Abstract
Glioma is the most prevalent primary brain tumors in adults. In addition to the high incidence and mortality rate, the 5-year survival rate of glioma is also extremely low. MicroRNAs (miRNAs), as a class of small non-coding RNAs, may play an important role in carcinogenesis. It was also proposed that miRNAs might also be associated with glioma diagnosis and prognosis. In this study, we aimed at investigating the predictive and prognostic values of miR-125b, miR-221, and miR-222 in glioma and, hopefully, to provide some evidence for novel therapy of glioma. Tissue specimens were obtained from tumor tissue and adjacent non-tumor tissue. RNA was extracted and qRT-PCR was performed with U6 being the internal control. Receiver-operating characteristic (ROC) curves were constructed, and the area under the ROC curves (AUC) was calculated to evaluate the significance of candidate miRNAs in distinguishing glioma tumor tissues and adjacent normal tissues. Survival curves of Kaplan-Meier method were constructed for both high expression group and low expression group, and the difference between curves was evaluated by log-rank test. All the statistical analyses were performed using Stata version 12.0 software, and graphs were generated by GraphPad Prism 5.0. The significance of miR-125b, miR-221, and miR-222 expression level in distinguishing glioma tumor from adjacent non-tumor tissues was further validated. Combination of miR-125b, miR-221, and miR-22 was significantly superior compared to the clinical standard of using these miRNAs alone. A clear demarcation was shown by survival analysis between patients with high miR-125b, miR-221, and miR-222 expression and patients with poor prognosis. Similarly, panel of these miRNAs could play a better prognostic role in glioma. In this study, we confirmed the significance of miR-125b, miR-221, and miR-222 in distinguishing glioma tumor from adjacent non-tumor tissues. Higher expressions of miR-125b and miR-222 have also been proved to be associated with glioma. Furthermore, glioma patients with higher miR-125b, miR-221, and miR-222 expression were manifested to have poorer prognostic status, which might be attributed to their attenuated sensitivity to chemotherapy and radiotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the most prevalent primary brain tumors in adults. They arise from glial or precursor cells, including astrocytoma, glioblastoma, oligodendroglioma, ependymoma, mixed glioma, and malignant glioma, not otherwise specified (NOS) and a few more rare histologies [1]. The annual incidence rate of glioma was 5/100,000, and approximately 18,000 new glioma cases and 13,000 deaths occur every year in the USA [2]. In addition to the high incidence and mortality rate, the 5-year survival rate of glioma is reported to be less than 3 %, ranking only second to pancreatic cancer and lung cancer [3]. Thus, novel biomarker for glioma diagnosis and prognosis is of vital importance.
MicroRNAs (miRNAs), as a class of small non-coding RNAs, may play an important role in cell proliferation and apoptosis and, further, in carcinogenesis. They are involved in the regulation of oncogene and tumor-suppressive gene expression [4], which facilitates them as a potential biomarker for cancer. According to recent studies, abnormal expression of miRNA was associated with various cancers, including lung cancer, gastric cancer, and breast cancer. MiRNAs were also confirmed to be able to serve as a biomarker in distinguishing cancer tissues from normal tissues [5]. It was also proposed that miRNAs might also be associated with cancer prognosis.
MiR-125b, miR-221, and miR-222 are three candidate miRNAs that have been confirmed to play an important role in glioma carcinogenesis [6–10]. In this study, we aimed at investigating the predictive and prognostic values of miR-125b, miR-221, and miR-222 in glioma patients and, hopefully, to provide some evidence for novel therapy of glioma.
Material and Methods
Subjects and Specimens
Forty-five glioma patients were enrolled in the present study. All enrolled patients were diagnosed and treated at Shengjing Hospital of China Medical University from 2011 to 2014. The diagnosis of glioma was confirmed by histological examination or magnetic resonance imaging. Clinicopathological features of patients, including gender, age, TNM stage, metastatic status, tumor size, T classification, and differentiation degree, was documented for further analysis. The glioma stages were determined according to the World Health Organization (WHO) classification and TNM classification system [11]. Patients with immune disease or previous history of cancer were excluded from the study. Tissue specimens were obtained through needle core biopsy or surgery specimens (1-mm thick) from both tumor tissue and adjacent non-tumor tissue. To investigate the prognostic value of target miRNAs in glioma, all selected glioma patients were followed up until death or on August 1, 2014.
All participants before the study approved an informed consent. The research protocol was approved by the Ethics Committee of Shengjing Hospital of China Medical University. The study was conducted in accordance with the Declaration of Helsinki and the REMARK guidelines for biomarker studies.
RNA Extraction and Quantitative Reverse-Transcriptase PCR
Total RNA was extracted from all specimens using miRNeasy Mini Kit (Qiagen, Mississauga, ON, Canada) according to manufacturer’s protocol. The concentration of extracted RNAs was determined by NanoDrop™ 1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE) with the absorbance at 260/280 (RNA/DNA) and 260/230 (RNA/Protein). All samples were stored at −80 °C for further requirement.
For reverse transcription reaction, Taqman® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was applied. A 5-μl mixture comprised of 0.5 μl of different primer, 0.05 μl of 100 mM dNTPs, 0.5 μl of 10× reverse transcription buffer, 0.063 μl of 20 U/μl RNase inhibitor, and 0.33 μl of 50 U/μl multiscribe reverse transcriptase, RNA sample, and RNase-free water was used for cDNA synthesis. The volume of RNA samples was determined by their concentration and normalized for the reaction. For miR-125b, the reaction solutions were incubated at 30 °C for 10 min, followed by 30-min incubation at 50 °C, 95 °C for 5 min, and held at 4 °C. For miR-221 and miR-222, the solutions were incubated at 37 °C for 30 min, 95 °C for 10 min, and held at 4 °C. All synthesized cDNA samples were diluted with RNase-free water for quantitative PCR reaction.
The qPCR reaction solution used in the present study contained 2 μl of cDNA solution, 5 μl of TaqMan® 2X Perfect Master Mix, 0.25 μl of specific primers for different miRNAs, and 2.75 μl of RNase-free water for a total volume of 10 μl. Bio-Rad IQ5 (Bio-Rad Laboratories Inc.) thermocycler was applied for qPCR reaction. U6 snRNA (Ambion, AM30303) was used as an internal control; specific primers for it were also added to the reaction solution. The procedure of PCR amplification was performed as follows: for miR-125b, an initial denaturation at 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min; the reaction was terminated by incubation at 95 °C for 15 s and held at 4 °C; for miR-221 and miR-222, the reaction was initialized by incubating at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s, and held at 4 °C. The expression of miRNAs was measured by the cycle threshold (Ct) values acquired from Bio-Rad iQ5 2.1 Standard Edition Optical System Software.
Follow-Up
Validated contact information was provided by selected patients, including phone numbers and e-mail address. Follow-up check was performed by direct phone call in most case, with e-mail being an alternative for unforeseen circumstances. The check was scheduled every month. The efficacy of treatment was assessed at the same time.
Data Analysis
To start with, paired t tests were applied to assess the difference in miR-125b, miR-221, and miR-222 expression between glioma tissue and adjacent normal controls. All samples were adjusted by gender and age. Receiver-operating characteristic (ROC) curves were constructed, and the area under the ROC curves (AUC) was calculated to evaluate the significance of candidate miRNAs in distinguishing glioma tumor tissues and adjacent normal tissues. Youden index was used to select the optimal cutoff values. Association of biomarker levels with glioma was evaluated using logistic regression analyses. Areas under the ROC curves were assessed for miR-125b, miR-221, and miR-222 and combinations of these biomarkers. For survival analysis, Cox regression analysis was performed. Survival curves of Kaplan-Meier method were constructed for both high expression group and low expression group of miR-125b, miR-221, and miR-222. The difference between curves was evaluated by log-rank test. The survival time was defined as the time between the first surgery until death or the end of the follow-up. All the statistical analyses were performed using Stata Version 12.0 software (Stata Corp, College Station, TX, USA), and graphs were generated by GraphPad Prism 5.0 (GraphPad Software Inc., CA, USA).
Result
Expression of miR-125b, miR-221, and miR-222 in Tumor Tissues of Glioma Patients
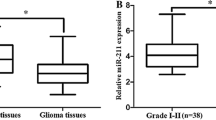
A total of 45 glioma patients, including 15 females and 30 males, were involved in the present study. Tumor tissues and adjacent normal tissues were collected and analyzed. As shown in Fig. 1, all candidate miRNAs were illustrated to be upregulated in glioma tissues. Significantly elevated miR-125b, miR-221, and miR-222 expression was observed in tumor tissues compared with adjacent normal tissues (miR-125b: P < 0.001, Fig. 1a; miR-221: P < 0.001, Fig. 1b; miR-222: P < 0.001, Fig. 1c).
Diagnostic performance of three candidate miRNAs in differentiating tumor and adjacent non-tumor tissue. a Relative expression levels of miR-125b; b relative expression levels of miR-221; c relative expression levels of miR-222; d receiver-operating characteristic (ROC) curve analyses of three candidate miRNAs and their combination in differentiating tumor from adjacent non-tumor tissue
Diagnostic Value of miR-125b, miR-221, and miR-222 in Glioma
Considering the significantly aberrant expression of miR-125b, miR-221, and miR-222, the significance of their expression level in distinguishing glioma tumor tissues from normal tissues was further validated by ROC analysis and assessed by AUC value. ROC curves (Fig. 1d) illustrated the significant value of candidate miRNAs expression level in discriminating tumor tissues (miR-125b: AUC = 0.86, 95 %CI, 0.78–0.93; miR-221: AUC = 0.74, 95 %CI, 0.63–0.84; miR-222: AUC = 0.80, 95 %CI, 0.71–0.89). The optimal cutoff value for miR-125b was 3.60, with a sensitivity of 0.78 and a specificity of 0.80. For miR-221, the optimal cutoff value was 2.17, a sensitivity of 0.73, and a specificity of 0.69 could be acquired from the cutoff value. As for miR-222, a sensitivity of 0.78 and a specificity of 0.76 were achieved with the optimal cutoff value of 3.23. Combination of miR-125b, miR-221, and miR-22 was significantly superior compared to the clinical standard of using these miRNAs alone, with AUC of 0.97 (95 %CI, 0.94–0.99).
Association Between miR-125b, miR-221, and miR-222 and Clinical Features of Glioma Patients
To further investigate the expression of miR-125b, miR-221, and miR-222 in relation to glioma progression and clinical characteristics of glioma patients, all enrolled patients were stratified by gender, age, TNM stage, metastatic status, tumor size, T classification, and differentiation degree. Student’s t test and one-way ANOVA test were applied to evaluate the difference of candidate miRNAs expression in different groups. The results were presented in Table 1. As can be seen from the table, the expression of all candidate miRNAs was not significantly altered by gender and age. The expression of miR-125b was significantly associated with glioma progression. As glioma progressed to advanced stages, the expression level of miR-125b was elevated (P = 0.037). Significantly upregulated miR-125b was also observed in patients with metastasis and low differentiation degree (P = 0.030, P < 0.001, respectively). Increased miR-125b was also related to larger tumor (P = 0.018). The expression of miR-222 was also illustrated to be significantly associated with glioma at advanced stages (P = 0.024) or serious symptoms (P = 0.008 for metastatic status; P = 0.002 for differentiation degree). However, miR-221 was not shown to be significantly related to glioma stages and metastatic status (P = 0.193, P = 0.107, respectively). Since the P value was 0.060 for tumor size and 0.049 for differentiation degree, the associations between miR-221 and tumor size/differentiation degree need to be further studied.
MiR-125b, miR-221, and miR-222 as Prognostic Marker for Glioma
To determine the prognostic value of miR-125b, miR-221, and miR-222 for glioma, Kaplan-Meier survival analysis of glioma patients was conducted, and log-rank test were performed to evaluate the statistical significance between stratified groups according to the expression of candidate miRNAs. High expression groups were defined as patients with higher expression level of candidate miRNA than average and vice versa. The results were plotted in Fig. 2. As illustrated in the figure, patients with higher expression of miR-125b, miR-221, and miR-222 showed significantly poor glioma prognosis (miR-125b: HR = 2.43, 95 %CI, 1.13–2.54, P = 0.023; miR-221, HR = 2.18; 95 %CI, 1.02–4.65, P = 0.044; miR-222: HR = 2.13, 95 %CI, 1.01–4.48, P = 0.043). In addition, panel of these miRNAs could play a better prognostic role in glioma (HR = 3.32, 95 %CI, 1.49–7.39, P = 0.001).
Survival analyses of glioma patients in different groups according to the relative expression of miR-125b, miR-221, and miR-222 and the panel of these miRNAs. a K-W curve of glioma patients in high/low miR-125b expression groups; b K-W curve of glioma patients in high/low miR-221 expression groups; c K-W curve of glioma patients in high/low miR-222 expression groups; d K-W curve of glioma patients in high/low the panel of three candidate miRNAs expression groups
Discussion
A total of 45 glioma patients with different clinical characteristics were involved in the present study. Our results manifested that miR-125b, miR-221, and miR-222 were upregulated in glioma tissues. MiR-125b and miR-222 had a high diagnostic value in distinguishing glioma tumor tissues from normal tissues. Combination of miR-125b, miR-221, and miR-22 was significantly superior compared to the clinical standard of using these miRNAs alone. Besides, all the three candidate miRNAs were proved to be an efficient prognostic marker for glioma. Patients with lower expression of miR-125b, miR-221, and miR-222 showed a better prognostic result compared with patients with higher expression level. Similarly, panel of these miRNAs could play a better prognostic role in glioma.
MiRNAs have been proved to be involved in the process of carcinogenesis in several aspects. MiRNAs target at certain genes participate in the regulation of cell cycle progression and proliferation and can regulate tumor invasion and the status of metastasis. It was also found that miRNAs played an important role in the mechanism of tumor resistance to chemo- and radiotherapy [12]. MiRNAs can be categorized into oncogenic miRNAs (oncomiRs) and tumor-suppressive miRNAs (tsmiRs). Aberrant expression of oncomiRs and tsmiRs can activate specific signaling pathways, which further have an impact on cell proliferation, cell invasion, and angiogenesis and inhibit or promote cancer progression (Fig. 3). For instance, miR-221/222 expression directly correlated with glioma grade and were prevalent in grade IV astrocytoma, where they targeted an important cell cycle regulator, namely p27/KIP1. These data support a critical role in the regulation of the cell cycle, proliferation, and invasion by miR-221/miR-222.
Although the mechanism of miR-125b in the tumorigenesis of glioma is still not clear, it has been demonstrated to be tumor-suppressive in glioblastoma (GBM). It activates carcinogenesis in several ways. Two binding sites for miR-125b have been identified in the 3′UTR of E2F2. It has been proved that miR-125b exert effects on glioma stem cell proliferation by directly targeting at E2F2 [7]. It has also been identified that miR-125b suppresses the expression of connexin43, the overexpression of which antagonizes the effects of miR-125b in cell growth and anti-apoptosis [13]. MiR-125b is also involved in a functional feed-forward loop in glioma angiogenesis. The expression of miR-125b is inhibited by VEGF, which leads to the elevated expression of Myc-associated zinc finger protein (MAZ). MAZ accumulation can attenuate endothelial cell migration and suppress angiogenesis and metastasis and, in its turn, results in the upregulation of VEGF. Thus, with a high level of miR-125b, the feed-forward loop is broken; cell invasion and angiogenesis are promoted [14]. Besides, miR-125b can regulate glioma stem cell proliferation by decreasing CDK6 and CDC25A expression, which leads to cell cycle arrest at the G1/S transition [15]. MiR-125b can interact with 3′UTR of cell apoptosis protein Bcl-2 modifying factor [16] and inhibits cell apoptosis through p53 and p38MAPK-independent pathways [6]. Collectively, overexpression of miR-125b promotes glioma cell proliferation and inhibits cell apoptosis, which contribute to the process of carcinogenesis; it also participates in cell invasion and angiogenesis, which further lead to metastasis. Moreover, miR-125b was found to be upregulated in temozolomide-resistant cells. MiR-125b inhibitor can significantly enhance the chemosensitivity of glioma stem cells to temozolomide, which is a common medicine in chemotherapy [17].
MiR-221 and miR-222 are paralogues that have been manifested to be associated with glioma procession. It has been proved by previous study that the upregulation of miR-221 and miR-222 can interfere with growth factor signaling pathways that stimulate cell proliferation [18]. They are also regulators of the tumor suppressor gene p27Kip1; thus, miR-221/222 can promote glioma cell proliferation by attenuating the expression of p27Kip1 both in vitro and in vivo [8]. Overexpression of miR-221 and miR-222 also results in the activation of Akt pathway, which enhances the glioma malignant phenotype [10]. MiR-221/222 can inhibit cell apoptosis as well. They target pro-apoptotic gene PUMA in glioma cells and reduce its expression [9]. Protein tyrosine phosphatase μ (PTPμ) has been proved to be involved in the carcinogenesis by regulating cell migration. Downregulated PTPμ induced by overexpression of miR-221 and miR-222 is related to glioma tumorigenesis [19]. MiR-221 can also regulate epithelial-mesenchymal transition by targeting related genes through PTEN and PI3-K/Akt pathways [20]. Their regulating function on connexin43 expression is also believed to be associated with glioma [21]. Dickkopf-2 gene (DKK2) is another direct target of miR-222. Overexpression of miR-222 results in constitutive activation of β-catenin through inhibition of DKK2 expression in glioma cells, which finally contributes to glioma procession [22]. Furthermore, miR-221 and miR-222 expression may exert effect on the sensitivity of glioma cells to chemotherapy and radiotherapy. It has been proved that downregulated miR-221 and miR-222 expression can sensitize glioma cells to temozolomide, which is similar to miR-125b [23]. It was suspected that chemotherapy of temozolomide led to increased DNA damage, which could be rescued by alkylating enzyme O(6)-methylguanine-DNA methyltransferase (MGMT). Meanwhile, downregulation of miR-221 and miR-222 attenuated the expression of MGMT. It rendered cells unable to repair DNA damage and contributed to temozolomide-mediated cell death [24]. Regarding the effect of miR-221/222 on radiotherapy, they have been proved to play an important part in mediating radio-induced DNA damage repair of glioma cells by activating Akt, independent of PTEN status. Knocking-down miR-221 and miR-222 can significantly increase the radiosensitivity of glioma cells [25].
Taken together, all the three candidate miRNAs are tightly linked to glioma carcinogenesis and contribute greatly to the procession of glioma. This is in accordance with our results that miR-125b, miR-221, and miR-222 were significantly elevated in glioma tissues, especially in tissues from advanced stages. It might be attributed to the inadequate sample size that we failed to confirm the association of miR-221 expression and glioma stages. The poor prognosis of patients with higher expression of miR-125b, miR-221, and miR-222 might be related to the sensitivity of glioma cells to chemotherapy and radiotherapy. Patients with a higher expression of candidate miRNAs have a low sensitivity to the therapy, and this may partially contribute to their poor prognosis. Nevertheless, higher expression of miRNAs might also indicate an advanced-staged glioma, which is usually characterized with poor diagnosis.
In summary, in this study, we confirmed the significance of miR-125b, miR-221, and miR-222 in distinguishing glioma tumor tissues and normal tissues. Higher expressions of miR-125b and miR-222 have also been proved to be associated with more advanced glioma. Furthermore, glioma patients with higher miR-125b, miR-221, and miR-222 expression were manifested to have poorer prognostic status, which might be attributed to their attenuated sensitivity to chemotherapy and radiotherapy.
References
Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS (2013) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro-Oncology 15(suppl 2):ii1–ii56
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M (2006) Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2(9):494–503. doi:10.1038/ncpneuro0289, quiz 491 p following 516
Fan Z, Wu Y, Shen J, Zhan R (2013) Glutathione S-transferase M1, T1, and P1 polymorphisms and risk of glioma: a meta-analysis. Mol Biol Rep 40(2):1641–1650. doi:10.1007/s11033-012-2213-8
Meister G (2007) miRNAs get an early start on translational silencing. Cell 131(1):25–28. doi:10.1016/j.cell.2007.09.021
Zen K, Zhang CY (2012) Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 32(2):326–348. doi:10.1002/med.20215
Wu N, Lin X, Zhao X, Zheng L, Xiao L, Liu J, Ge L, Cao S (2013) MiR-125b acts as an oncogene in glioblastoma cells and inhibits cell apoptosis through p53 and p38MAPK-independent pathways. Br J Cancer 109(11):2853–2863
Wu N, Xiao L, Zhao X, Zhao J, Wang J, Wang F, Cao S, Lin X (2012) miR-125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS Lett 586(21):3831–3839
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, Wang G, Zhang A, Jia Z, Han L, Jiang H (2009) Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol 34(6):1653–1660
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9:229. doi:10.1186/1476-4598-9-229
Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C (2010) miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 36(4):913–920
Sobin LH, Fleming ID (1997) TNM classification of malignant tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer 80(9):1803–1804
Koshkin PA, Chistiakov DA, Chekhonin VP (2013) Role of microRNAs in mechanisms of glioblastoma resistance to radio- and chemotherapy. Biochem Biokhimiia 78(4):325–334. doi:10.1134/s0006297913040019
Jin Z, Xu S, Yu H, Yang B, Zhao H, Zhao G (2013) miR-125b inhibits connexin43 and promotes glioma growth. Cell Mol Neurobiol 33(8):1143–1148
Smits M, Wurdinger T, van het Hof B, Drexhage JA, Geerts D, Wesseling P, Noske DP, Vandertop WP, de Vries HE, Reijerkerk A (2012) Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. Faseb J 26(6):2639–2647
Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu N, Fu Z, You Y (2010) MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res 2:120–126
Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, Ma X (2009) MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem 23(4–6):347–358
Chen J, Fu X, Wan Y, Wang Z, Jiang D, Shi L (2014) miR-125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting Bak1. Tumour Biol 35(7):6293–6302
Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS (2008) MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res 68(8):2773–2780. doi:10.1158/0008-5472.can-07-6754
Quintavalle C, Garofalo M, Zanca C, Romano G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato M, Nuovo G, Croce CM, Condorelli G (2012) miR-221/222 overexpression in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene 31(7):858–868. doi:10.1038/onc.2011.280
Xie Q, Huang Z, Yan Y, Li F, Zhong X (2014) miR-221 mediates epithelial-mesenchymal transition-related gene expressions via regulation of PTEN/Akt signaling in drug-resistant glioma cells. Nan fang yi ke da xue xue bao J South Med Univ 34(2):218–222
Hao J, Zhang C, Zhang A, Wang K, Jia Z, Wang G, Han L, Kang C, Pu P (2012) miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol Rep 27(5):1504–1510. doi:10.3892/or.2012.1652
Li Q, Shen K, Zhao Y, He X, Ma C, Wang L, Wang B, Liu J, Ma J (2013) MicroRNA-222 promotes tumorigenesis via targeting DKK2 and activating the Wnt/beta-catenin signaling pathway. FEBS Lett 587(12):1742–1748. doi:10.1016/j.febslet.2013.04.002
Chen L, Zhang J, Han L, Zhang A, Zhang C, Zheng Y, Jiang T, Pu P, Jiang C, Kang C (2012) Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep 27(3):854–860. doi:10.3892/or.2011.1535
Quintavalle C, Mangani D, Roscigno G, Romano G, Diaz-Lagares A, Iaboni M, Donnarumma E, Fiore D, De Marinis P, Soini Y, Esteller M, Condorelli G (2013) MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One 8(9):e74466. doi:10.1371/journal.pone.0074466
Li W, Guo F, Wang P, Hong S, Zhang C (2014) miR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr Mol Med 14(1):185–195
Acknowledgments
This work is supported by grants from the Natural Science Foundation of China (81372484): Molecular mechanisms of EMAP-II suppressing glioma by inducing GSC autophagy, and (81172197): EMAP-II regulates the PKC subtype increased the permeability of blood tumor barrier and inhibited the proliferation of glioblastoma via down-regulating miR-330.
Conflict of Interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, X., Zheng, J., Chen, L. et al. Predictive and Prognostic Roles of Abnormal Expression of Tissue miR-125b, miR-221, and miR-222 in Glioma. Mol Neurobiol 53, 577–583 (2016). https://doi.org/10.1007/s12035-014-9017-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-9017-x