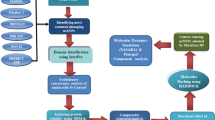
Abstract
Ataxia telangiectasia-mutated (ATM) protein kinase, a key player in cellular integrity regulation, is known for its role in DNA damage response. This study investigates the broader impact of ATM on cellular processes and potential clinical manifestations arising from mutations, aiming to expand our understanding of ATM’s diverse functions beyond conventional roles. The research employs a comprehensive set of computational techniques for a thorough analysis of ATM mutations. The mutation data are curated from dbSNP and HuVarBase databases. A meticulous assessment is conducted, considering factors such as deleterious effects, protein stability, oncogenic potential, and biophysical characteristics of the identified mutations. Conservation analysis, utilizing diverse computational tools, provides insights into the evolutionary significance of these mutations. Molecular docking and dynamic simulation analyses are carried out for selected mutations, investigating their interactions with Y2080D, AZD0156, and quercetin inhibitors to gauge potential therapeutic implications. Among the 419 mutations scrutinized, five (V1913C, Y2080D, L2656P, C2770G, and C2930G) are identified as both disease causing and protein destabilizing. The study reveals the oncogenic potential of these mutations, supported by findings from the COSMIC database. Notably, Y2080D is associated with haematopoietic and lymphoid cancers, while C2770G shows a correlation with squamous cell carcinomas. Molecular docking and dynamic simulation analyses highlight strong binding affinities of quercetin for Y2080D and AZD0156 for C2770G, suggesting potential therapeutic options. In summary, this computational analysis provides a comprehensive understanding of ATM mutations, revealing their potential implications in cellular integrity and cancer development. The study underscores the significance of Y2080D and C2770G mutations, offering valuable insights for future precision medicine targeting-specific ATM. Despite informative computational analyses, a significant research gap exists, necessitating essential in vitro and in vivo studies to validate the predicted effects of ATM mutations on protein structure and function.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Several DNA damage events ensue in the human body every day as a result of exposure to diverse environments [1]. These conditions effect the DNA by simple base alterations, base incongruities, inter-strand crosslinks, intra-strand crosslinks, bulky DNA adducts, DNA–protein crosslinks, single-stranded break (SSB), and double stranded break (DSB) [2]. When normal cells are stressed and their DNA is damaged, the damage can be repaired utilising intact DNA repair pathways until the stress becomes severe enough to cause cell death or senescence [3]. The ataxia telangiectasia mutated (ATM) protein is one of the most important unit of the DNA damage response system, acting as an intra-cellular sensor for DSB [4]. It is generally found in cells in the dimeric forms and undergoes auto-phosphorylation in response to DNA damage, resulting in the separation of the inactive complex. The following activation of a signalling cascade linking the phosphorylation of several substrates, which leads to two critical responses to DNA damage: the cell-cycle checkpoints activation and the beginning of DNA repair. Therefore, when DNA repair mechanism fails, apoptosis gets triggered [5]. ATM substrates comprise Mdm2, c-Abl, and p53, which impact the G1 checkpoint; Rad51, NbsS1, FANCD2, and BRCA1 that plays role in the transient IR-induced S-phase arrest; besides Chk1, Chk2, and BRCA1 that control the G2 checkpoint [5, 6]. It modulates networks participating in DNA repair, insulin-like growth factor, stress response and other metabolic pathways, with approximately phosphorylating 700 targets, as a result of DSBs. The large number of ATM targets during DNA repair or genomic stress is most likely a method of coordinating many pathways. ATM and other members of the PIKK family, such as the catalytic subunit of DNA-dependent protein kinase (DNA-PKc) and ATM-related (ATR), exhibit redundancy and collaborate in response to various forms of genotoxic stress (Fig. 1).
The structure of ATM is characterized by a butterfly-shaped dimer, formed by the combination of the FAT and KD domains into a dimeric body referred to as FATKD (Fig. 2). Emerging from this body is the N-terminal α–α solenoids, spanning approximately 1900 residues, identified as Spiral and Pincer domains. The Spiral domain covers residues 1–1166, followed by the Pincer domain encompassing residues 1167–1898 [7, 8]. Moving along the sequence, the FAT domain, named after FRAP, ATM, TRRAP, extends from residues 1899–2613, while the Kinase domain occupies residues 2614–3056. Similar to other PIKKs, the Kinase domain comprises an N-terminal lobe (residues 2614–2770) and a C-terminal lobe (residues 2771–2957), with the catalytic cleft situated between them. The C lobe concludes with the FAT C-terminal domain (residues 3027–3056), a distinctive feature within the PIKK family, absent in canonical kinases [9, 10]. Maintaining structural integrity is crucial, as mutations in key residues of the ATM protein can potentially alter its structure, thereby leading to significant functional changes.
Understanding the significance of single-nucleotide polymorphisms (SNPs) in human genetic phenotypic variation will help us better understand human genetic phenotypic variability, particularly in complex illnesses. Additionally, SNPs in the ATM gene can disturb all of the above-mentioned interactions, which are necessary for the kinase’s normal function, and several studies have connected SNPs in the ATM gene to a range of diseases [11,12,13]. SNPs in the biologically important regions of ATM can alter its normal function. Despite the fact that ATM is an important kinase linked to DNA repair and a diversity of malignancies, only a few computational studies have been demonstrated to be involved in detecting disease-associated mutations and their role in structure and function change.
Several computational analyses have already been carried out in past to find harmful SNPs in the gene linked to human diseases [14,15,16,17]. As a result, the goal of this study was to assess the potential impacts of SNPs on distinct structural regions of ATM that might affect its function and perhaps play a role in cancer progression. To accompany this, we primarily used several computational algorithms such MetaSNP, Pmut, and Provean to evaluate the deleterious/disease-causing potential of SNPs. Additionally, the web servers like I-Mutant 2.0, mCSM, SDM2, CUPSAT, and MUpro were used to evaluate the effect of SNPs on protein stability. Later the cancer-promoting potentials and residual conservation of SNPs were evaluated by FATHMM-cancer and ConSurf server, respectively. Following that, we presented modelled protein structures for the mutations using PyMOL mutagenesis plugin. The molecular-docking analysis of wild type and mutants was performed against ATM inhibitors such as AZD0156 and AZD1390 along with the natural compound quercetin and best docked possess were analysed and represented. Lastly, to validate the docking experiments, the molecular dynamics simulation was performed.
Materials and Methods
The workflow that was followed is depicted in Fig. 3.
Data Collection
The ATM kinase mutations (SNPs) list was gleaned through online mutational databases such as HuVarBase (https://www.iitm.ac.in/bioinfo/huvarbase/), and dbSNP (https://www.ncbi.nlm.nih.gov/snp/). The UniProt KB (https://www.uniprot.org/) a protein sequences database was used to obtain the protein sequence information of ATM kinase [UniProt Id: Q13315 (ATM_HUMAN)]. The 3D coordinates of ATM kinase protein were obtained from the Protein Data Bank (RCSB PDB) PDB Id: 5PN0 (http://www.rcsb.org/) for the study.
Deleterious Mutation Analysis
We used a number of publicly available tools for this research, which are briefly described below.
A web-based tool called MetaSNP that aids in identifying polymorphic missense SNPs associated with disease is based on a random forest binary classifier. It primarily incorporates four widely used techniques, SIFT, PhD-SNP, PANTHER, and SNAP, which aid MetaSNP in more effectively detecting harmful variants. SV-2009 dataset was used to train and test this tool using a 20-fold cross-validation procedure (https://snps.biofold.org/meta-snp/index.html) [18].
A protein’s biological function can be predicted using the online tool PROVEAN v1.1.3 (Protein Variation Effect Analyzer), which predicts how an amino acid substitution or indel will impact a protein. The scores generated both within and between clusters are averaged to produce the PROVEAN score. The tool’s default threshold score is “− 2.5,” and if the variant is predicted to be less than that score, it is predicted to be “deleterious,” while if it is predicted to be more than that score, it is predicted to be “neutral” (http://provean.jcvi.org/about.php) [19].
A neural network algorithm is used by the online server PMut to forecast the pathological nature of missense mutations. SwissVar is a variation database that has been manually curated to train this tool. It primarily functions on two levels; first, it retrieves data from a local database of mutational hotspots, and then it assesses a specific SNP in a particular protein. It foresees that the mutation score will range from 0 to 1. If mutations scoring 0 to 0.5 are considered neutral mutations and mutations scoring 0.5 to 1 are considered disease-causing mutations (http://mmb.irbbarcelona.org/PMut/) [20].
Protein Stability Check
A novel programme called mCSM uses a graph-based approach to examine the effects of missense mutations on protein stability. As a result of the atomic distance pattern of various residues, it has been trained in a particular environment. mCSM provides a better understanding of mutations and their relationship to diseases for a large number of proteins. For evaluating mutation stability, this programme has a unique cutoff (scoring pattern). When a mutation’s Gibbs free energy is predicted to be greater than zero, it is said to be “stabilising,” and vice versa if the mutation’s Gibbs free energy is below zero (http://biosig.unimelb.edu.au/mcsm/) [21].
Site-directed mutator 2, or SDM2, is a computer programme that assesses the variation in protein stability brought on by mutations. Following the environment-specific amino acid substitutions tables based on density packing and residue length, it evaluates the effects of mutations. Over 130 different proteins have been tested using this tool’s nearly 2690 different amino acid substitutions. If the Gibbs free energy is above “0,” it is predicted to be stabilising, and if it is below “0,” it is predicted to be destabilising (http://marid.bioc.cam.ac.uk/sdm2) [22].
A web server called iSTABLE is used to forecast the stability of proteins. It establishes whether a mutation has made a protein more or less stable. Support vector machines are used as integrators by this server. The two primary input options for this tool are structural and sequential. A stabilising mutation is indicated by a positive Gibbs free energy value, while a destabilising mutation is indicated by a negative number (http://predictor.nchu.edu.tw/istable/) [23].
Cologne University Protein Stability Analysis Tool (CUPSAT) is a computer programme that analyses the effects of point mutations on protein stability. It predicts the difference in Gibbs free energy between wild-type/normal and mutant proteins. The findings include information on the mutation’s location, structure, and the specific effects of 19 different amino acid substitutions on protein stability. A positive Gibbs free energy value indicates a stabilising mutation, whereas a negative number indicates a destabilising mutation (http://cupsat.tu-bs.de/) [24].
I-Mutant 3.0 is a machine-learning-based technique that considers altered residues’ spatial surroundings in terms of surrounding residue types and surface accessibility. I-Mutant 3.0 has been trained to perform the following tasks: (I) Predict the direction of protein stability changes as a result of mutations (a classification task); (II) Predict the Gibbs free energy as a result of mutations (a function approximation task) (https://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) [25].
MUpro predicts the effect of a mutation on protein stability using a suite of machine learning systems. The results are centred on two machine learning methodologies, support vector machines, and neural networks. It calculates the effect of mutation on protein stability using the value of the Gibbs free energy change. It also forecasts the direction of energy change using neural networks and support vector machines. Furthermore, it predicts protein stability without knowing the protein’s tertiary structure (http://mupro.proteomics.ics.uci.edu/) [26].
Cancer-Causing Potential
FATHMM-cancer is a web-based high-throughput tool for predicting the functional consequences of mutations. It forecasts the cancer-causing potential of specific mutations. Based on the default threshold score of “− 0.75”, this tool generates a prediction. A predicted score less than “− 0.75” indicates that the mutation is “cancer-promoting”, whereas a score greater than “− 0.75” indicates that the mutation is a “passenger” (http://fathmm.biocompute.org.uk/cancer) [27].
Biophysical Characteristics
The biophysical properties were examined using the Align GVGD server. The prediction analysis was given the mutation list and a multiple sequence alignment as inputs. The information is arranged by Class, which ranges from 0 (most likely neutral) to 65 (most likely deleterious) (http://agvgd.hci.utah.edu/agvgd_input.php).
Conservation Analysis
The conservation of amino acids is critical for understanding protein evolution and function. The ConSurf server is a computational tool that uses multiple sequence alignment to assess amino acid conservation in a protein based on phylogenetic relationships between homologous sequences. It has a scoring scale of “1 to 9”, with 1 indicating little or no conservation, 5 indicating moderate conservation, and 9 indicating high conservation. Furthermore, buried amino acids with a high conservation value are considered structural residues, whereas exposed amino acids with a high conservation score are considered functional residues (https://consurf.tau.ac.il) [28, 29].
Mutant Protein Modelling and Quality Assessment
Using the mutagenesis plugin embedded in PyMOL (www.pymol.org), Y2080D and C2770G mutant models were created using the wild-type ATM as a reference model. Subsequently, the SwissPDB viewer was employed to mitigate high-energy configurations, employing the GROMOS 43B1 force field for energy minimization in both mutant and wild-type ATM structures. This involved adjusting their coordinate geometries to release internal constraints and diminish the overall potential energy.
Drug-Likeness Property and ADME Check
The ADMETlab 2.0 server (https://admetmesh.scbdd.com/service/evaluation/cal) was used to evaluate the drug likeness and pharmacokinetic property of two known ATM kinase inhibitors, AZD0156 and AZD1390, as well as a natural chemical compound “quercetin”, which has previously exhibited to have anticancer characteristics.
Molecular Docking Analysis
The AutoDock software was used to perform molecular-docking studies with AZD0156, AZD1390, and quercetin for wild type and mutants [30]. The wild-type ATM and mutants were given all of the necessary polar hydrogen, solvation parameters, and were assigned Kollman United Atom charges. Grid (affinity) maps with 100 (X), 100 (Y), and 100 (Z) grid points, plus a spacing of 0.375, were created for the protein’s active site using the AutoGrid programme. The Lamarckian Genetic Algorithm (LGA) was used to perform the molecular docking, with each experiment containing ten distinct runs [31]. Finally, using the Discovery studio visualizer and Pymol software, the structure of the docked complexes with the highest binding affinity was visualised.
Molecular Dynamic (MD) Simulation
MD simulations were conducted for docked complexes involving wild-type ATM, Y2080D, and C2770G as protein targets, along with the ligands AZD0156, AZD1390, and quercetin. GROMACS 2021 and the PROGRG server were employed to generate ligand and complex topologies. The complexes were solvated with simple point charge (SPC) water molecules, and NA+ and Cl− ions were added for neutralization. The system underwent initial equilibration in the NVT ensemble, addressing particle number, volume, and temperature, followed by equilibration in the NPT ensemble, which considered particle number, pressure, and temperature. Subsequently, 10,000 picoseconds (ps) of MD simulation were conducted for the complexes.
Post MD Analysis
The analysis of MD simulations results involved the utilization of trajectory files, including computations for Root-Mean-Square Deviation (RMSD), Radius of Gyration (Rg), and Solvent-Accessible Surface Area (SASA). Additionally, Principal Component Analysis (PCA) was performed using various built-in scripts in GROMACS. The graphical representation of all trajectory files was generated using the QtGRACE visualization software.
MM-PBSA Assessment
The g_mmpbsa package was employed in conjunction with GROMACS 2021 to assess the molecular mechanics Poisson Boltzmann surface area (MM-PBSA) and analyze the free binding energy of wild-type ATM, Y2080D, and C2770G proteins in complex with ligands (AZD0156, AZD1390, and quercetin). The binding energy was computed based on the final 1000 ps from the 10,000 ps MD simulation production. The estimation of binding affinity considered both bonded and non-bonded interactions in the solvent stage, distinguishing between interactions in the vacuum. To calculate polar and non-polar solvation energy, the Poisson Boltzmann equation and solvent-accessible surface area (SASA) were utilized. The binding free energy (ΔG binding) was determined using the following equation:
Results
Distribution of ATM SNPs
For our analysis, we used a list of 419 ATM kinase SNPs found in public databases which are positioned in different coding regions of the protein.
Analysis of Pathogenicity
The impact of missense SNPs on the amino acids they alter can be used to estimate their pathogenicity. Therefore, this investigation was mainly focused on ATM kinase missense mutations and their pathogenic/deleterious effect. A total of 419 SNPs were analysed by Provean, Pmut, and MetaSNP (PANTHER, PhD-SNP, SIFT, SNAP) which resulted 167, 89, 250, 311, 240, 283, and 269 as deleterious SNPs, respectively, and is represented in the graphical manner (Fig. 4). In addition, a detailed dataset of predicted results is presented in Supplementary Table 1. Overall results achieved from this investigation exhibited “54” SNPs as deleterious/pathogenic which are residing on different domains of ATM from the large pool of mutations. The 54 deleterious SNPs details sheet obtained, replete with score and server predictions, is displayed in Table 1. Later, these 54 deleterious SNPs were further analysed for protein stability check.
Analysis of Protein Stability
The impact of the 54 most deleterious mutations on protein stability was predicted using Mupro, iStable, iMutant 3.0, mCSM, SDM, and CUPSAT. Out of 54 mutations, 5 (V1913C, Y2080D, L2656P, C2770G, C2930G) were found to be destabilising SNPs based on the examination of all 6 stated web-based algorithms; details related to the score and predictions are listed in Table 2 and are graphically depicted in Fig. 5.
Analysis of Oncogenic Nature of ATM Mutants
FATHMM-cancer was used to check the cancer-causing potentials of the V1913C, Y2080D, L2656P, C2770G, and C2930G mutations. The scores for V1913C, Y2080D, L2656P, C2770G, and C2930G derived from this study were − 2.87, − 1.5, − 2.4, − 1.69, and − 2.64, respectively, and were predicted to have cancer-promoting potential (Table 3). Overall, the results of this prediction indicated that these mutations have a role in cancer and subjected for further analysis.
Analysis of Biophysical Characteristics
The V1913C, Y2080D, L2656P, C2770G, and C2930G mutations were subjected to Align GVGD server to assess the biophysical characteristics. The results obtained from the server showed that all of the mutations belong to the class 65 (most likely deleterious) (Table 4).
Prevalence of Mutations in Cancer
We investigated the cancer incidence of the mutations in the COSMIC database based on the FATHMM-cancer and Align GVGD prediction results and discovered that Y2080D was reported in Haematopoietic and lymphoid cancer, and C2770G was reported in squamous cell carcinomas. Therefore, these two mutations were taken for further analysis.
Conservation Analysis
The level of residue conservation gives an approximate notion of the structural and functional impact that deleterious mutations can have on the protein. A damaging mutation at a highly conserved residue are always harmful in nature. The ConSurf server was given the protein sequence and structure as input, and the conservation level of residue was calculated using the Bayesian technique. The conservation indices of the residues can range from 1 to 9. The score of “1” for a residue suggests that it is extremely variable, whereas a score of “9” for a residue suggests that it is highly conserved. The results of this analysis revealed that Y2080D mutations were at a highly conserved location with a score of “8” and C2770G mutations were at a highly conserved position with a score of “9,” as shown in Fig. 6.
Drug-Likeness Property and ADME Check
The results obtained from the ADMETlab 2.0 server exhibited that AZD0156, AZD1390, and quercetin follow the Lipinski rule of five and other pharmacokinetic factors are shown in Fig. 7; therefore, they were used as plausible ligands for molecular-docking analysis.
Molecular Docking Analysis
The molecular-docking studies were accomplished for the wild-type ATM kinases and mutants (Y2080D, C2770G) against the two ATM inhibitors AZD0156, AZD1390, and natural compound quercetin. The docking poses of wild type and both mutants against all three inhibitors used were analysed, and it was revealed that the Y2080D mutant forms two hydrogen bonds with quercetin with binding energy of − 8.8 kcal/mol, whereas the C2770G mutant forms one hydrogen bond with quercetin with binding energy of − 8.8 kcal/mol. Docking poses for all interaction are depicted in Figs. 8A–C, 9A–C, 10A–C; additionally, the binding energy and hydrogen bond interactions for all the docked complexes are shown in Figs. 8D, 9D, and 10D (Table 5). The overall results obtained from molecular-docking analysis suggest that quercetin has better binding affinity for the mutant Y2080C when compared to AZD0156 and AZD1390. In addition, quercetin interacts better with C2770G mutant when compared to AZD1390. Therefore, quercetin can be used as potent inhibitors against the Y2080C and C2770G mutants as it is a natural compound with less side effects.
Molecular docking analysis of ATM wild-type protein represented in 3D and 2D form; A AZD0156 interacts with ATM wild type; B AZD1390 interacts with ATM wild type; C quercetin interacts with ATM wild type; D Graph represents the binding energy and number of hydrogen bonds formed between ATM wild type and AZD1390, AZD0156, and quercetin
Molecular docking analysis of ATM-mutant (Y2080D) protein represented in 3D and 2D form; A AZD0156 interacts with ATM mutant; B AZD1390 interacts with mutant; C quercetin interacts with ATM mutant; D Graph represents the binding energy and number of hydrogen bonds formed between ATM mutant and AZD1390, AZD0156, and quercetin
Molecular docking analysis of ATM mutant (C2770G) protein represented in 3D and 2D form; A AZD0156 interacts with ATM mutant; B AZD1390 interacts with mutant; C quercetin interacts with ATM mutant; D Graph represents the binding energy and number of hydrogen bonds formed between ATM mutant and AZD1390, AZD0156, and quercetin
Molecular Dynamic Simulations
MD simulation, a frequently utilized technique in computer-aided drug design, is employed to evaluate the kinetic and thermodynamic characteristics of biological systems under-defined physiological conditions. Consequently, MD simulations were conducted for all the docked complexes, and the resulting data were analysed for RMSD, Rg, SASA, and PCA.
Root-Mean-Square Deviations (RMSD)
To assess protein stability, RMSD was calculated over a 10,000 ps simulation for all docked complexes (Fig. 11A–C). Higher RMSD values suggest less stability, while lower values indicate greater stability in the complex. Therefore, analysis revealed that the Y2080D-quercetin complexes exhibited the lowest RMSD, ranging from ~ 0.1 to ~ 0.45 nm in the C-alpha backbone. Conversely, the Y2080D-AZD1390 and Y2080D-AZD0156 complexes showed RMSD values of ~ 0.1 to ~ 0.65 nm and ~ 0.1 to ~ 0.55 nm, respectively (Fig. 11B). The C2770G-AZD0156 complex demonstrated an RMSD of ~ 0.1 to ~ 0.15, indicating higher stability compared to C2770G-AZD1390 and C2770G-quercetin with RMSD values of ~ 0.1 to ~ 0.4 and ~ 0.1 to ~ 0.25, respectively (Fig. 11C).
Radius of Gyration (Rg)
In a stably folded protein, Rg values remain constant, signifying structural stability. Conversely, as the protein undergoes unfolding, Rg values exhibit temporal fluctuations. A lower Rg value suggests a more compact protein structure. Therefore, Rg analysis was conducted for all the complexes, as illustrated in Fig. 12A–C. The Rg values of the Y2080D-quercetin complexes were ~ 2.0 nm, indicating structural compactness. In contrast, the Y2080DAZD1390 and Y2080D-AZD0156 complexes displayed higher Rg values of ~ 2.2 nm and ~ 2.15 nm, suggesting a loss of compactness during unfolding (Fig. 12B). Similarly, the C2770G-AZD0156 complex exhibited an Rg value of ~ 2.05 nm, while C2770G-AZD1390 and C2770G-quercetin showed higher Rg values of ~ 2.07 nm and ~ 2.15 nm, respectively, indicating a loss of compactness in these complexes as well (Fig. 12C).
Solvent-Accessible Surface Area (SASA)
SASA evaluates solvent behaviour accessibility, distinguishing between hydrophilic and hydrophobic regions within protein molecules, while also investigating how ligand-binding sites contribute to the solvent effect on the protein molecule. The SASA values for the complexes are depicted in Fig. 13A–C. For the Y2080D-quercetin complexes, SASA values were approximately ~ 127 nm2, indicating the accessibility of solvent behaviour. In contrast, the Y2080D-AZD1390 and Y2080D-AZD0156 complexes displayed higher SASA values of ~ 139 nm2 and ~ 132 nm2, respectively (as shown in Fig. 13B), suggesting increased exposure of solvent-accessible areas. Similarly, the C2770G-AZD0156 complex demonstrated an SASA value of ~ 165 nm2, while C2770G-AZD1390 and C2770G-quercetin exhibited higher SASA values of ~ 176 nm2 and ~ 180 nm2, respectively (Fig. 13C), indicating a greater accessibility of solvent behaviour in these complexes.
Principal Component Analysis (PCA)
The collective motions of atoms were assessed by considering the principal components PC1 and PC2. PC1 captures the primary direction of variations, while PC2 records the second most significant variations. The eigenvector calculates the Cα motion within the ligand complex. In Fig. 14A–C, we illustrate the 2D projection of the trajectory, with the X-axis representing the projection of eigenvector 1 and the Y-axis representing the projection of eigenvector 2 for all ATM, Y2080D, and C2770G proteins with their respective ligand complexes. The shared space occupancy by the protein and ligand complexes in the plot indicates a higher likelihood of stability. Conversely, when they occupy ample space, it suggests lesser stability during their dynamic interaction. The Y2080D-quercetin complexes demonstrate a more confined conformational space, indicating a comparatively more stable conformation when contrasted with the Y2080D-AZD1390 and Y2080D-AZD0156 complexes (Fig. 14B). Conversely, in PCA analysis, the C2770G-AZD0156 complex shows a limited conformational space coverage compared to C2770G-AZD1390 and C2770G-quercetin, suggesting a less stable state for these complexes (Fig. 14C).
MM-PBSA Assessment for Binding Stability
In addition, we performed binding free energy calculations utilizing the MM-PBSA technique for all complexes to augment the information on molecular interaction energy. Table 6 provides a breakdown of the energy components contributing to the binding free energy in each complex. The end-state-free approach serves to illustrate the strength of the interaction between the bound molecule and receptor, a crucial aspect in drug development assessment. Notably, the Y2080D-quercetin complex exhibited the lowest binding free energy at − 128.913 kJ/mol, in contrast to Y2080D-AZD1390 and Y2080D-AZD0156. Similarly, the C2770G-AZD0156 complex demonstrated a low binding free energy of − 139.904 kJ/mol compared to C2770G-AZD1390 and C2770G-quercetin (Table 6).
Discussion
The ATM kinase is the major transducer of DSB-induced signalling and is a member of the Phosphatidylinositol 3 kinase-related kinases (PIKK) family [32]. It phosphorylates p53, Chk2, BRCA1, RPAp34, H2AX, SMC1, FANCD2, Rad17, Artemis, and Nbs1, among other proteins involved in cell-cycle checkpoint control, apoptotic response, and DNA repair. All of these substrates must be phosphorylated for cell-cycle arrest at the G1/S, intra-S, and G2/M checkpoints, as well as for DNA repair [6, 13, 33]. ATM kinase has potential to auto-phosphorylate itself at specific sites of its own, which is significant for monomerization and association of chromatin. Mutations in ATM gene are responsible for various diseases which includes rare autosomal recessive disorder ataxia-telangiectasis that is considered by cerebellar degeneration, immunodeficiency, and can be the leading cause of an increased risk of cancer [32, 34]. Recent advances in computing resources and their use in biomedical sciences have enabled researchers to look into the impact of mutations on protein stability and function [35, 36]. Therefore, in this study, we retrieved SNPs details from various mutations databases such as HuVarBase and dbSNP. To assess the pathogenic potential of 419 mutation of ATM kinase, 13 different algorithms were used. As noted in the results section, the MetaSNP, Provean, and Pmut results highlighted “54” mutations as the most deleterious mutations. These 54 mutations were then run via several servers including CUPSAT, Mupro, iStable, mCSM, SDM2, and I-mutant, revealing 5 mutations (V1913C, Y2080D, L2656P, C2770G, C2930G) that have the potential to induce protein destability. The cancer-promoting potentials of the V1913C, Y2080D, L2656P, C2770G, and C2930G mutations were further investigated, and the results revealed that all of the variants have cancer-promoting potential, as shown in Table 3. After using the AVGD server to examine the biophysical parameters, it was discovered that all of the V1913C, Y2080D, L2656P, C2770G, and C2930G belonged to the class65. Additionally, the prevalence of V1913C, Y2080D, L2656P, C2770G, and C2930G mutations in cancer was assessed with the help of COSMIC database which resulted; Y2080D (sample name: 325-01-1TD) has been found in patient with Haematopoietic & lymphoid cancer whereas C2770G (sample name: P-0005509-T01-IM5) was found in Skin cancer; therefore, based on these results, we took these two mutations for further analysis. The wild-type ATM kinase protein structure was used as reference model to create Y2080D and C2770G mutations using mutagenesis plugin embedded in PyMOL, and energy minimization was performed with the help SwissPDB viewer tool. Later AZD1390 and AZD0156 two inhibitors of ATM kinase along with the natural compound quercetin were taken as inhibitors. The most important reason behind considering quercetin as inhibitor in this study was its safety and is widely used as dietary supplement additionally its numerous biological activities. Several researches on quercetin have shown to have a potential role in medical application. Further, the molecular-docking analysis of AZD1390, AZD0156, and quercetin was performed against ATM wild type, ATM mutant (Y2080D), and ATM mutant (C2770G) with the help of AutoDock Vina tool. Thus, the results obtained from molecular-docking analysis showed that quercetin has better binding affinity with ATM wild type, and ATM mutant (Y2080D) than AZD1390 and AZD0156, whereas AZD0156 showed good binding affinity with ATM mutant (C2770G). In addition, we employed MD simulations, a widely employed technique in computer-aided drug design, to probe the kinetic and thermodynamic characteristics of biological systems under specific physiological conditions. Comprehensive analyses, including RMSD, Rg, SASA, and PCA, were conducted for all docked complexes. The RMSD analysis revealed that the Y2080D-quercetin complexes exhibited the lowest deviations, suggesting a more stable conformation compared to Y2080D-AZD1390 and Y2080D-AZD0156. Conversely, the C2770G-AZD0156 complex demonstrated consistently low RMSD values, indicating enhanced stability compared to C2770G-AZD1390 and C2770G-quercetin. In terms of Rg, the Y2080D-quercetin complexes displayed lower values, indicative of structural compactness, while the Y2080D-AZD1390 and Y2080D-AZD0156 complexes exhibited higher Rg values, implying a loss of compactness during unfolding. Similarly, the C2770G-AZD0156 complex demonstrated an Rg value suggesting structural compactness, whereas C2770G-AZD1390 and C2770G-quercetin showed higher Rg values, indicating potential instability. SASA analysis further highlighted variations in solvent behaviour accessibility among complexes. For the Y2080D-quercetin complexes, SASA values indicated solvent behaviour accessibility, while the Y2080D-AZD1390 and Y2080D-AZD0156 complexes displayed higher SASA values, suggesting increased exposure of solvent-accessible areas. Similarly, the C2770G-AZD0156 complex exhibited an SASA value indicative of solvent behaviour accessibility, while C2770G-AZD1390 and C2770G-quercetin exhibited higher SASA values, indicating greater accessibility of solvent behaviour. PCA illustrated collective motions of atoms, indicating shared space occupancy for stable complexes. Conversely, limited conformational space coverage in the C2770G-AZD0156 complex suggested decreased stability. Finally, the MM-PBSA assessment for binding stability highlighted the Y2080D-quercetin complex with the lowest binding free energy, underscoring its potential in drug development. Similarly, the C2770G-AZD0156 complex exhibited a low binding free energy compared to its counterpart. These comprehensive analyses offer valuable insights into the dynamic behaviour and stability of the studied complexes, providing essential information for drug development considerations. Therefore, to bridge the existing research gap, further in vitro and in vivo studies are imperative. These studies would contribute to a deeper understanding of the inhibiting potentials of AZD0156 and quercetin against the specified ATM mutants, providing crucial insights for the development of effective therapeutic interventions.
Conclusion
In this current study, a computational analysis was conducted on nsSNPs within the ATM kinase associated with various diseases. The strength of computational analysis lies in its efficiency, saving both resources and time compared to traditional experimental approaches. Among the 419 identified SNPs in ATM, the Y2080D and C2770G mutants were identified as highly deleterious mutations with potential roles in cancer development. Molecular docking and dynamic simulations unveiled quercetin as a potent inhibitor against the ATM-mutant Y2080D, while AZD0156 exhibited favourable binding affinity with the ATM-mutant C2770G. These computational predictions provide a basis for experimental validation in future studies. The outcomes of this research not only shed light on the deleterious impact of Y2080D and C2770G ATM mutations in cancer formation but also offer insights for the development of targeted therapeutic strategies. This study serves as a guide for further investigations aimed at understanding the molecular implications of these ATM mutations and lays the groundwork for potential therapeutic interventions.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ATR:
-
ATM- and Rad3-related
- ATM:
-
Ataxia telangiectasia mutated
- BRCA1:
-
Breast cancer type 1 susceptibility protein
- c-Abl:
-
Abelson tyrosine-protein kinase 1
- Chk1:
-
Checkpoint kinase 1
- Chk2:
-
Checkpoint kinase 2
- ConSurf:
-
Conservation of amino acid residues in proteins
- DNA:
-
Deoxyribonucleic acid
- DNA-PKc:
-
DNA-dependent protein kinase catalytic subunit
- DSB:
-
Double-stranded break
- FANCD2:
-
Fanconi anemia group D2 protein
- FATHMM-cancer:
-
Functional analysis through hidden Markov models
- IR:
-
Ionizing radiation
- MetaSNP:
-
Metastability-based SNP predictor
- Mdm2:
-
Murine double minute 2
- MDS:
-
Molecular dynamics simulation
- NbsS1:
-
Nijmegen breakage syndrome 1
- p53:
-
Tumour protein 53
- PIKK:
-
Phosphatidylinositol 3-kinase-related kinase
- Provean:
-
Protein variation effect analyzer
- Pmut:
-
Pathogenicity prediction software for missense variants
- Rad51:
-
DNA repair protein RAD51 homolog 1
- SDM2:
-
Site-directed mutator 2
- SNPs:
-
Single-nucleotide polymorphisms
- mCSM:
-
Mutations of computational saturation mutagenesis
- PyMOL:
-
Python molecular graphics system
References
Jackson, S. P., & Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature, 461(7267), 1071–1078. https://doi.org/10.1038/nature08467
Roos, W. P., Thomas, A. D., & Kaina, B. (2016). DNA damage and the balance between survival and death in cancer biology. Nature Reviews Cancer, 16(1), 20–33. https://doi.org/10.1038/nrc.2015.2
O’Connor, M. J. (2015). Targeting the DNA damage response in cancer. Molecular Cell, 60(4), 547–560. https://doi.org/10.1016/j.molcel.2015.10.040
Savitsky, K., Sfez, S., Tagle, D. A., Ziv, Y., Sartiel, A., Collins, F. S., Shiloh, Y., & Rotman, G. (1995). The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Human Molecular Genetics, 4(11), 2025–2032. https://doi.org/10.1093/hmg/4.11.2025
Bakkenist, C. J., & Kastan, M. B. (2003). DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature, 421(6922), 499–506. https://doi.org/10.1038/nature01368
Mochan, T. A., Venere, M., DiTullio, R. A., & Halazonetis, T. D. (2003). 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Research, 63(24), 8586 LP–8591.
Wang, X., Chu, H., Lv, M., Zhang, Z., Qiu, S., Liu, H., Shen, X., Wang, W., & Cai, G. (2016). Structure of the intact ATM/Tel1 kinase. Nature Communications. https://doi.org/10.1038/ncomms11655
Baretić, D., Pollard, H. K., Fisher, D. I., Johnson, C. M., Santhanam, B., Truman, C. M., Kouba, T., Fersht, A. R., Phillips, C., & Williams, R. L. (2017). Structures of closed and open conformations of dimeric human ATM. Science Advances, 3(5), e1700933. https://doi.org/10.1126/sciadv.1700933
Yang, H., Jiang, X., Li, B., Yang, H. J., Miller, M., Yang, A., Dhar, A., & Pavletich, N. P. (2017). Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature, 552(7685), 368–373. https://doi.org/10.1038/nature25023
Yang, H., Rudge, D. G., Koos, J. D., Vaidialingam, B., Yang, H. J., & Pavletich, N. P. (2013). MTOR kinase structure, mechanism and regulation. Nature, 497(7448), 217–223. https://doi.org/10.1038/nature12122
Putti, S., Giovinazzo, A., Merolle, M., Falchetti, M. L., & Pellegrini, M. (2021, November 1). ATM kinase dead: From ataxia telangiectasia syndrome to cancer. Cancers. https://doi.org/10.3390/cancers13215498
Smith, J., Tho, L. M., Xu, N., & Gillespie, D. A. (2010). The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Advances in Cancer Research, 108, 73–112. https://doi.org/10.1016/B978-0-12-380888-2.00003-0
Menolfi, D., & Zha, S. (2020). ATM, ATR and DNA-PKcs kinases—The lessons from the mouse models: Inhibition ≠ deletion. Cell & Bioscience, 10(1), 8. https://doi.org/10.1186/s13578-020-0376-x
Dash, R., & Munni, Y. A. (2020). Computational SNP analysis and molecular simulation revealed the most computational SNP analysis and molecular simulation revealed the most deleterious missense variants in the NBD1 domain of human ABCA1 transporter. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms21207606
Panchal, N. K., Bhale, A., Verma, V. K., & Beevi, S. S. (2020). Computational and molecular dynamics simulation approach to analyze the impact of XPD gene mutation on protein stability and function. Molecular Simulation, 46(15), 1200–1219. https://doi.org/10.1080/08927022.2020.1810852
Panchal, N. K., Mohanty, S., & Prince, S. E. (2023). Computational insights into NIMA-related kinase 6: Unraveling mutational effects on structure and function. Molecular and Cellular Biochemistry. https://doi.org/10.1007/s11010-023-04910-0
Kumar, A., Rajendran, V., Sethumadhavan, R., & Purohit, R. (2012). In silico prediction of a disease-associated STIL mutant and its affect on the recruitment of centromere protein J (CENPJ). FEBS Open Bio, 2, 285–293. https://doi.org/10.1016/j.fob.2012.09.003
Capriotti, E., Altman, R. B., & Bromberg, Y. (2013). Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics, 14(Suppl 3), S2. https://doi.org/10.1186/1471-2164-14-s3-s2
Choi, Y., & Chan, A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics, 31(16), 2745–2747. https://doi.org/10.1093/bioinformatics/btv195
López-Ferrando, V., Gazzo, A., De La Cruz, X., Orozco, M., & Gelpí, J. L. (2017). PMut: A web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Research, 45(W1), W222–W228. https://doi.org/10.1093/nar/gkx313
Pires, D. E. V., Ascher, D. B., & Blundell, T. L. (2014). MCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics, 30(3), 335–342. https://doi.org/10.1093/bioinformatics/btt691
Worth, C. L., Preissner, R., & Blundell, T. L. (2011). SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Research, 39(SUPPL. 2), 215–222. https://doi.org/10.1093/nar/gkr363
Chen, C. W., Lin, M. H., Liao, C. C., Chang, H. P., & Chu, Y. W. (2020). iStable 2.0: Predicting protein thermal stability changes by integrating various characteristic modules. Computational and Structural Biotechnology Journal, 18, 622–630. https://doi.org/10.1016/j.csbj.2020.02.021
Parthiban, V., Gromiha, M. M., & Schomburg, D. (2006). CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Research, 34(WEB. SERV. ISS.), 239–242. https://doi.org/10.1093/nar/gkl190
Capriotti, E., Fariselli, P., & Casadio, R. (2005). I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Research, 33(SUPPL. 2), 306–310. https://doi.org/10.1093/nar/gki375
Cheng, J., Randall, A., & Baldi, P. (2006). Prediction of protein stability changes for single-site mutations using support vector machines. Proteins, 62(4), 1125–1132. https://doi.org/10.1002/prot.20810
Rogers, M. F., Shihab, H. A., Mort, M., Cooper, D. N., Gaunt, T. R., & Campbell, C. (2018). FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics, 34(3), 511–513. https://doi.org/10.1093/bioinformatics/btx536
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., & Ben-Tal, N. (2016). ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research, 44(W1), W344–W350. https://doi.org/10.1093/nar/gkw408
Celniker, G., Nimrod, G., Ashkenazy, H., Glaser, F., Martz, E., Mayrose, I., Pupko, T., & Ben-Tal, N. (2013). ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Israel Journal of Chemistry, 53(3–4), 199–206. https://doi.org/10.1002/ijch.201200096
Hemalatha, K., & Girija, K. (2016). Evaluation of drug candidature of some benzimidazole derivatives as biotin carboxylase inhibitors: Molecular docking and insilico studies. Asian Journal of Research in Pharmaceutical Science, 6(1), 15–20. https://doi.org/10.5958/2231-5659.2016.00002.3
Seeliger, D., & De Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design, 24(5), 417–422. https://doi.org/10.1007/s10822-010-9352-6
Shiloh, Y., & Ziv, Y. (2013). The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nature Reviews Molecular Cell Biology, 14(4), 197–210. https://doi.org/10.1038/nrm3546
Lee, J.-H., & Paull, T. T. (2007). Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene, 26(56), 7741–7748. https://doi.org/10.1038/sj.onc.1210872
Maréchal, A., & Zou, L. (2013). DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspectives in Biology, 5(9), a012716. https://doi.org/10.1101/cshperspect.a012716
Solayman, M., Saleh, M. A., Paul, S., Khalil, M. I., & Gan, S. H. (2017). In silico analysis of nonsynonymous single nucleotide polymorphisms of the human adiponectin receptor 2 (ADIPOR2) gene. Computational Biology and Chemistry, 68, 175–185. https://doi.org/10.1016/j.compbiolchem.2017.03.005
Jia, P., & Zhao, Z. (2017). Impacts of somatic mutations on gene expression: An association perspective. Briefings in Bioinformatics, 18(3), 413–425. https://doi.org/10.1093/bib/bbw037
Acknowledgements
The authors are thankful to the Vellore Institute of Technology, Vellore, India for providing the necessary facilities to carry out this work.
Funding
No funding was received for this work.
Author information
Authors and Affiliations
Contributions
Nagesh Kishan Panchal: idea for the article, literature survey, formal analysis, investigation, manuscript writing. Poorva Samdani: formal analysis, manuscript writing. Tiasa Sengupta: formal analysis, manuscript writing. Sabina Evan Prince: project administration, idea for the article, formal analysis, investigation, manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest for this work.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable: no human/animal subjects were involved in study.
Consent to Publish
Not applicable: no human/animal subjects were involved in study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Panchal, N.K., Samdani, P., Sengupta, T. et al. Computational Analysis of Non-synonymous SNPs in ATM Kinase: Structural Insights, Functional Implications, and Inhibitor Discovery. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01120-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01120-x