Abstract
Glioma is the most common type of primary malignant tumor in the central nervous system (CNS) with a high incidence and a high mortality rate, as well as an extremely low 5-year survival rate. As a class of small non-coding RNAs, microRNAs (miRNAs) may be closely involved in carcinogenesis and might also be connected with glioma diagnosis and prognosis. In this study, we aimed at investigating the expression level of microRNA-183 (miR-183) in 105 cases of glioma tissues of four World Health Organization (WHO) grades and 10 cases of normal brain tissues and its potential predictive and prognostic values in glioma. We found that the expression levels of miR-183 were significantly higher in glioma tissues than that in normal brain tissues, and also higher in high-grade gliomas (WHO grade III and IV) compared with low-grade gliomas (WHO grade I and II). The miR-183 expression level was classified as low or high according to the median value. High expression of miR-183 was found to significantly correlate with larger tumor size, higher WHO grade, and worse Karnofsky performance score (KPS). Kaplan–Meier survival analysis showed that patients with high miR-183 expression had worse overall survival (OS) and progression-free survival (PFS) than patients with low miR-183 expression. Moreover, univariate and multivariate analyses indicated that miR-183 expression level was an independent prognostic parameter of a patient’s OS and PFS. In conclusion, our study indicated that miR-183 was upregulated in glioma, and that it may be used as a potential biomarker of poor prognosis in patients with glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas account for the most prevalent malignancy of the brain, especially glioblastoma multiforme (GBM), which is characterized by a rapid infiltrative growth pattern, making complete surgical resection impossible and leading to a poor prognosis [1]. Based on the degree of malignancy according to the World Health Organization (WHO) classification, gliomas are divided into four histopathologic grades: low-grade astrocytomas (WHO grade I–II) and high-grade gliomas, including anaplastic astrocytomas (WHO grade III) and GBM (WHO grade IV). Although great improvements have been made in neurosurgical techniques, exploitation of accurate radiotherapy, and development of new chemotherapeutic agents, extremely poor prognosis of malignant gliomas remains unchanged over the last three decades [2, 3]. The 5-year survival rate of low-grade gliomas is approximately 30–70 %, while the median survival time of GBM is reported to range from 9 to 12 months due to its rapid and infiltrative growth, as well as its high, rapid rate of recurrence after operation [4]. Therefore, it is vitally important to further explore precise prognostic biomarkers and more effective therapeutic targets for this deadly disease [5].
MiRNAs, as small, non-coding RNA molecules, are involved in a variety of biological processes, such as cell development, differentiation, proliferation, apoptosis, metabolism, and cell-cycle control [6]. Many studies have shown that aberrant miRNA expression is often associated with the occurrence and development of various human cancers. In addition, deregulated miRNAs can be used as biomarkers for cancer diagnosis and prognosis [7]. As a member of an evolutionarily conserved miRNA family (miR-96, miR-182 and miR-183), miR-183 has been proved to act as either a suppressor or an oncogene in various types of human cancers. For example, miR-183 inhibits cell migration and invasion of lung [8], breast [9], and gastric [10] cancer by targeting Ezrin. In addition, miR-183 is significantly upregulated in synovial sarcoma, colon cancer, and rhabdomyosarcoma, promoting tumor cell migration by targeting the transcription factor early growth response gene 1 (EGR1) [11]. However, the role of miR-183 in glioma and its clinical significance have not been reported. In this study, we investigated the association between miR-183 expression and the clinicopathological parameters as well as the prognosis of patients with glioma.
Materials and methods
Patients and specimens
This study was approved by the First Affiliated Hospital, Wannan Medical College, Wuhu, Anhui Province, People’s Republic of China. Informed consents in this study were obtained from all subjects or the next of kin. A total of 105 glioma tissue specimens were obtained from patients who underwent surgical treatment at the Department of Neurosurgery of the First Hospital Affiliated to Wannan Medical College between March 2007 and May 2010. Normal brain tissues used as controls were obtained from 10 patients with severe traumatic brain injury who received surgical treatment for internal decompression. All specimens were collected right after resection in the operating room and quickly frozen in liquid nitrogen and stored at −80 °C. All specimens were made anonymous and were handled according to the ethical and legal standards. Patients with the previous history of cancer or immune disease and patients without complete clinical and follow-up data were excluded from the study. Any different conclusions were settled by rigorous study and discussion. A total of 60 men and 45 women were enrolled in this study, and the median age was 52 years (range 18–78 years). Of the 105 gliomas, 41 were classified as low-grade gliomas [15 pilocytic astrocytoma (WHO grade I), 26 diffuse astrocytomas (WHO grade II)], 64 as high-grade gliomas [29 anaplastic astrocytomas (WHO grade III), and 35 glioblastomas (WHO grade IV)]. The characteristics of the patients are shown in Table 1. All patients were under 5-year close follow-up observation for disease recurrence at no less than 3-month intervals during the first two postoperative years and no less than every 6 months thereafter. Overall survival (OS) time was counted from the date of the initial surgery to death. Progression-free survival (PFS) time was defined as the time from the date of the initial surgery until the first evidence of local, regional, or distant tumor progression of disease.
RNA isolation and realtime-PCR
Total RNA was extracted using TRIzol according to the manufacturer’s protocol. The miR-183 stem-loop reverse-transcription (RT) primer was designed as 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTGAA′-3. RT reactions contained 10 ng of total RNAs, 50-nmol/l stem-loop RT primer, 1× RT buffer, 0.25 mmol/l each of deoxynucleotide triphosphate (dNTP), 3.33U/l MultiScribe reverse transcriptase, and 0.25U/μl RNase Inhibitor. Volumes of 7.5 μl were incubated in Bio-Rad i-Cycler (Bio-Rad Laboratories, Hercules, CA, USA) in a 96-well plate for 30 min at 16 °C, 30 min at 42 °C, 5 min at 85 °C, and then held at 4 °C. Targeted DNAs’ amplification was performed using an Applied Biosystems 7500 Real-Time PCR system. The 10-μl PCR reaction system included 0.6 μl of RT products, 1× TaqMan Universal PCR Master Mix, 1 μl of primers, and a probe-mix of the TaqMan MicroRNA Assays. The PCR primers were designed as follows: miR-183 sense, 5′-CGCGGTATGGCACTGGTAGA′-3, and reverse, 5′-AGTGCAGGGTCCGAGGTATTC′-3; U6 sense, 5′-CTCGCTTCGGCAGCACA′-3, and reverse, 5′-AACGCTTCACGAATTTGCGT′-3. Relative change in miR-183 expression to U6 was determined by the equation: fold change = 2−ΔΔCt, ΔΔCt = (CtmiR-183−CtU6)Glioma−(CtmiR-183−CtU6) Control. Ct value is the cycle number at which fluorescence signal crosses the threshold. Mean-normalized gene expression ± standard deviation (SD) was calculated from independent experiments.
Statistical analysis
All data analyses were carried out using SPSS software, version 19.0 for Windows (SPSS Inc., IL, USA) and GraphPad Prism 5 (GraphPad Software Inc., CA, USA). Data were expressed as mean ± SD. The Chi-square test was used to assess miR-183 expression with respect to clinicopathological parameters. One-way ANOVA Tukey’s multiple comparisons test or student’s t test was used to analyze the difference of miR-183 expression between groups. The survival curves of the patients were determined using the Kaplan–Meier method and Cox regression analysis. The log-rank test was used for statistical evaluations. Univariate Cox regression was performed on each clinical covariate to examine its influence on patient survival. Final multivariate models were based on a stepwise addition. Differences were considered statistically significant when P was less than 0.05.
Results
Increased expression of miR-183 in human glioma tissues
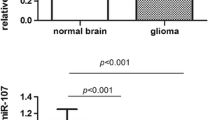
All data of target miRNA, including 105 cases of glioma and 10 cases of normal brain tissues, are shown in Fig. 1. MiR-183 expression levels were significantly higher in glioma tissues than that in normal brain tissues (mean ± SD, 2.731 ± 0.110 vs. 1.026 ± 0.034; P < 0.01), and the miR-183 expression levels in glioma tissues of each WHO grade were all significantly higher than that in normal brain tissues (WHO grade I tumor vs. normal: 1.535 ± 0.071 vs. 1.026 ± 0.034; WHO grade II tumor vs. normal: 1.730 ± 0.069 vs. 1.026 ± 0.034; WHO grade III tumor vs. normal: 3.313 ± 0.151 vs. 1.026 ± 0.034, both P < 0.05; and WHO grade IV tumor vs. normal: 3.691 ± 0.132 vs. 1.026 ± 0.034, both P < 0.01; as shown in Fig. 1a). Moreover, the miR-183 expression levels of high-grade (III–IV) glioma tissues were distinctly higher than that of low-grade (I–II) glioma tissues (3.512 ± 0.102 vs. 1.650 ± 0.051, P < 0.01; as shown in Fig. 1b). However, there was no difference of miR-183 expression between grade I and grade II (P = 0.162), as well as between grade III and IV (P = 0.064).
Increased expression of miR-183 in human glioma tissues. a qRT-PCR results of 105 cases of glioma (grade I–IV) and 10 cases of normal brain tissues. *P < 0.05, **P < 0.01 vs. control group, ns P > 0.05. b Comparison of relative miR-183 expression between low-grade gliomas (grade I and II) and high-grade gliomas (grade III and IV). **P < 0.01. Data were normalized to U6
The association of miR-183 expression with clinicopathological parameters in glioma
The miR-183 expression level was classified as low or high according to the median value. High expression of miR-183 was found to significantly correlate with tumor size (P = 0.0318), WHO grade (P < 0.01), and Karnofsky performance score (KPS; P < 0.01). However, no significant difference in miR-183 expression was determined with the extent of resection, location, age, or gender (both P > 0.05; as shown in Table 1).
High miR-183 expression predicts poor prognosis in patients with glioma
Kaplan–Meier analysis with the log-rank test indicated that high miR-183 expression significantly correlated with shorter OS and PFS (both P < 0.001; as shown in Fig. 2a, b, respectively). Univariate and multivariate analyses were used to determine whether miR-183 expression levels, and various clinicopathological features were independent prognostic parameters of patient prognosis. Multivariate analysis indicated that miR-183 expression levels [hazard ratio (HR) = 7.34 (95 % confidence interval (CI), 2.11–12.70); P = 0.007], extent of resection [HR = 6.46 (95 % CI, 0.31–7.86); P = 0.011], KPS score [HR = 4.57 (95 % CI, 0.15–8.49); P = 0.004], WHO grading [HR = 3.18 (95 % CI, 0.36–6.94); P < 0.001], tumor size [HR = 2.51 (95 % CI, 0.36–4.14); P = 0.029], chemotherapy [HR = 1.56 (95 % CI, 0.12–6.34); P = 0.048], and age [HR = 4.41 (95 % CI, 0.39–5.97); P = 0.036] were independent prognostic factors for OS, and that miR-183 expression [HR = 7.28 (95 % CI, 1.10–9.73); P = 0.006], extent of resection [HR = 9.07 (95 % CI, 0.29–13.77); P = 0.003], tumor size [HR = 3.17 (95 % CI, 0.42–5.15); P = 0.037], location [HR = 3.41 (95 % CI, 0.52–6.03); P = 0.026], chemotherapy [HR = 2.01 (95 % CI, 0.36–4.35); P = 0.031], radiotherapy [HR = 1.84 (95 % CI, 0.54–5.11); P = 0.042], and WHO grading [HR = 2.28 (95 % CI, 0.25–7.19); P = 0.038] were independently associated with the PFS (details in Table 2).
High miR-183 expression predicts poor prognosis in patients with glioma. a Kaplan–Meier curves of the overall survival of 105 glioma patients. Overall survival rate in patients with high miR-183 expression was significantly lower than that in patients with low miR-183 expression. b Kaplan–Meier curves of the progression-free survival of 105 glioma patients. Progression-free survival rate in patients with high miR-183 expression was significantly lower than that in patients with low miR-183 expression
Discussion
Gliomas are the most prevalent primary brain neoplasms with extremely high morbidity and mortality rates. Despite tremendous efforts in glioma diagnosis and treatment, including surgery, chemotherapy, and radiation therapy, prognosis of patients with glioma, especially with high-grade gliomas, remains unfavorable due to the rapid recurrence and symptomless progression in the early stage of these tumors. Therefore, it is of great importance and significance to identify powerful and valid biomarkers for gliomas, and to understand the complicated underlying molecular mechanisms of these malignancies.
As single stranded and noncoding RNAs, miRNAs play important roles in a series of biological processes [12] and have been proved to regulate cell proliferation, survival, as well as tumor angiogenesis, invasion, and metastasis during the initiation and progression of human malignancies [13, 14]. Dysregulation of miRNA expression has been found in many types of human cancers, including cancers in colon, breast, lung, and malignant gliomas [15–18]. Though the underlying mechanisms of miRNA-mediated gene modulation are still under investigation, increasing studies suggest that signatures of miRNA expression can be diagnostically and/or prognostically indicative for human cancers [19].
MiR-183 is one member of the miR-183-96-182 cluster located in the 7q31-34 locus [20]. Recently, many studies have reported that miR-183 is a potential inhibitor of cancer cell invasion and metastasis [8–10, 21] by targeting Ezrin, which has been identified to be a key component in tumor metastasis [22]. Previously, Xu et al. [23] identified that miR-183 was declined in gastric cancer cells and tissues, and that it inhibited gastric cancer cell proliferation and invasion by targeting Bmi-1. Thus, it is plausible to postulate that miR-183 may function as a suppressor in various types of human cancers. However, increasing evidence suggests that miR-183 may also act as an oncogene via different mechanisms. Li et al. [24] reported that miR-183 was significantly upregulated in hepatocellular carcinoma (HCC) and inhibited TGF-β1-induced apoptosis in human HCC cells by repressing programmed cell death 4 (PDCD4), which has been originally identified as a human tumor-related gene [25] and loss of which contributes to cell growth by several means and, consequently, facilitates the development of cancer [26, 27]. Recent studies have also demonstrated that overexpression of miR-183 not only enhances proliferation, migration, and invasion, but also inhibits apoptosis in papillary thyroid carcinoma and pancreatic cancer cells by negatively regulating PDCD4 [28, 29]. In addition, Sarver et al. [11] showed that downregulation of miR-183 inhibited migration in synovial sarcoma and colon cancer cell lines. Decrease in miR-183 leads to increase in EGR1 protein as well as its transcriptional target PTEN, both of which are tumor suppressors, suggesting that miR-183 may act as an oncogene by targeting the transcriptional factor EGR1 via a miR-183-EGR1-PTEN pathway. MiR-183 is also reported to promote growth of non-small cell lung cancer cells through FOXO1 inhibition [30]. Taken together, the effect of miR-183 as a suppressor or an oncogene seems to be cell- and/or tissue-type dependent. Therefore, the role of miR-183 remains a complex area of study that needs further investigation in tumors with a highly complex microenvironment.
What we concerned was the role of miR-183 in gliomas, and to the best of our knowledge, there are few studies regarding the role of miR-183 in glioma. Tang et al. reported that knockdown or inhibition of the entire miR-183/96/182 cluster or knockdown of individual components of this miRNA cluster induces apoptosis and decreases survival of glioma cells via regulating ROS-mediated AKT/survival pathway and p53/apoptosis signaling. In addition, knockdown of the entire cluster enhances the anticancer effect of Temozolomide on glioma cells [31]. Zhang et al. [32] demonstrated that the miR-183/96/182 cluster increases proliferation of medulloblastoma cells in a mouse model, which was dependent on hedgehog signaling activation. In addition, gain-of-function studies showed that by targeting isocitrate dehydrogenase 2 (IDH2) directly, overexpression of miR-183 in glioma cells augmented hypoxia-inducible factor 1α (HIF-1α) [33], a subset of HIF-1, which activates the transcription of genes involved in crucial aspects of cancer biology, including cell survival, proliferation, apoptosis, invasion, angiogenesis, and glucose metabolism [34, 35]. Thus, we inclined to consider miR-183 mainly as an “oncomiR” in glioma according to the limited relevant studies as described above.
With that assumption, we conducted a preliminary study on the expression level of miR-183 in glioma and its relationship with clinicopathological parameters and prognosis of patients with this malignancy, so as to gain clinical evidence for further mechanistic research. In this study, we identified that tissues of high-grade gliomas had much higher miR-183 expression levels than that of low-grade gliomas and normal brain tissues. Importantly, the results of a 60-month follow-up in these patients demonstrated that miR-183 expression levels significantly correlated with the survival of patients with glioma. At the same time, glioma patients with high miR-183 expression levels had a distinctly shorter OS and PFS. In addition, univariate and multivariate analyses showed that the miR-183 expression level was an independent prognostic parameter of OS and PFS in glioma patients. Therefore, we present evidence here for miR-183 as a potential prognostic indicator for human brain glioma.
Similarly, two previous clinical investigations have also reported that miR-183 is upregulated in both prostate and colorectal cancer and high miR-183 level correlated with high tumor malignancy and poor prognosis of patients [36, 37]. Combined with these findings, we speculate that increased expression level of miR-183 in glioma, especially in high-grade gliomas, might involve in carcinogenesis and tumor progression by regulating a panel of genes that regulate cell survival, proliferation, migration, and invasion, such as PDCD4, EGR1/PTEN, FOXO1, IDH-2/HIF-1α, and ROS-mediated genes as described above, eventually contributing to the degree of malignancy and patients’ poor prognosis. Hence, additional studies are needed to investigate the precise underlying molecular mechanisms of both the cause and the effects of elevated expression of miR-183 in glioma.
To the best of our knowledge, this is the first report to investigate the miR-183 expression and its clinical significance in human brain glioma; however, we must report that limitations of this study should be noted. First, when we evaluated the expression level of miR-183 in glioma tissues, we used normal brain tissues obtained from patients with severe traumatic brain injury who received surgical treatment for internal decompression as controls, instead of the paired peritumoral normal brain tissues, which may be a more scientific control group. Second, this is a preliminary single-center study and the factors used in multivariate analyses might be not enough. Thus, our results should be interpreted with caution and multicenter and large-scale studies may be warranted.
In conclusion, our study demonstrates that upregulation of miR-183 in glioma correlates with prognosis of patients with this devastating disease, suggesting the potential role of miR-183 as a biomarker in human brain glioma. However, it is very unlikely that a single biomarker will accurately predict prognosis of glioma, thus a combination of biomarkers is still needed to achieve a more precise outcome prediction, and further mechanistic studies are needed to provide deep insights into molecular mechanisms of miR-183 in glioma.
References
McCarthy BJ, Shibui S, Kayama T, Miyaoka E, Narita Y, Murakami M et al (2012) Primary CNS germ cell tumors in Japan and the United States: an analysis of 4 tumor registries. Neuro Oncol 14:1194–1200
Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21:2683–2710
Meyer MA (1850) Malignant gliomas in adults. N Engl J Med 2008(359):1850
Zhang X, Yang H, Gong B, Jiang C, Yang L (2012) Combined gene expression and protein interaction analysis of dynamic modularity in glioma prognosis. J Neurooncol 107:281–288
Liang Z, Gao Y, Shi W, Zhai D, Li S, Jing L et al (2013) Expression and significance of microRNA-183 in hepatocellular carcinoma. Sci World J 2013:1–6
Yates LA, Norbury CJ, Gilbert RJ (2013) The long and short of microRNA. Cell 153:516–519
Uchino K, Ochiya T, Takeshita F (2013) RNAi therapeutics and applications of microRNAs in cancer treatment. Jpn J Clin Oncol 43:596–607
Wang G, Mao W, Zheng S (2008) MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett 582:3663–3668
Lowery AJ, Miller N, Dwyer RM, Kerin MJ (2010) Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 10:502
Cao LL, Xie JW, Lin Y, Zheng CH, Li P, Wang JB et al (2014) MiR-183 inhibits invasion of gastric cancer by targeting Ezrin. Int J Clin Exp Pathol 7:5582–5594
Sarver AL, Li L, Subramanian S (2010) MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res 70:9570–9580
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N et al (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299:425–436
Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S et al (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13:48–57
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070
Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G et al (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334:1351–1358
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D (1998) Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 58:2170–2175
Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X et al (2012) MiR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med 30:1013–1020
Hunter KW (2004) Ezrin, a key component in tumor metastasis. Trends Mol Med 10:201–204
Xu L, Li Y, Yan D, He J, Liu D (2014) MicroRNA-183 inhibits gastric cancer proliferation and invasion via directly targeting Bmi-1. Oncol Lett 8:2345–2351
Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J et al (2010) MiR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer 10:354
Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y et al (2006) Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 25:6101–6112
Bohm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer KH (2003) The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene 22:4905–4910
Jansen AP, Camalier CE, Colburn NH (2005) Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 65:6034–6041
Lu YY, Zheng JY, Liu J, Huang CL, Zhang W, Zeng Y (2015) MiR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed Pharmacother 70:151–157
Wei C, Song H, Sun X, Li D, Song J, Hua K et al (2015) MiR-183 regulates biological behavior in papillary thyroid carcinoma by targeting the programmed cell death 4. Oncol Rep 34:211–220
Zhang L, Quan H, Wang S, Li X, Che X (2015) MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol 36:8121–8126
Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q et al (2013) The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets 13:221–231
Zhang Z, Li S, Cheng SY (2013) The miR-183 approximately 96 approximately 182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J Biomed Res 27:486–494
Tanaka H, Sasayama T, Tanaka K, Nakamizo S, Nishihara M, Mizukawa K et al (2013) MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J Neurooncol 111:273–283
Targeting Semenza GL (2003) HIF-1 for cancer therapy. Nat Rev Cancer 3:721–732
Semenza GL (2010) HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev 20:51–56
Zhou T, Zhang GJ, Zhou H, Xiao HX, Li Y (2014) Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol 26:229–233
Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL et al (2013) MicroRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer 108:1659–1667
Acknowledgments
This study was supported by National Natural Science Foundation of China (Nos. 81371294, 81401026, 81400980 and 81401029) and Jiangsu Provincial Natural Science Foundation of China (No. BK20141375).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Z. Ye and Z. Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Ye, Z., Zhang, Z., Wu, L. et al. Upregulation of miR-183 expression and its clinical significance in human brain glioma. Neurol Sci 37, 1341–1347 (2016). https://doi.org/10.1007/s10072-016-2599-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-016-2599-5