (Fig. 5b, c). Our data strongly suggest that GATA-3 may play an important role in autism. Overall, our data show that Th17-, Th1-, and Th2-related transcription factors are upregulated and Treg-related transcription factor is downregulated in PBMCs of autistic children as compared to typically developing children.
a Flow cytometric analysis of GATA-3+ transcription factor production by all cells and specifically by CD4+ T cells in peripheral blood mononuclear cells (PBMCs). b Quantitative RT-PCR of GATA-3 mRNA expression in PBMCs. c Western blot analysis of GATA-3 protein expression in lysates generated from PBMCs. Peripheral blood was incubated with PMA/ionomycin (10 μg/ml) for 4 h. d Dot plots represent events from PBMCs taken from one typically developing (TD) control child and one autistic (AU) child. Statistical analyses were performed using Student’s t test. The level of significance was set at *p < 0.05 compared with the TD group
Discussion
Autistic disorders are complex neurodevelopmental conditions principally characterized by dysfunctions linked to mental development [38]. Recent studies have shown that there are more than 1000 genes likely involved in autism, which are highly interconnected among themselves and expressed mainly in the brain [39]. There are also many reports of peripheral immune abnormalities in autistic individuals [40–42]. Immune aberrations consistent with a dysregulated immune response that have been reported in autistic subjects include abnormal or skewed Th1 and Th2 cytokine profiles, decreased lymphocyte numbers, and an imbalance in serum immunoglobulin levels [43, 44]. Recently, a number of studies have examined the role played by Treg cells in human immune-mediated diseases and their findings suggest that an acquired defect in Foxp3 expression may be a contributing factor [45, 46]. Treg cells play an important role in the formation of immunological self-tolerance and thereby in the prevention of autoimmunity [47]. Moreover, Foxp3 expression has been shown to be reduced in individuals who develop inflammatory neurologic diseases, and reduced functionality of Treg cells is associated with upregulation of RORγt [48, 49]. In the present investigation, we found a significant downregulation of Foxp3 specifically within CD4+ T cells in the peripheral blood of autistic children. These results suggest that a reduction in Foxp3 expression is associated with autism. Together, these observations confirm that immune dysfunction, and specifically deficits in Treg cells, correlates with the modulation of behaviors and core features of autism.
A recent study reported higher expression of GATA-3 in lymphoblastic cell lines derived from the lymphocytes of autistic subjects as compared to their non-autistic siblings [50], suggesting the possible use of GATA-3 as a disease biomarker. In addition, in a neuron-like PC-12 cell culture, GATA-3 expression increased after treatment with valproate, thalidomide, or alcohol, three teratogens known to cause autism [51]. Our data confirms that the GATA-3 expression is higher in children with autism as compared to TD controls. It is hypothesized that an excessive differentiation of neurons expressing GATA-3 could cause anomalies in the development of brain resulting in autistic symptoms. Thus, GATA-3 may represent a molecular link between immune and neuronal dysfunction in autism and further study is warranted.
STAT3 and RORγt are thought to collaboratively regulate the transcriptional profile of Th17 cells, and both are known to be critical for Th17 cell differentiation [23]. The underlying mechanism is believed to involve the translocation of activated STAT3 into the nucleus where it promotes transcription of RORγt, the essential transcription factor for Th17 cell differentiation [52]. The prominent expression of IL-17A has been reported in children with autism [15, 16]. There are several STAT protein variants, which lead to an array of signaling that can result either in neuronal death or growth and differentiation [53]. The activation and regulation of STAT3 signal is known to be critically involved during neurodevelopment [54]. In addition, earlier results showed that blocking the STAT3 signaling pathway significantly opposes maternal immune activation (MIA)-induced abnormal behavior and neuropathological abnormalities in MIA/adult offspring [54]. Similarly, children with autism demonstrated noticeable upregulation of STAT3 mRNA and protein expression. Therefore, our study suggests the involvement of STAT3 in autism and further exploration of this key signaling pathway is warranted.
T-bet, a master transcription factor, controls lineage commitment of CD4+ T cells and promotes Th1 differentiation [55–57]. Our study shows increased T-bet expression in CD4 T cells in children with autism. Th1 cells are known to be involved in the pathogenesis of neurological disorders such as multiple sclerosis and depression and Th1 cells, along with Th17 cells, regulate neuroinflammation [58, 59]. Our study shows increases in both of these cells, which suggests involvement of T-bet as well as RORγt in autism. Overall, our study shows immune dysregulation of transcription factors related to Th1/Th2/Th17 cells and Treg cells in autistic children. These findings suggest that the correction of transcription factor signaling could potentially be beneficial for treatment of autism.
Conclusion
While it is well known that autism disturbs brain function, it is becoming clear that the neuroimmune axis is involved in many neurological disorders. In this manuscript, we described the modification of the immune system of autistic children as assessed through alterations in transcriptional signaling pathways. The impact of transcription factor imbalance in autistic children and the association with behavior impairment is intriguing and warrants further consideration. Future studies will provide a better understanding of the underlying mechanisms responsible for the pathogenesis of autism and may improve diagnosis and therapy. Furthermore, the correction of the immune system dysfunction in autism may signify a novel target for treatment.
Abbreviations
- ASD:
-
Autism spectrum disorders
- PBMCs:
-
Peripheral blood mononuclear cells
- PMA:
-
Phorbol-12-myristate 13-acetate
- AU:
-
Children with autism
- TD:
-
Typically-developing control children
- MIA:
-
Maternal immune activation
- Tregs:
-
Regulatory T cells
- CD4:
-
Cluster of differentiation 4
- Th cells:
-
T helper cells
- IL:
-
Interleukin
- Foxp3:
-
Forkhead box P3
- STAT3:
-
Signal transducer and activator of transcription 3
- RORγt:
-
Retinoid-acid receptor-related orphan receptor gamma t
- T-bet:
-
T-box transcription factor
- GATA3:
-
GATA Binding Protein 3
- mRNA:
-
Messenger RNA
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- FITC:
-
Fluoro isothiocyanate
- PE:
-
Phycoerythrin
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- CARS:
-
Childhood Autism Rating Scale
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- HBSS:
-
Hanks balanced salt solution
- cDNA:
-
Complementary DNA
- PBS:
-
Phosphate-buffered saline
References
Lord C, Cook EH, Leventhal BL, Amaral DG (2000) Autism spectrum disorders. Neuron 28(2):355–363
American Psychiatric Association (2015) Diagnostic and statistical manual of mental disorders, 5th edn. American Psychiatric Association, Arlington, VA
Herbert MR (2010) Contributions of the environment and environmentally vulnerable physiology to autism spectrum disorders. Curr Opin Neurol 23:103–110
Enstrom AM, Lit L, Onore CE, Gregg JP, Hansen RL, Pessah IN, Hertz-Picciotto I, Van de Water JA, Sharp FR, Ashwood P (2009) Altered gene expression and function of peripheral blood natural killer cells in children with autism. Brain Behav Immun 23(1):124–133
AL-Ayadhi LY, Mostafa GA (2014) Serum antinucleosome-specific antibody as a marker of autoimmunity in children with autism. J Neuroinflammation 3:11–69
Le Couteur A, Haden G, Hammal D, McConachie H (2008) Diagnosing autism spectrum disorders in pre-school children using two standardised assessment instruments: the ADI-R and the ADOS. J Autism Dev Disord 38:362–372
Dawson G, Jones EJ, Merkle K, Venema K, Lowy R, Faja S, Kamara D, Murias M et al (2012) Early behavioral intervention is associated with normalized brain activity in young children with autism. J Am Acad Child Adolesc Psychiatry 51:1150–1159
Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah I, Van de Water J (2011) Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun 25:40–45
Goines PE, Croen LA, Braunschweig D, Yoshida CK, Grether J, Hansen R, Kharrazi M, Ashwood P et al (2011) Increased mid-gestational IFN-γ, IL-4, and IL-5 in women giving birth to a child with autism: a case- control study. Mol Autism 2:13
Tsarovina K, Pattyn A, Stubbusch J, Müller F, van der Wees J, Schneider C, Brunet JF, Rohrer H (2004) Essential role of GATA transcription factors in sympathetic neuron development. Development 131(19):4775–4786
Frisullo G, Angelucci F, Caggiula M, Nociti V, Iorio R, Patanella AK, Sancricca C, Mirabella M et al (2006) pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J Neurosci Res 84(5):1027–1036
Zhang L, Zhao Y (2007) The regulation of Foxp3 expression in regulatory CD4(+)CD25(+)T cells: multiple pathways on the road. J Cell Physiol 211:590–597
Matejuk A, Bakke AC, Hopke C, Dwyer J, Vandenbark AA, Offner H (2004) Estrogen treatment induces a novel population of regulatory cells, which suppresses experimental autoimmune encephalomyelitis. J Neurosci Res 77(1):119–126
Gupta S, Aggarwal S, Rashanravan B, Lee T (1998) Th1- and Th2-like cytokines in CD4+ and CD8+ T cells in autism. J Neuroimmunol 85(1):106–109
Bettelli E, Oukka M, Kuchroo VK (2007) T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol 8(4):345–350
Al-Ayadhi LY, Mostafa GA (2012) Elevated serum levels of interleukin-17A in children with autism. J Neuroinflammation 2(9):158
Molloy CA, Morrow AL, Meinzen-Derr J, Schleifer K, Dienger K, Manning-Courtney P, Altaye M, Wills-Karp M (2006) Elevated cytokine levels in children with autism spectrum disorder. J Neuroimmunol 172(1-2):198–205
Patterson PH (2011) Maternal infection and immune involvement in autism. Trends Mol Med 17:389–394
Akintunde ME, Rose M, Krakowiak P, Heuer L, Ashwood P, Hansen R, Hertz-Picciotto I, Van de Water J (2015) Increased production of IL-17 in children with autism spectrum disorders and co-morbid asthma. J Neuroimmunol 15(286):33–41
Ranganath S, Murphy KM (2001) Structure and specificity of GATA proteins in Th2 development. Mol Cell Biol 21:2716–2725
He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D et al (2005) A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci 8:616–625
Khan D, Fernando P, Cicvaric A, Berger A, Pollak A, Monje FJ, Pollak DD (2014) Long-term effects of maternal immune activation on depression-like behavior in the mouse. Transl Psychiatry 18(4):e363
Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C (2007) STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 282:9358–9363
Schopler E, Reichler RJ, Renner BR (1986) The childhood autism rating scale (CARS): for diagnostic screening and classification of autism. Irvington, New York
Ahmad SF, Zoheir KM, Ansari MA, Nadeem A, Bakheet SA, Al-Hoshani AR, Al-Shabanah OA, Al-Harbi MM et al (2015) Histamine 4 receptor promotes expression of costimulatory B7.1/B7.2 molecules, CD28 signaling and cytokine production in stress-induced immune responses. J Neuroimmunol 15(289):30–42
Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah IN, Van de Water J (2011) Altered T cell responses in children with autism. Brain Behav Immun 25(5):840–849
Wei J, Zheng M, Liang P, Wei Y, Yin X, Tang Y, Xue Y (2013) Apolipoprotein E and its mimetic peptide suppress Th1 and Th17 responses in experimental autoimmune encephalomyelitis. Neurobiol Dis 56:59–65
Ahmad SF, Zoheir KM, Bakheet SA, Ashour AE, Attia SM (2014) Poly(ADP-ribose) polymerase-1 inhibitor modulates T regulatory and IL-17 cells in the prevention of adjuvant induced arthritis in mice model. Cytokine 68(2):76–85
Drulovic J, Savic E, Pekmezovic T, Mesaros S, Stojsavljevic N, Dujmovic-Basuroski I, Kostic J, Vasic V (2009) Expression of Th1 and Th17 cytokines and transcription factors in multiple sclerosis patients: does baseline T-bet mRNA predict the response to interferon-beta treatment? J Neuroimmunol 215(1-2):90–95
Sthoeger Z, Zinger H, Sharabi A, Asher I, Mozes E (2013) The tolerogenic peptide, hCDR1, down-regulates the expression of interferon-α in murine and human systemic lupus erythematosus. PLoS ONE 8(3), e60394
Chen YC, Tsai WJ, Wu MH, Lin LC, Kuo YC (2007) Suberosin inhibits proliferation of human peripheral blood mononuclear cells through the modulation of the transcription factors NF-AT and NF-kappaB. Br J Pharmacol 150(3):298–312
Seevaratnam R, Patel BP, Hamadeh MJ (2009) Comparison of total protein concentration in skeletal muscle as measured by the Bradford and Lowry assays. J Biochem 145(6):791–797
Maussion G, Diallo AB, Gigek CO, Chen ES, Crapper L, Théroux JF et al (2015) Investigation of genes important in neurodevelopment disorders in adult human brain. Hum Genet 134(10):1037–1053
Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126(6):1121–1133
Yin L, Chen Y, Qu Z, Zhang L, Wang Q, Zhang Q, Li L (2014) Involvement of JAK/STAT signaling in the effect of cornel iridoid glycoside on experimental autoimmune encephalomyelitis amelioration in rats. J Neuroimmunol 274(1-2):28–37
Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O’Shea JJ, Strober W (2006) T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med 203:755–766
Van Doorninck JH, Van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M et al (1999) GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neurosci 19(12):RC12
Abrahams BS, Geschwind DH (2008) Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 9:341–355
Moreno-Ramos OA, Olivares AM, Haider NB, de Autismo LC, Lattig MC (2015) Whole-exome sequencing in a South American cohort links ALDH1A3, FOXN1 and retinoic acid regulation pathways to autism spectrum disorders. PLoS ONE 10(9):e0135927
Hsiao EY, McBride SW, Chow J, Mazmanian SK, Patterson PH (2012) Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci 109:12776–12781
Onore C, Careaga M, Ashwood P (2012) The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun 26:383–392
Lucchina L, Depino AM (2014) Altered peripheral and central inflammatory responses in a mouse model of autism. Autism Res 7(2):273–289
Ashwood P, Wills S, Van de Water J (2008) The immune response in autism: a new frontier for autism research. J Leukoc Biol 80(1):1–15
Al-Ayadhi LY, Mostafa GA (2013) Elevated serum levels of macrophage-derived chemokine and thymus and activation-regulated chemokine in autistic children. J Neuroinflammation 19(10):72
Lu L, Lan Q, Li Z, Zhou X, Gu J, Li Q, Wang J, Chen M et al (2014) Critical role of all-trans retinoic acid in stabilizing human natural regulatory T cells under inflammatory conditions. Proc Natl Acad Sci U S A 111(33):E3432-40
Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SC, Erdos MR, Davis SR, Roychoudhuri R et al (2015) Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520(7548):558–562
Vignali DA, Collison LW, Workman CJ (2008) How regulatory T cells work. Nat Rev Immunol 8:523–532
Yamano Y, Takenouchi N, Li HC, Tomaru U, Yao K, Grant CW, Maric DA, Jacobson S (2005) Virus-induced dysfunction of CD4 + CD25+ T cells in patients with HTLV-I-associated neuroimmunological disease. J Clin Invest 115:1361–1368
Yu F, Sharma S, Edwards J, Feigenbaum L, Zhu J (2015) Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol 16(2):197–206
Hu VW, Nguyen A, Kim KS, Steinberg ME, Sarachana T, Scully MA, Soldin SJ, Luu T et al (2009) Gene expression profiling of lymphoblasts from autistic and nonaffected sib pairs: altered pathways in neuronal development and steroid biosynthesis. PLoS ONE 4(6), e5775
Rout UK, Clausen P (2009) Common increase of GATA-3 level in PC-12 cells by three teratogens causing autism spectrum disorders. Neurosci Res 64(2):162–169
Park JS, Kwok SK, Lim MA, Kim EK, Ryu JG, Kim SM, Oh HJ et al (2014) STA-21, a promising STAT-3 inhibitor that reciprocally regulates Th17 and Treg cells, inhibits osteoclastogenesis in mice and humans and alleviates autoimmune inflammation in an experimental model of rheumatoid arthritis. Arthritis Rheumatol 66(4):918–929
Giunta B, Obregon D, Hou H, Zeng J, Sun N, Nikolic V, Ehrhart J, Shytle D et al (2006) EGCG mitigates neurotoxicity mediated by HIV-1 proteins gp120 and Tat in the presence of IFN-gamma: role of JAK/STAT1 signaling and implications for HIV-associated dementia. Brain Res 1123(1):216–225
Parker-Athill EC, Tan J (2010) Maternal immune activation and autism spectrum disorder: interleukin-6 signaling as a key mechanistic pathway. Neurosignals 18:113–128
Lazarevic V, Glimcher LH (2011) T-bet in disease. Nat Immunol 12(7):597–606
Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100:655–669
Bouchery T, Kyle R, Ronchese F, Le Gros G (2014) The Differentiation of CD4+ T-Helper Cell Subsets in the Context of Helminth Parasite Infection. Front Immunol 5:487
Maes M, Song C, Lin A, De Jongh R, Van Gastel A, Kenis G, Bosmans E, De Meester I et al (1998) The effects of psychological stress on humans: increased production of pro-inflammatory cytokines and a Th1-like response in stress-induced anxiety. Cytokine 10(4):313–318
McWilliams IL, Rajbhandari R, Nozell S, Benveniste E, Harrington LE (2015) STAT4 controls GM-CSF production by both Th1 and Th17 cells during EAE. J Neuroinflammation 30(12):128
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award Number (12-MED2910-02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Ahmad, S.F., Zoheir, K.M.A., Ansari, M.A. et al. Dysregulation of Th1, Th2, Th17, and T regulatory cell-related transcription factor signaling in children with autism. Mol Neurobiol 54, 4390–4400 (2017). https://doi.org/10.1007/s12035-016-9977-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9977-0