Abstract
One major pathological hallmark of Alzheimer’s disease (AD) is the accumulation of senile plaques mainly composed of neurotoxic amyloid beta-peptide (Aβ) in the patients’ brains. Sonic hedgehog (SHH) is a morphogen critically involved in the embryonic development of the central nervous system (CNS). In the present study, we tested whether Aβ may induce SHH expression and explored its underlying mechanisms. We found that both Aβ25-35 and Aβ1-42 enhanced SHH expression in the primary cortical neurons derived from fetal rat brains. Immunohistochemistry revealed heightened expression of SHH in the cortex and hippocampus of aged (9 and 12 months old) AD transgenic mouse brains as compared to age-matched littermate controls. Chromatin immunoprecipitation (ChIP) assay demonstrated that Aβ25-35 enhanced binding of hypoxia-inducible factor-1 (HIF-1) to the promoter of the Shh gene in primary cortical cultures; consistently, Aβ25-35 induction of SHH was abolished by HIF-1α small interfering RNA (siRNA). Aβ25-35 also time-dependently induced inhibitor of differentiation-1 (Id1) that has been shown to stabilize HIF-1α; further, Aβ25-35-mediated induction of HIF-1α and SHH was both suppressed by Id1 siRNA. Pharmacological induction of HIF-1α by cobalt chloride and application of the cell-permeable recombinant Id1 proteins were both sufficient to induce SHH expression. Finally, both the SHH pathway inhibitor cyclopamine and its neutralizing antibody attenuated Aβ cytotoxicity, albeit to a minor extent. These results thus established a signaling cascade of “Aβ → Id1 → HIF-1 → SHH” in primary rat cortical cultures; furthermore, SHH may in part contribute to Aβ neurotoxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia in the elderly. One of the major pathological hallmarks of AD is the accumulation of extracellular senile plaques containing aggregation of amyloid beta-peptide (Aβ), the neurotoxic peptide fragment derived from the cleavage of amyloid precursor protein (APP) sequentially by β- and γ-secretase [1, 2]. In addition to causing neuronal demise, Aβ is also known to induce the expression of specific genes via activation of such transcription factors as hypoxia-inducible factor-1 [3] and nuclear factor-kappaB (NF-κB) [4] in neurons. Aβ-dependent activation of transcription factors with resultant expression of specific target genes may either contribute to Aβ neurotoxicity [5] or, alternatively, represent an endogenous protective response under the stressful conditions of Aβ exposure [6].
The sonic hedgehog (Shh) gene is a mammalian member of the hedgehog family known to regulate polarity of the central nervous system (CNS) [7]. Physiologically, SHH is renowned for its function as a morphogen that directs ventral patterning in the spinal cord [8]. Once secreted, the biologically active N-terminal fragment of SHH protein (SHH-N) exerts its actions through binding to the Patched [9], a 12-pass transmembrane protein that, in the absence of SHH-N, suppresses another 7-pass transmembrane protein called Smoothened [10]. Activation of Smoothened by SHH-N then stimulates its downstream transcription factors of the Gli family [11], leading to the expression of specific target genes. These include N-Myc [12], Bcl-2 [13], and Bmi1 [14] that participate respectively in the regulation of proliferation, survival, and self-renewal.
Inhibitor of differentiation (Id) is a family of helix-loop-helix (HLH) proteins containing four members, Id1 to Id4, in mammalian cells [15]. These proteins do not contain the essential motif for specific DNA binding as in other bHLH transcription factors [16]. Instead, Id inhibits bHLH proteins via formation of a nonfunctional complex in a dominant negative fashion, thus negatively regulating these bHLH factors [15, 16]. Because the Id substrates, including MyoD, E12, and E47 [16], all participate in differentiation, Id proteins play a pivotal role in cell fate determination and cell cycle progression [15, 17].
Hypoxia-inducible factor-1 (HIF-1), a key regulator of mammalian oxygen homeostasis, is a heterodimeric protein complex consisting of an oxygen-sensing alpha (HIF-1α) subunit and a constitutively expressed beta (HIF-1β) subunit [18, 19]. In response to hypoxia, HIF-1 dimers enter the nucleus and bind to the consensus sequence 5′-RCGTG-3′ within the promoters of its target genes [20]. Under both physiological and pathological conditions, activation of HIF-1 may contribute to the modulation of multiple cellular adaptive responses to hypoxia including angiogenesis, cell proliferation, glucose catabolism and energy metabolism, synthesis of fatty acids and glycogen, as well as pH regulation [21, 22]. In addition to being regulated by cellular oxygen tension under hypoxia, HIF-1α may also be induced by cobalt chloride under a normoxic condition [23].
AD is a chronic neurodegenerative disorder that afflicts mostly the aging population. SHH is a morphogen that plays a critical role in the embryonic development of the nervous system. The potential correlation between Aβ, the main neurotoxic component of senile plaques in the AD brains, and SHH has been elusive. In this study, we characterized the molecular mechanisms underlying Aβ-induced SHH and explored the potential SHH effects on Aβ toxicity in primary cortical neurons.
Materials and Methods
Reagents and Preparation of Aβs
The HIF-1 inhibitor 2-methoxyestradiol (2ME2; Cat. No. M6383; Sigma, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO; Cat. No. JTB-9033-04; Capitol Scientific, Inc., Austin, TX, USA) as a stock solution of 10 mM. Cobalt chloride (CoCl2; Cat. No. C8661; Sigma) was dissolved in autoclaved ddH2O as a stock solution of 0.5 M. The recombinant human Id1 protein (amino acids 2–155) with an 11-arginine tag on its C-terminus was purchased from Abcam (Cat. No. ab134896; Cambridge, MA, USA) as a stock solution of 1 mg/ml. The Smoothened inhibitor cyclopamine (CPM; Cat. No. GR-334; Biomol International, LP, Plymouth Meeting, PA, USA) was prepared as a 10-mM stock solution in DMSO. The rabbit polyclonal antibody of SHH capable of neutralizing its biological functions was prepared as a stock solution of 200 μg/ml (Cat. No. sc-9024; Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Preparation of Aβ was based on our previously published paper [24]. Aβ25-35 (Cat. No. A4559; Sigma) was dissolved in autoclaved ddH2O to make a stock solution of 2 mM, dispensed into aliquots, and immediately stored at −80 °C until use. One day prior to experimentation, aliquots of Aβs were incubated at 37 °C for 24 h to allow aggregation. Aβ1-42 (Cat. No. 20276; AnaSpec, Inc., San Jose, CA, USA) was prepared based on a previous report [25]. Briefly, Aβ1-42 was first resuspended in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP; Cat. No. 100528; Sigma) to make a stock solution of 1 mM and then dispensed into aliquots. Thereafter, HFIP was allowed to evaporate in the lamina flow overnight and stored at −80 °C until use. Prior to experimentation, Aβ1-42 was reconstituted in dry DMSO (Cat. No. 1029310500; Merck, Darmstadt, Germany) to make a stock solution of 5 mM, diluted to 100 μM in 1× phosphate-buffered saline (PBS), and then incubated at 4 °C for 24 h to allow aggregation.
Primary Cortical Culture and AD Transgenic Mice
All the procedures for animal care and preparation of fetal rat cortical cultures were performed humanely in accordance with the guidelines described in the “User Manual of Laboratory Animal Center at National Yang-Ming University.” Primary cortical cultures were prepared from embryonic day 18 (E18) fetal Sprague–Dawley (SD) rat brains as previously described [26]. The cells were maintained in Neurobasal medium supplemented with B27 (GIBCO/Life Technologies of Thermo Fisher Scientific Corporation) at 37 °C in a humidified incubator with 5 % CO2 for 7–9 days in vitro (DIV) to allow growth of dendrites before experimentation. The APPswe/PS1dE9 mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA) and were imported by BioLASCO Taiwan Co. (Taipei, Taiwan). The mice were housed in a specific pathogen-free (SPF) environment in the Animal Center of National Yang-Ming University, Taipei, Taiwan.
Western Blotting
Western blotting was performed as previously described [27, 28]. The rabbit antibody against SHH (1:500; Cat. No. 2207; Cell Signaling Technology, Danvers, MA, USA) was diluted in signal enhancer HIKARI solution 1 (Cat. No. NT08044-71R; Nacalai Tesque, Kyoto, Japan). The rabbit antibody against Id1 (1:400; Cat. No. sc-488; Santa Cruz Biotechnology, Inc.) was diluted in 1× blocking reagent (Cat. No. B6429; Sigma). The mouse antibody against β-actin (1:7000; Cat. No. MAB1501; Merck Millipore, Billerica, MA, USA), rabbit antibody against Sox2 (1:1000; Cat. No. GTX101507; GeneTex, Irvine, CA, USA), rabbit antibody against HIF-1α (1:600; Cat. No. NB100-134; Novus Biologicals, Littleton, CO, USA), and mouse antibody against NeuN (1:1000; Cat. No. ABN78; Merck Millipore) were all diluted in blocking buffer (5 % nonfat dry milk in TBST buffer containing 0.05 % Tween 20, 137 mM NaCl, and 20 mM Tris–HCl, pH 7.5). The horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse secondary antibodies were applied in the fresh blocking buffer at 1:5000 to detect the respective primary antibodies. Immunoreactive signal was detected using ECL-Plus Western blotting detection reagents (Merck Millipore). The blots were examined under the luminescence/fluorescence imaging system LAS-4000 (FUJIFILM, Tokyo, Japan). Quantification of protein expression was accomplished by using Multi Gauge analysis software (FUJIFILM). In all the Western blotting experiments, the signal intensity of β-actin served as a control for equal loading of proteins in each lane.
Immunocytochemistry and Immunohistochemistry
For live-labeling SHH, cells were incubated with the SHH antibody (1:100; Cat. No. 2207; Cell Signaling Technology) in blocking buffer at room temperature for 10 min prior to fixation. After washing with PBS, cells were fixed with 4 % paraformaldehyde and then incubated with the fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:150, Cat. No. 02-15-06; Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD, USA) to recognize the rabbit primary antibodies. The samples were observed under a laser scanning confocal microscope Olympus FV1000 (Olympus, Tokyo, Japan) equipped with filter sets to detect FITC (excitation/emission: 494 nm/518 nm) fluorescence signal. For double immunofluorescence staining of SHH and microtubule-associated protein-2 (MAP-2), cells grown on coverslips were hybridized with the rabbit antibody against SHH (1:50; Cat. No. sc-9024; Santa Cruz Biotechnology, Inc.) and the mouse antibody against MAP-2 (1:150; Cat. No. MAB378; CHEMICON International, Inc., Temecula, CA, USA) in blocking buffer at 4 °C overnight. After one wash with PBS, the goat anti-mouse IgG Dylight 594-conjugated secondary antibody (1:100; excitation 596 nm/emission 617 nm; Cat. No. GTX76719; GeneTex, Inc., Irvine, CA, USA) and goat anti-rabbit IgG HylightFluor 488-labeled secondary antibody (1:100; excitation 499 nm/emission 523 nm; Cat. No. 61056-H488; AnaSpec, Inc.) were applied to respectively recognize the MAP-2 and SHH primary antibodies.
For immunohistochemical staining of SHH, the brain slices of 30-μm thickness were rinsed with TBS buffer (137 mM NaCl and 20 mM Tris–Base, pH 7.5), incubated in TBS containing 0.1 % H2O2 to block intrinsic peroxidase activity, and washed twice with TBS buffer containing 0.3 % Triton X-100 (TBST). Sections were then incubated in the blocking solution containing 6 % normal donkey serum and 4 % bovine serum albumin (BSA) in TBST buffer for 1 h. This was followed by incubation with the rabbit antibody against SHH (1:100; Cat. No. 2207; Cell Signaling Technology) at 4 °C overnight and then biotinylated anti-rabbit IgG (1:200) at room temperature for 1 h. The signal was amplified with avidin–horseradish peroxidase complex (ABC Kit; Cat. No. PK-6100; Vector Labs, Burlingame, CA, USA). Specific binding was visualized with 0.5 mg/ml diaminobenzidine (DAB; Cat. No. D5637; Sigma) plus nickel and 0.03 % H2O2 in TBS.
To detect Id1 expression in vivo, the mouse brain slices of 20-μm thickness were washed twice with TBS buffer before bathing in citric acid (pH 8.0) at 80 °C for 10 min, cooled down to room temperature, and subsequently washed twice with TBS. After antigen retrieval, the sections were incubated in 0.2 % H2O2 in TBS followed by two washes in TBST buffer. Sections were then incubated in the blocking solution containing 1 % normal goat serum and 4 % BSA in TBST for 1 h at room temperature. This was followed by incubation with the mouse antibody against Id1 (1:100; Cat. No. sc-133104; Santa Cruz Biotechnology, Inc.) at 4 °C overnight. Afterwards, brain slices were washed three times in TBST and then incubated with goat anti-mouse IgG secondary antibodies conjugated with Dylight 594 (1:100; excitation 596 nm/emission 617 nm; Cat. No. GTX76719; GeneTex, Inc.) at room temperature for 1 h. After three washes with TBST, sections were stained with Hoechst 33258 (2.5 μg/ml; Cat. No. H1398; Molecular Probes/Life Technologies of Thermo Fisher Scientific Corporation) in TBST at room temperature for 15 min. Finally, sections were mounted and sealed for confocal microscopy. The samples were observed under a laser scanning confocal microscope (Zeiss LSM700, Oberkochen, Germany).
Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated by RNeasy mini kit from QIAGEN Inc. (Valencia, CA, USA) before reverse transcription into complementary DNA (cDNA) by the SuperScript III RNase H− Reverse Transcriptase kit (Invitrogen/Life Technologies of Thermo Fisher Scientific Corporation). For synthesis of the first-strand cDNA, 2 μg total RNA and 0.5 μg oligo(dT) primer were preheated at 75 °C for 15 min. The reverse transcription buffer mixture with a final volume of 20.5 μl contained 2 U/μl RNaseOUT recombinant ribonuclease inhibitor, 0.5 mM dNTP, 5 mM DTT, 1× First-Strand Buffer, and 5 U/μl SuperScript III RNase H− Reverse Transcriptase. The mixture was incubated at 42 °C for 90 min to allow reverse transcription. Real-time PCR primers were designed using PRIMER EXPRESS software (Version 2.0, Applied Biosystems/Life Technologies of Thermo Fisher Scientific Corporation). The primers used were as follows: 5′-TGACTGAGGGCTGGGATGA-3′ (forward) and 5′-CCCTG TCAGACGTGGTGATG-3′ (reverse) for SHH, and 5′-AGAGACAGCCGCATCTTCTTG-3′ (forward) and 5′-CGACCTTCACCATCTTGTCTATGA-3′ (reverse) for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Reaction mixture contained 4 μl of 100× diluted cDNA samples, forward and reverse primers (200 nM for each primer), and 1× SYBR Green PCR Master Mix (containing nucleotides, HotStart DNA polymerase, and optimized buffer components; Protech Technology Enterprise Co., Ltd., Taipei, Taiwan). The real-time PCR assays were performed on the Applied Biosystems Prism 7000 sequence detection system by the following program: 50 °C for 2 min, 95 °C for 10 min, and then followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Predicted cycle threshold (CT) values were exported directly into EXCEL worksheets for analysis. At the end of chain reaction, a melting curve was produced by slow denaturation of the PCR end products to validate the specificity of amplification. GAPDH served as an internal reference for normalization of amplification efficiency.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was accomplished by Manga ChIP™ G (Cat. No. 17-611; Merck Millipore) according to the manufacturer’s protocols. Cells were washed with 1× PBS and then crosslinked with 1 % paraformaldehyde at room temperature for 15 min. Following addition of glycine supplied in the kits, cells were placed on ice for 5 min, washed twice in PBS, and then collected in PBS containing the protease inhibitor supplied in the kit. After the nuclei were extracted, chromatin DNA was subjected to sonication through 60 cycles of 2-s pulse on and 1-s pulse off on ice. Equal amounts of nuclear proteins were incubated with 20 μl beads and 16 μg normal mouse IgG (Cat. No. 12-371; Merck Millipore) or 16 μg mouse primary antibody against HIF-1α (Cat. No. NB100-105; Novus Biologicals) at 4 °C overnight. Thereafter, the samples were sequentially washed by low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer for 10 min each at room temperature before addition of elution buffer with 10 μg protease K. Samples were incubated at 62 °C for 2 h with shaking followed by incubation at 95 °C for 10 min. The immunoprecipitated DNA was extracted from the supernatant by the spin column provided in the kits for amplification of the target sequences. The PCR reaction mixtures containing 1 μl of DNA samples, forward and reverse primers (200 nM for each primer), and 1× PCR Master Mix (containing nucleotides, DNA polymerase, and optimized buffer components; Protech Technology Enterprise Co., Ltd.) were subjected to the following thermocycle program: 94 °C for 3 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The primers used were as follows: 5′-GCCACCTTG CCCACTCTCG-3′ (forward) and 5′-ACGGCTGCTTCCCCCTTACTTT-3′ (reverse). The PCR products were then analyzed by gel electrophoresis through agarose gels (Cat. No. UR-AGA001; UniRegion Bio-Tech, Taiwan) in 0.5× TBE buffer (Cat. No. UR-TBE-002-1L; UniRegion Bio-Tech, Taiwan).
siRNA Transfection
Transfection of small interfering RNA (siRNA) into primary cortical neurons was performed as previously described [29]. All Accell SMART pool siRNAs were purchased from Thermo Scientific Dharmacon Inc. (Lafayette, CO, USA). Cells were incubated with 0.1 μM scrambled siRNA (sc siRNA) or target siRNA for 72 h in Accell siRNA Deliver media (Cat. No. B-005000-100; Thermo Scientific Dharmacon Inc.) supplemented with B27 and additional 28 mM d-(+)-glucose (Cat. No. G7021; Sigma; the final glucose concentration was 33 mM). At the completion of siRNA transfection, cells were washed once with Neurobasal medium containing B27 before proceeding to experimental manipulations. The four target sequences for the HIF-1α Accell SMARTpool siRNA mixture were as follows: 5′-GUAUUAUUCAGCACGACUU-3′, 5′-GCGAUAUGGUCAAUGUAUU-3′, 5′-UUACUGAGUUGAUGGGU UA-3′, and 5′-CUCUGAUUUAGCAUGUAGA-3′. The Id1 siRNA mixture (Id: 25261) contained the following four sequences: 5′-GUUUU GUAUUGUAUAUUAC-3′, 5′-CA AACACUUUAGAUAACGU-3′, 5′-GGCUGAGAAUAUUGUUUUA-3′, and 5′-CCUC AGAACCGCAAAGUGA-3′. A non-targeting Accell siRNA pool (Cat. No. D-001950-01-05) was used as the negative control in all siRNA transfection experiments.
Cell Survival Assays
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) reduction assay was performed as previously described [27]. Hoechst staining to assess the extents of cell survival was performed as described in our previous publications [30, 31]. Cells were grown on coverslips in 24-well culture dishes prior to experimental manipulations. For counting of Hoechst-stained surviving cells with normal nuclear morphology, at least three vision fields were randomly selected to obtain the averaged numbers of nuclei per vision field on each coverslip. The “cell survival (%)” was defined as the mean numbers of surviving cells per vision field in the experimental groups divided by those of the control cultures and then multiplied by 100 %.
Statistical Analysis
Results are expressed as mean ± SEM from the sample number (N). For Western blotting, each N represents the results derived from one independent experiment using one different culture. Representative blots are shown in the upper panels; combined results derived from quantification of the signal intensities on these blots are shown in the lower panels. For immunohistochemical staining of the brain slices from wild-type and AD transgenic mouse brains, each N represents one animal. For real-time reverse transcription polymerase chain reaction (RT-PCR) assay, each N represents the data collected from one independent experiment using one different culture. For ChIP assay, each N represents the results derived from one independent experiment using one different culture. Representative images of gel electrophoresis after PCR reaction are shown in the upper panels; combined results derived from quantification of signal intensities on these images are shown in the lower panels. For MTT assays, each N represents the data collected from one independent experiment using one different culture. In each experiment, the assays were performed in triplicates or quadruplicates to obtain the averaged reading values, which were later calculated into cell survival in percentage, for each experimental condition. Combined results from these replicated experiments are shown. For Hoechst staining, each N represents data collected from a single coverslip. In any given experiment, mean values of cell survival in percentage were derived from the combined results of 3–4 replicated experiments, each using one independent culture, comprising a total of 9–14 coverslips.
Two groups were compared by Student’s t test. Multiple groups were first analyzed by one-way analysis of variance (ANOVA) followed by a post hoc Student–Newman–Keuls test. A P value of less than 0.05 was considered significant.
Results
Aβs Induce SHH Expression in Primary Cortical Neurons
We first determined whether Aβ may induce expression of SHH in vitro. Results shown in Fig. 1a indicated that exposure of primary cortical culture to Aβ25-35 at 10–20 μM for 48 h significantly enhanced SHH expression. Quantification of signal intensities on the Western blots revealed that Aβ25-35 at 10 and 20 μM both achieved similar extents of SHH induction (2.41 ± 0.25-fold versus 2.51 ± 0.23-fold; 10 μM Aβ25-35 versus 20 μM Aβ25-35; not significant; Fig. 1a). Therefore, 10 μM Aβ25-35 was selected as the experimental condition to test the time dependency of Aβ25-35 effects on SHH induction. Results indicated that Aβ25-35 at 10 μM markedly induced expression of SHH at 24–48 h (Fig. 1b). In addition to Aβ25-35, we also examined the effects of Aβ1-42, a pathologically more relevant neurotoxic peptide, on SHH induction in primary cortical cultures. Consistent with the previous findings, Aβ1-42 at 5 μM also significantly increased SHH expression at 48 h (Fig. 1c). To observe SHH induction by Aβ in primary cortical neurons, immunofluorescence staining was conducted. Based on direct labeling of the live cells, we demonstrated an increased expression level of SHH in the cortical cells with neuronal morphology after an episode of 48-h exposure to 10 μM Aβ25-35 (Fig. 1d). Confirming this observation, double immunofluorescence confocal microscopy revealed Aβ-induced expression of SHH in MAP-2-positive cells, indicative of neuronal expression of this protein (Fig. 1e). Together, our findings indicate that both Aβ25-35 and Aβ1-42 are capable of SHH induction, at least in the primary cortical neurons in vitro.
Induction of SHH proteins by Aβs in primary cortical neurons. a, b Primary cortical cultures were treated with Aβ25-35 at indicated concentrations for 48 h (a) or 10 μM Aβ25-35 for indicated times (b) before detection of SHH by Western blotting. Mean ± SEM from N = 3 in a and N = 6 in b. *P < 0.05 compared with the corresponding control cultures without Aβ25-35 treatment. c Cells were treated with 5 μM Aβ1-42 for 24 or 48 h before detection of SHH. Mean ± SEM from N = 3. *P < 0.05 compared with the control cultures without Aβ1-42 treatment. d Cells were exposed to 10 μM Aβ25-35 for 48 h before live immunostaining of SHH (green) to reveal its expression. Hoechst 33258 served as counterstaining (blue). Scale bar = 20 μm. Note the heightened SHH expression upon Aβ25-35 exposure. e Cells were exposed to 10 μM Aβ25-35 for 24 h before double immunofluorescence confocal microscopy to detect SHH (green) and MAP-2 (red). Hoechst 33258 served as counterstaining (blue). Scale bar = 10 μm. Note the heightened SHH expression in the MAP-2-positive primary cortical neurons upon Aβ25-35 exposure
Expression of SHH in AD Transgenic Mice
Because primary cortical cultures were prepared from fetal rat brains that may not truly reflect the pathological conditions of AD despite of their exposure to Aβs, we further characterized the expression pattern of SHH in the APPswe/PS1dE9 transgenic mouse brains in vivo. These mice, which overexpress human APP with Swedish double mutation and human presenilin-1 (PS1) with exon 9 deletion in the brains, show Aβ deposits at 6 months of age [32] and commit more errors in the water maze test at 13 months [33]. We therefore examined SHH expression in these AD mice at 6, 9, and 12 months old. Results derived from immunohistochemistry indicated that, as compared with wild-type littermate controls of the same ages, SHH expression levels were substantially higher in the hippocampi and cortices of the 12-month-old APPswe/PS1dE9 mice; in contrast, expression of SHH was similar between APPswe/PS1dE9 mice and wild-type littermate controls at the age of 6 months (Fig. 2a). Furthermore, heightened expression of SHH was also observed in the cortices and hippocampi of the 12-month-old wild-type mouse brains as compared to the younger wild-type animals, suggesting an increased expression level of SHH in these brain regions during normal aging, albeit progressing at a slower pace as compared to AD mice (Fig. 2a). Detailed examination into specific regions of the hippocampus from the 12-month-old AD mouse brain at a higher magnification revealed prominent SHH staining in the dentate gyrus, CA1, CA3, and subgranular zone (Fig. 2b). Thus, SHH level is increased in both hippocampus and cortex, two brain regions most vulnerable to AD, in the aged APPswe/PS1dE9 mouse brains.
SHH expression in AD transgenic mouse brains. a AD transgenic (AD-Tg) mice at the ages of 6, 9, and 12 months were sacrificed and brains removed for immunohistochemistry to assess the expression levels of SHH in the cortex (CTX) and hippocampus (HIP). Wild-type littermates (WT) at the same ages served as controls. Representative micrographs from three different animals at 6 and 9 months old and two animals at 12 months old are shown. Scale bar = 400 μm. b Micrographs of higher magnification showing heightened extents of SHH expression in various brain regions of AD transgenic mice (AD-Tg) at 12 months old as compared to wild-type littermates (WT) at the same age. CA1 cornu ammonis 1, CA3 cornu ammonis 3, DG dentate gyrus, SGZ subgranular zone. Representative micrographs from two different animals are shown. Scare bar = 100 μm. Note significantly higher levels of SHH expression in the cortex, CA1, CA3, and dentate gyrus derived from AD-Tg mouse brains as compared to those from the WT controls
HIF-1 Mediates Aβ-Induced SHH Expression
Prior to characterization of the transcription factor mediating Aβ induction of SHH, we first examined whether Aβ may stimulate SHH expression at the transcriptional level. Our data indicated that Aβ25-35 at 10 μM time-dependently increased expression of SHH mRNA based on real-time RT-PCR; statistical significance was achieved beginning at 24 h (Fig. 3a). This experimental condition was therefore selected for ChIP assay for transcription factor binding to the Shh gene promoter upon Aβ exposure. Among several candidates, it was recently reported that HIF-1 may mediate SHH expression in response to hypoxia [34]; HIF-1 has also been shown to be activated by Aβ in primary cortical culture [3]. We therefore tested the potential roles of HIF-1 in Aβ-dependent SHH expression by ChIP assays using a HIF-1α antibody. Figure 3b depicts a diagram showing the predicted HIF-1 binding sites on the Shh promoter as well as the positions of the primer pair designed to amplify the immunoprecipitated DNA fragment that contains the HIF-1 binding sites. Results shown in Fig. 3c revealed that Aβ exposure led to a heightened extent of HIF-1 binding to the Shh promoter, thus directly demonstrating that Shh is under the transcriptional control of HIF-1.
Involvement of HIF-1 in Aβ-mediated SHH induction. a Primary cortical cultures were exposed to 10 μM Aβ25-35 for indicated times before real-time RT-PCR to determine SHH expression at mRNA levels. Mean ± SEM from N = 5. *P < 0.05 compared with control cultures without Aβ25-35 treatment. b A diagram showing the promoter region of the Shh gene and the positions of PCR primers used in ChIP assay. c Cells were treated with 10 μM Aβ25-35 or empty medium for 24 h before ChIP assay. Immunoprecipitation was conducted using 16 μg HIF-1α antibody (HIF-1α Ab). The same amounts of normal mouse IgG (Mouse IgG) served as the negative control for immunoprecipitation. Genomic DNA without prior immunoprecipitation was also amplified using the same set of primers to serve as the positive control (PC) for PCR reaction. Mean ± SEM from N = 3. *P < 0.05. d Cells were exposed to 10 μM Aβ25-35, 10 μM 2ME2, or both for 48 h before detection of SHH by Western blotting. Mean ± SEM from N = 3. *P < 0.05, #P < 0.05. e Cells were transfected with 0.1 μM HIF-1α siRNA (HIF-1α siRNA) or scrambled siRNA (sc siRNA) for 72 h before detection of HIF-1α expression. Mean ± SEM from N = 4. *P < 0.05. f Cells were transfected with 0.1 μM HIF-1α siRNA (HIF-1α siRNA) or scrambled siRNA (sc siRNA) for 72 h. This was followed by exposure to 10 μM Aβ25-35 for additional 24 h before detection of HIF-1α expression. Mean ± SEM from N = 4. *P < 0.05, #P < 0.05. g The experimental conditions were the same as in f except that SHH expression was assessed 48 h after exposure to 10 μM Aβ25-35. Mean ± SEM from N = 5. *P < 0.05, #P < 0.05. h, i Primary cortical cultures were treated with 400 μM cobalt chloride (CoCl 2 ) for indicated times before detection of HIF-1α (h) and SHH (i). Cells incubated in the empty medium without cobalt chloride for the same periods of time served as corresponding negative controls. Mean ± SEM from N = 4–7 in h and N = 3–6 in i. *P < 0.05 as compared to the corresponding control cultures without cobalt chloride treatment
We then examined the causal relationship between HIF-1α expression and SHH induction mediated by Aβ. It has been shown that 2ME2 can downregulate HIF-1α expression at the post-transcriptional level [35], thereby functioning as a HIF-1 inhibitor. Results derived from Western blotting showed that Aβ-induced SHH expression was partially abolished by 2ME2 (Fig. 3d). To firmly establish the crucial roles of HIF-1 in mediating Aβ-dependent SHH induction, siRNA-mediated gene-specific knockdown strategy was applied. We found that transfection of HIF-1α siRNA led to an approximately 22 % reduction in the expression of HIF-1α proteins in primary cortical cultures (Fig. 3e); upon Aβ25-35 exposure, this siRNA also markedly inhibited the expression of HIF-1α proteins (Fig. 3f). More importantly, HIF-1α siRNA partially, but significantly, attenuated SHH expression by Aβ25-35 (Fig. 3g). Results shown in Fig. 3c–g together firmly establish the critical roles of HIF-1 in mediating Aβ25-35 induction of SHH.
While Aβ-mediated SHH induction requires HIF-1 activity, whether HIF-1 activation alone is sufficient to induce SHH remained to be determined. To address this issue, we treated primary cortical cultures with cobalt chloride, which is known to stabilize HIF-1α [23], and determined the expression level of both HIF-1α and SHH. Results demonstrated that cobalt chloride exposure transiently increased expression of HIF-1α at 8–24 h, which declined at later time points of 40–48 h (Fig. 3h). Notably, the enhanced HIF-1α expression induced by cobalt chloride was followed by a progressively heightened induction of SHH beginning at 16 h and sustained until 48 h (Fig. 3i). These results confirmed that pharmacological induction of HIF-1 by cobalt chloride is sufficient to induce SHH in primary cortical cultures.
Id1 Induction by Aβ25-35 in Differentiated Primary Cortical Cultures and Id1 Expression in AD Transgenic Mice
Id1 is a member of the Id protein family critically involved in the regulation of cellular differentiation, cell cycle progression, senescence, and apoptosis in cancer cells [36]. In the nervous system, Id proteins function as a negative regulator of differentiation that also increase self-renewal and proliferation potential of cortical neural stem cells (NSCs) while inhibiting neuronal differentiation [37]. However, its potential roles in AD have never been explored. Interestingly, Id1 has been shown to upregulate HIF-1 activity via stabilization of HIF-1α proteins [38, 39]. We therefore tested the possibility that Id1 may act upstream of HIF-1 to induce SHH expression associated with Aβ exposure. Because our primary cortical cultures consist of the proliferating NSCs at DIV 1 and mostly the differentiated cortical neurons at DIV 9, we first examined the expression profile of Id1 in the cortical cultures during the process of differentiation in vitro. Using NeuN as a marker protein for mature neurons, we observed progressively declined expression of Id1 from DIV 1 to 9; this was accompanied by an increased cellular extent of NeuN proteins (Fig. 4a), indicating downregulation of Id1 during differentiation of our primary cortical neurons in vitro. In addition to Id1, we also examined the expression pattern of HIF-1α during in vitro differentiation. We found that, unlike the continued decrease in the expression level of Id1, a transient increase in the cellular contents of HIF-1α was noted from DIV 4 to 6, achieving maximal extent at DIV 5 and declining thereafter at DIV 9 (Fig. 4b). These results indicated that, at least in the fully differentiated cortical neurons, downregulation of Id1 during differentiation is accompanied by an attenuated expression level of endogenous HIF-1α from DIV 5 to 9.
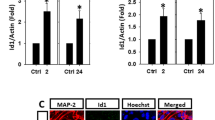
Aβ induction of Id1 and expression of Id1 in AD transgenic mouse brains. a, b Proteins were extracted from the primary cortical cultures during DIV 1 to 9 before detection of NeuN and Id1 in a and HIF-1α in b. Mean ± SEM from N = 3 in a and N = 4 in b. *P < 0.05 as compared to DIV 1. c, d Primary cortical cultures were treated with Aβ25-35 at indicated concentrations for 24 h (c) or with 10 μM Aβ25-35 for indicated times (d) before detection of Id1. Mean ± SEM from N = 3 in both c and d. *P < 0.05 as compared to the corresponding controls without Aβ25-35 treatment. e AD transgenic (AD-Tg) mice and wild-type (WT) littermates at the age of 12 months were sacrificed and their brains removed for immunohistochemistry to assess the expression levels of Id1 (red) in the hippocampus (HIP). Hoechst 33258 served as counterstaining (blue). The white arrows indicate the Id1-positive cells in the dentate gyrus (DG) of the hippocampus derived from the 12-month-old AD mouse brains. Scale bar = 200, 50, and 10 μm in the upper, middle, and lower panels, respectively. Representative images from three WT and three AD-Tg mice are shown
Once the expression profiles of Id1 and HIF-1α were established during differentiation of primary cortical neurons, we then examined the Aβ effects on the expression of Id1 protein, which has never been reported before. When the primary cortical cultures at DIV 9 were exposed to 1–20 μM Aβ25-35 for 24 h, we detected a dose-dependent increase of Id1 expression in these cells (Fig. 4c). At 10 μM, Aβ25-35 also enhanced Id1 expression in the primary cortical cultures at DIV 9 after an episode of 24- or 48-h exposure (Fig. 4d). In the AD transgenic mouse brains, heightened expression of Id1 was detected, as compared to the age-matched wild-type littermate controls, in the dentate gyrus of the hippocampus derived from the 12-month-old APPswe/PS1dE9 mice (Fig. 4e); similar results were observed in the cortices of 12-month-old AD mice (data not shown). These results were consistent with our observation that SHH was induced in the same brain regions of the aged AD mice (Fig. 2b). Together, these findings indicate an increased expression level of Id1 in the differentiated primary cortical cultures upon Aβ exposure and in the aged AD transgenic mouse brains, similar to those observed for SHH.
Id1 Acts Upstream of HIF-1 in Mediating SHH Induction by Aβ
To demonstrate the causative relationship between induction of Id1 and SHH by Aβ, we first tested the knockdown efficacy of Id1 siRNA. We found that transfection of Id1 siRNA resulted in an approximately 35 % reduction in the expression level of Id1 proteins (Fig. 5a). Upon Aβ25-35 exposure, this siRNA suppressed Id1 induction by Aβ25-35 down to the basal level (Fig. 5b). More importantly, Id1 siRNA also completely inhibited the induction of SHH mediated by Aβ25-35 (Fig. 5c). We then determined whether Aβ-induced Id1 may act upstream of HIF-1 in primary rat cortical cultures. Western blots reveal that Id1 siRNA, but not sc siRNA, markedly suppressed HIF-1α induction by Aβ25-35; quantitative analyses of signal intensities on the immunoblots confirmed this finding (Fig. 5d). These results thus established the causal relationship between Aβ-induced Id1 and HIF-1α. However, it has been shown that HIF-1 may mediate the expression of Id2, another Id family member, in hypoxic neuroblastoma cells [40]. To explore the possibility of HIF-1-dependent Id1 induction in the primary cortical cultures exposed to Aβ, we also tested whether HIF-1α siRNA was capable of blocking Id1 expression induced by Aβ. Results shown in Fig. 5e appeared to negate this possibility because HIF-1α siRNA failed to suppress Aβ25-35-dependent Id1 induction (2.33 ± 0.40-fold versus 2.75 ± 0.30-fold; Aβ25-35 + sc siRNA versus Aβ25-35 + HIF siRNA; not significant). This finding ruled out the possible positive feedforward regulation between Aβ-induced Id1 expression and HIF-1 activation, thus firmly establishing the notion that Aβ-induced Id1 acts upstream of HIF-1, but not vice versa. In addition to HIF-1, another transcription factor known to regulate SHH expression is Sox2 [41]. However, Sox2 is unlikely to be involved in Aβ-dependent SHH induction. As shown in Fig. 5f, Aβ25-35 failed to enhance Sox2 expression (0.84 ± 0.33-fold in Aβ25-35 + sc siRNA group; not significant as compared to the sc siRNA group); further, Id1 siRNA did not show any significant effects in suppressing Sox2 expression (0.84 ± 0.33-fold versus 0.98 ± 0.34-fold; Aβ25-35 + sc siRNA versus Aβ25-35 + Id1 siRNA; not significant). Potential roles of Sox2 were therefore not further investigated.
Involvement of Id1 in Aβ-mediated SHH induction. a Primary cortical cultures were transfected with 0.1 μM Id1 siRNA (Id1 siRNA) or scrambled siRNA (sc siRNA) for 72 h before detection of Id1 expression. Mean ± SEM from N = 3. *P < 0.05. b–d Cells were transfected with 0.1 μM Id1 siRNA (Id1 siRNA) or scrambled siRNA (sc siRNA) for 72 h. This was followed by exposure to 10 μM Aβ25-35 for additional 24 h before detection of Id1 (b), SHH (c), or HIF-1α (d). Mean ± SEM from N = 3. *P < 0.05, #P < 0.05. e Cells were transfected with 0.1 μM HIF-1α siRNA (HIFnn1α siRNA) or scrambled siRNA (sc siRNA) for 72 h. This was followed by exposure to 10 μM Aβ25-35 for additional 24 h before detection of Id1. Mean ± SEM from N = 3. *P < 0.05 compared to the cells transfected with sc siRNA but without exposure to Aβ25-35. The “ns” denotes “not significant.” f Cells were transfected with 0.1 μM Id1 siRNA (Id1 siRNA) or scrambled siRNA (sc siRNA) for 72 h. This was followed by exposure to 10 μM Aβ25-35 for additional 24 h before detection of Sox2. Mean ± SEM from N = 5. The “ns” denotes “not significant.” Note that Aβ25-35 failed to induce Sox2 and Id1 siRNA has no significant effects on Sox2 expression. g Primary cortical cultures were treated with or without 4 μg/ml recombinant Id1 protein tagged with 11 arginines (Id1-Tag) for 2 or 8 h before Western blotting to detect the expression level of Id1. Note the strong signal of Id1-Tag (molecular weight 18.5 kD) in the cells exposed to the recombinant Id1 proteins; endogenous Id1 proteins (molecular weight 14 kD) were also detectable, albeit to a much lesser extent, in all the cortical cultures. h, i Cells were treated with 4 μg/ml Id1-Tag for 8 h before Western blotting to detect the expression levels of HIF-1α (h) and SHH (i). Mean ± SEM from N = 4 in h and N = 3 in i. *P < 0.05
Because pharmacological stabilization of HIF-1α by cobalt chloride (Fig. 3h) was sufficient for SHH induction (Fig. 3i), we also tested whether exogenous recombinant Id1 proteins may exert similar effects. To achieve this, a commercially available human Id1 protein tagged with a peptide containing 11 arginines (Id1-Tag), thus rendering it cell-permeable, was used in the present study. We first confirmed the permeability of these Id1-Tag proteins through the plasma membrane of cells. Western blots demonstrated that, after incubation for 2 or 8 h with the recombinant proteins at 4 μg/ml, significantly higher amounts of Id1-Tag with a predicted molecular weight of 18–19 kD were detectable in the protein lysates derived from primary cortical cultures; in addition, a much weaker signal of endogenous Id1 proteins with a molecular weight of 14 kD was also observed in the cultures with or without Id1-Tag treatment (Fig. 5g). In accordance with the notion that Id1 acts upstream of HIF-1 with induction of SHH, Id1-Tag at 4 μg/ml markedly enhanced the expression levels of both HIF-1α (Fig. 5h) and SHH (Fig. 5i) in primary cortical cultures. These findings indicate that Id1 protein alone is sufficient to induce expression of both HIF-1α and SHH, at least in this experimental system. Taken together, our results support the signaling cascade of “Aβ → Id1 → HIF-1 → SHH” in primary cortical cultures derived from fetal rat brains.
SHH Induced by Aβ Contributes to Aβ Cytotoxicity
We have delineated the underlying mechanisms mediating Aβ induction of SHH. However, the potential biological functions of the Aβ-induced SHH remained obscure. The steroidal alkaloid cyclopamine is the most commonly used SHH inhibitor that abolishes its Smoothened-dependent activity [42]. We therefore first examined the effects of cyclopamine on the cytotoxicity of Aβ25-35 in primary rat cortical cultures. Results derived from MTT reduction assay (Fig. 6a) and counting of Hoechst-stained surviving cells with normal nuclear morphology (Fig. 6b) both revealed partial, but statistically significant, protective effects of cyclopamine against Aβ25-35 toxicity. In addition to a Smoothened-dependent action, SHH may also trigger Patched-dependent signaling pathways independent of Smoothened that affect apoptosis [43]. Therefore, we applied the neutralizing antibody against SHH to knock down its biological functions. Similar to cyclopamine, results derived from the MTT assay revealed that suppression of SHH activity exerted a modest (percent cell survival: 49 % in Aβ25-35 group versus 61 % in Aβ25-35 + CPM group), but statistically significant, protective effect (Fig. 6c). Together, results shown in Fig. 6 suggest that SHH induced by Aβs appears to mediate, at least partially, its cytotoxic effects.
Effects of SHH induced by Aβ25-35 on cell survival. a, b Primary cortical cultures were treated with 10 μM Aβ25-35 with or without 20 μM CPM for 48 h before determination of cell survival by MTT assay (a) or Hoechst staining (b). Mean ± SEM from N = 4 in a and N = 9 in b. c Cells were treated with 10 μM Aβ25-35 with 1 μg/ml SHH neutralizing antibody or 1 μg/ml normal rabbit IgG for 48 h before determination of cell survival by Hoechst staining. Mean ± SEM from N = 4. *P < 0.05, #P < 0.05
Discussion
In the current study, we have demonstrated heightened SHH expression in aged AD transgenic mice in vivo and further established a signaling cascade of Aβ → Id1 → HIF-1 → SHH in primary cortical neurons in vitro. While the potential pathophysiological roles of SHH in AD remain to be fully defined, it is intriguing to note that a morphogen like SHH critical for embryonic development can be induced during progression of AD, a chronic neurodegenerative disorder often afflicting aging populations. Recently, increased SHH signaling was reported in the hippocampi derived from aged AD transgenic mice overexpressing human APP with the Swedish double mutations (APP23 mice) and human AD patients [44]. The authors further demonstrated that, in hippocampal progenitor cells, Aβ1-42 upregulates SHH while decreasing Patched1–Gli1 signaling that together lead to impaired neurogenesis accompanied by heightened astrogenesis in AD brains [44]. Despite the elevated SHH levels in the hippocampus derived from aged APP23 mice and in the Aβ-treated hippocampal neural stem cells/glial precursor cells (NSCs/GPCs), direct impacts of the increased SHH proteins on the differentiated hippocampal neurons were not examined in that report [44]. In our in vitro cultures mainly composed of post-mitotic cortical neurons from DIV 7 to 9, we found that both Aβ25-35 and Aβ1-42 were capable of SHH induction in differentiated neurons based on immunocytochemistry (Fig. 1d, e). Intriguingly, inhibition of Aβ-induced SHH signaling, either by pharmacological inhibition using cyclopamine or by the SHH-N neutralizing antibody, abolished Aβ neurotoxicity, albeit to a minor extent (Fig. 6). This finding was initially unexpected given that SHH has previously been shown to protect primary cortical neurons against mitochondrial inhibition [45] and confer resistance to cardiomyocytes against oxidative stress induced by hydrogen peroxide [46]. At present, we are unable to provide a plausible explanation to this seemingly contradictory result. One possibility is SHH-dependent stimulation of cell cycle re-entry in fully differentiated post-mitotic neurons, thereby causing neuronal apoptosis. Additional studies are required to confirm this contention.
SHH has been shown to be expressed in embryonic brain cortices [47], consistent with our finding that basal level of SHH was detected in primary cortical neurons derived from E-18 fetal rat brains (Fig. 1a, b). Initially, we speculated that Aβ-induced SHH expression in the neurons derived from fetal rat cortices may represent an artifact irrelevant to AD. However, results derived from immunohistochemistry using APPswe/PS1dE9 mice of various ages revealed that SHH was indeed expressed at higher levels in hippocampus and cortex, two brain regions vulnerable to AD, derived from the 12-month-old AD mouse brains as compared to those from the wild-type littermate controls at the same ages (Fig. 2). Actually, heightened expression of SHH was observed in the cortices and hippocampi of the 12-month-old wild-type mouse brains as compared to the younger wild-type animals, suggesting an increased expression level of SHH during normal aging, albeit progressing at a slower pace as compared to AD mice (Fig. 2a). Interestingly, in the hippocampi of APP23 mice, elevated SHH levels were observed at 3, 12, and 24 months old, in which the most prominent elevation in SHH expression was also detected at the age of 12 months [44]. Together, these previous results and our own findings thus confirm that Aβ may induce SHH expression in vitro and increased SHH level can be observed in vivo in two distinct lines of AD transgenic mice, namely APPswe/PS1dE9 and APP23.
After confirmation of Aβ-induced SHH in vitro and in vivo, we sought to identify the transcription factor responsible for this effect. Thus far, at least three transcription factors have been reported to activate SHH, including HIF-1 [34, 48], Sox2 [41], and nuclear factor-κB (NF-κB) [49, 50]. In the current study, we provided evidence supporting the critical roles of HIF-1 in mediating Aβ-dependent SHH induction; indeed, cobalt chloride-dependent activation of HIF-1α is sufficient to induce SHH (Fig. 3h, i). Although Sox2 has been shown to induce SHH expression in NSCs and mediate neurogenesis in the hippocampus [41], at least in this experimental paradigm, we did not observe Aβ25-35-dependent Sox2 induction; moreover, Id1 siRNA failed to affect endogenous Sox2 expression (Fig. 5f). Therefore, potential involvement of Sox2 in Aβ induction of SHH was not further pursued, which requires siRNA-mediated knockdown of Sox2. Despite this, we cannot rule out the possibility of other transcription factors being responsible for the observed Aβ effects. For example, Aβ may also induce NF-κB in cultured cerebellar neurons [4]. Possible roles of this transcription factor in SHH induction require further investigation.
Previously, Id1 has been shown to enhance the stability and activity of HIF-1α in human endothelial and breast cancer cells, which involves reduced association of the HIF-1α protein with von Hippel–Lindau protein [38]. In human umbilical vascular endothelial cells, overexpression of Id1 also stabilizes HIF-1α protein [39]. These reports appear to place Id1 upstream of HIF-1 to regulate the expression of its target genes, consistent with our observation that Id1 acts upstream of HIF-1 in the regulation of SHH expression induced by Aβ (Fig. 5). Conversely, Id2 can be transcriptionally upregulated by HIF-1 in hypoxic neuroblastoma cells [40]. In our experimental paradigm, we have ruled out the possibility of Aβ-induced HIF-1 regulating Id1 because siRNA of HIF-1α failed to attenuate Aβ25-35-induced Id1 expression (Fig. 5e). Another interesting finding was that the expression profiles of endogenous Id1 and HIF-1α during in vitro differentiation of primary cortical cultures in the absence of Aβ appeared different. Unlike the progressively declined expression of Id1 from DIV 1 to 9 (Fig. 4a), a transient increase in the expression level of HIF-1α was observed from DIV 4 to 6, reaching the maximal extent at DIV 5 (Fig. 4b). Although the rationale behind this distinctive expression pattern between endogenous Id1 and HIF-1α during in vitro differentiation remains unknown, Id1 is certainly not the only modulator controlling the stability of HIF-1α, which is regulated by multiple factors including oxygen tension [18] and cellular redox status [51]. Therefore, despite our observations of Aβ-induced Id1 acting upstream of HIF-1α in the more differentiated neurons, potential impacts of Id1 on the regulation of endogenous HIF-1α in less mature cortical cells in the absence of Aβ remain to be confirmed.
Overall, in this study, we demonstrated a novel mechanism by which Aβ may induce SHH expression via Id1-dependent HIF-1 activation. Our results firmly establish a novel signal cascade of Aβ → Id1 → HIF-1 → SHH in primary cortical neurons; furthermore, the SHH induced by Aβs in part contributes to Aβ toxicity. Detailed molecular mechanisms by which SHH may affect Aβ toxicity in differentiated neurons are currently under investigation. Knowledge in this regard may contribute to the future development of therapeutic regimen for AD, a devastating neurodegenerative disorder.
References
Palop JJ, Mucke L (2010) Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci 13(7):812–818
O’Brien RJ, Wong PC (2011) Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci 34:185–204
Soucek T, Cumming R, Dargusch R, Maher P, Schubert D (2003) The regulation of glucose metabolism by HIF-1 mediates a neuroprotective response to amyloid beta peptide. Neuron 39(1):43–56
Kawamoto EM, Lepsch LB, Boaventura MF, Munhoz CD, Lima LS, Yshii LM, Avellar MC, Curi R, Mattson MP, Scavone C (2008) Amyloid beta-peptide activates nuclear factor-kappaB through an N-methyl-D-aspartate signaling pathway in cultured cerebellar cells. J Neurosci Res 86(4):845–860
Akhter R, Sanphui P, Biswas SC (2014) The essential role of p53 upregulated modulator of apoptosis and its regulation by FoxO3a transcription factor in beta-amyloid induced neuron death. J Biol Chem 289(15):10812–10822
Chen YS, Chen SD, Wu CL, Huang SS, Yang DI (2014) Induction of sestrin2 as an endogenous protective mechanism against amyloid beta-peptide neurotoxicity in primary cortical culture. Exp Neurol 253:63–71
Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP (1993) Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75(7):1417–1430
Fuccillo M, Joyner AL, Fishell G (2006) Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 7(10):772–783
Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A (1996) The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384(6605):129–134
Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE (1996) The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell 86(2):221–232
Riobo NA, Manning DR (2007) Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem J 403(3):369–379
Kenney AM, Cole MD, Rowitch DH (2003) Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130(1):15–28
Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, Esterbauer H, Hauser-Kronberger C, Frischauf AM, Aberger F (2004) Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res 64(21):7724–7731
Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, Van Lohuizen M, Marino S (2004) Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428(6980):337–341
Zebedee Z, Hara E (2001) Id proteins in cell cycle control and cellular senescence. Oncogene 20(58):8317–8325
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61(1):49–59
Yokota Y, Mori S (2002) Role of Id family proteins in growth control. J Cell Physiol 190(1):21–28
Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92(12):5510–5514
Wang GL, Semenza GL (1995) Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270(3):1230–1237
Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem 271(51):32529–32537
Semenza GL (2011) Oxygen sensing, homeostasis, and disease. N Engl J Med 365(6):537–547
Semenza GL (2013) HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 123(9):3664–3671
Wang GL, Semenza GL (1993) General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A 90(9):4304–4308
Chen SD, Wu CL, Lin TK, Chuang YC, Yang DI (2012) Renin inhibitor aliskiren exerts neuroprotection against amyloid beta-peptide toxicity in rat cortical neurons. Neurochem Int 61(3):369–377
Stine WB Jr, Dahlgren KN, Krafft GA, LaDu MJ (2003) In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem 278(13):11612–11622
Ju TC, Yang YT, Yang DI (2004) Protective effects of S-nitrosoglutathione against neurotoxicity of 3-nitropropionic acid in rat. Neurosci Lett 362(3):226–231
Ju TC, Chen SD, Liu CC, Yang DI (2005) Protective effects of S-nitrosoglutathione against amyloid beta-peptide neurotoxicity. Free Radic Biol Med 38(7):938–949
Wu CL, Chen SD, Yin JH, Hwang CS, Yang DI (2010) Erythropoietin and sonic hedgehog mediate the neuroprotective effects of brain-derived neurotrophic factor against mitochondrial inhibition. Neurobiol Dis 40(1):146–154
Wu CL, Yin JH, Hwang CS, Chen SD, Yang DY, Yang DI (2012) c-Jun-dependent sulfiredoxin induction mediates BDNF protection against mitochondrial inhibition in rat cortical neurons. Neurobiol Dis 46(2):450–462
Wu CL, Hwang CS, Yang DI (2009) Protective effects of brain-derived neurotrophic factor against neurotoxicity of 3-nitropropionic acid in rat cortical neurons. Neurotoxicology 30(4):718–726
Wu MF, Yin JH, Hwang CS, Tang CM, Yang DI (2014) NAD attenuates oxidative DNA damages induced by amyloid beta-peptide in primary rat cortical neurons. Free Radic Res 48(7):794–805
Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR (2004) Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 13(2):159–170
Volianskis A, Kostner R, Molgaard M, Hass S, Jensen MS (2010) Episodic memory deficits are not related to altered glutamatergic synaptic transmission and plasticity in the CA1 hippocampus of the APPswe/PS1deltaE9-deleted transgenic mice model of β-amyloidosis. Neurobiol Aging 31(7):1173–1187
Bijlsma MF, Groot AP, Oduro JP, Franken RJ, Schoenmakers SH, Peppelenbosch MP, Spek CA (2009) Hypoxia induces a hedgehog response mediated by HIF-1alpha. J Cell Mol Med 13(8B):2053–2060
Mabjeesh NJ, Escuin D, LaVallee TM, Pribluda VS, Swartz GM, Johnson MS, Willard MT, Zhong H, Simons JW, Giannakakou P (2003) 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 3(4):363–375
Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5(8):603–614
Jung S, Park RH, Kim S, Jeon YJ, Ham DS, Jung MY, Kim SS, Lee YD, Park CH, Suh-Kim H (2010) Id proteins facilitate self-renewal and proliferation of neural stem cells. Stem Cells Dev 19(6):831–841
Kim HJ, Chung H, Yoo YG, Kim H, Lee JY, Lee MO, Kong G (2007) Inhibitor of DNA binding 1 activates vascular endothelial growth factor through enhancing the stability and activity of hypoxia-inducible factor-1alpha. Mol Cancer Res 5(4):321–329
Lee TK, Poon RT, Yuen AP, Ling MT, Wang XH, Wong YC, Guan XY, Man K, Tang ZY, Fan ST (2006) Regulation of angiogenesis by Id-1 through hypoxia-inducible factor-1alpha-mediated vascular endothelial growth factor up-regulation in hepatocellular carcinoma. Clin Cancer Res 12(23):6910–6919
Lofstedt T, Jogi A, Sigvardsson M, Gradin K, Poellinger L, Pahlman S, Axelson H (2004) Induction of ID2 expression by hypoxia-inducible factor-1: a role in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem 279(38):39223–39231
Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK (2009) Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci 12(10):1248–1256
Chen JK, Taipale J, Cooper MK, Beachy PA (2002) Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 16(21):2743–2748
Mille F, Thibert C, Fombonne J, Rama N, Guix C, Hayashi H, Corset V, Reed JC, Mehlen P (2009) The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat Cell Biol 11(6):739–746
He P, Staufenbiel M, Li R, Shen Y (2014) Deficiency of Patched 1-induced Gli1 signal transduction results in astrogenesis in Swedish mutated APP transgenic mice. Hum Mol Genet 23(24):6512–6527
Wu CL, Chen SD, Hwang CS, Yang DI (2009) Sonic hedgehog mediates BDNF-induced neuroprotection against mitochondrial inhibitor 3-nitropropionic acid. Biochem Biophys Res Commun 385(1):112–117
Kusano KF, Pola R, Murayama T, Curry C, Kawamoto A, Iwakura A, Shintani S, Ii M, Asai J, Tkebuchava T, Thorne T, Takenaka H, Aikawa R, Goukassian D, von Samson P, Hamada H, Yoon YS, Silver M, Eaton E, Ma H, Heyd L, Kearney M, Munger W, Porter JA, Kishore R, Losordo DW (2005) Sonic hedgehog myocardial gene therapy: tissue repair through transient reconstitution of embryonic signaling. Nat Med 11(11):1197–1204
Ruiz i Altaba A, Palma V, Dahmane N (2002) Hedgehog-Gli signalling and the growth of the brain. Nat Rev Neurosci 3(1):24–33
Wang G, Zhang Z, Xu Z, Yin H, Bai L, Ma Z, Decoster MA, Qian G, Wu G (2010) Activation of the sonic hedgehog signaling controls human pulmonary arterial smooth muscle cell proliferation in response to hypoxia. Biochim Biophys Acta 1803(12):1359–1367
Kasperczyk H, Baumann B, Debatin KM, Fulda S (2009) Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. FASEB J 23(1):21–33
Nakashima H, Nakamura M, Yamaguchi H, Yamanaka N, Akiyoshi T, Koga K, Yamaguchi K, Tsuneyoshi M, Tanaka M, Katano M (2006) Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res 66(14):7041–7049
Guo S, Bragina O, Xu Y, Cao Z, Chen H, Zhou B, Morgan M, Lin Y, Jiang BH, Liu KJ, Shi H (2008) Glucose up-regulates HIF-1 alpha expression in primary cortical neurons in response to hypoxia through maintaining cellular redox status. J Neurochem 105(5):1849–1860
Acknowledgments
This study was supported by the National Science Council/Ministry of Science and Technology in Taiwan (NSC 101-2314-B-010-042MY2 and MOST 103-2314-B-010-013MY3 to Ding-I Yang; NSC 102-2314-B-038-024 to Liang-Yo Yang), Ministry of Education in Taiwan Aim for the Top University Plan (103AC-B5 to Ding-I Yang), Department of Health in Taipei City Government (10201-62-067 and 10301-62-003 to Ding-I Yang and Chi-Shin Hwang), and Cheng Hsin General Hospital (102F218C12 and 103F003C16 to Ding-I Yang and Jiu-Haw Yin).
Conflict of Interest
All authors declare no actual or potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yu-Hsing Hung and Shih-Hsin Chang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hung, YH., Chang, SH., Huang, CT. et al. Inhibitor of Differentiation-1 and Hypoxia-Inducible Factor-1 Mediate Sonic Hedgehog Induction by Amyloid Beta-Peptide in Rat Cortical Neurons. Mol Neurobiol 53, 793–809 (2016). https://doi.org/10.1007/s12035-014-9046-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-9046-5