Abstract
Human umbilical cord mesenchymal stem cell (hucMSC)-derived exosomes (Exo) have been frequently investigated for disease control. This study was designed to explore the effects of hucMSC-Exo carrying lncRNA family with sequence similarity 99-member B (Exo-lncRNA FAM99B) on hepatocellular carcinoma (HCC) cell behaviour. The expression of lncRNA FAM99B in HCC cells was measured by reverse-transcription quantitative polymerase chain reaction. Protein levels of exosomal markers were quantified using western blotting. Flow cytometry analyses were performed to detect surface markers of hucMSCs and to measure the effects of Exo-lncRNA FAM99B on HCC cell cycle progression and cell apoptosis. Nanoparticle tracking analysis was used to measure the particle size of the exosomes. Additionally, cell viability was evaluated using methyl thiazolyl tetrazolium assays, and Transwell assays were performed to measure cell migration and invasion. Xenograft tumor models were established to explore the role of Exo-lncRNA FAM99B in vivo. Experimental results revealed that lncRNA FAM99B was downregulated in HCC cell lines, and low level of FAM99B is associated with poor survival rates in patients with HCC according to bioinformatics analysis. HucMSCs were identified in a good morphology with positively expressed CD105, CD29, and CD44 as well as negatively expressed CD31, CD14, and HLA-DR. High protein levels of exosomal markers (Alix, CD63 and TSG101) identified the existence of HucMSC-Exo. Importantly, the hucMSCs-Exo could enter HCC cells and exerted a suppressive effect on malignant cell activities. Moreover, overexpression of Exo-lncRNA FAM99B enhanced cell cycle arrest and cell apoptosis while suppressing cell viability, migration, and invasion in HCC. Exo-siRNA-FAM99B exerted the opposite effects on HCC cell process. In vivo experiments verified that Exo-lncRNA FAM99B inhibited tumorigenesis in HCC. In summary, lncRNA FAM99B derived from hucMSC-Exo inhibited malignant cellular phenotypes and tumorigenesis in HCC, which might provide a novel therapeutic strategy for HCC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related mortality worldwide [1]. It has been reported that the main risk factors for HCC are aflatoxin B1 exposure, alcohol consumption and infection of viral hepatitis B and C [2]. The main therapeutic methods for patients diagnosed with HCC are liver transplantation and surgical resection, but only approximately 20% patients are eligible for resection [3]. In addition, the inapparent clinical symptoms at early stages may lead to delayed diagnosis, resulting in poor outcomes in HCC [4]. Therefore, it is important to explore the complex molecular mechanisms underlying HCC initiation and progression, thus improving early diagnosis and developing efficient therapeutic strategies for patients with HCC.
Long noncoding RNA (lncRNA) is considered as a “dark matter” in the transcription process [5], which has more than 200 nt in length and can alter the expression of cancer-related genes in different ways [6]. Neighbor of breast-cancer susceptibility gene 1 lncRNA 2 [7], lncRNA NIFK antisense RNA 1 [8], and lncRNA tubulin alpha 1b antisense RNA 1 [9] have been recently reported to participate in HCC tumorigenesis and progression by regulating cellular process. LncRNA family with sequence similarity 99-member B (FAM99B) shows a high expression level in hepatic tissues (mean RPKM value: 12) according to RNA-sequencing analysis of tissue-specific genes in major human organs [10]. As previously revealed, lncRNA FAM99B plays a suppressive role in regulating HCC cell proliferation, migration, and invasion [11]. Different from the previous study, the current study focuses on the roles of human umbilical cord mesenchymal stem cell (hucMSC)-derived exosomes (hucMSC-Exo) and exosomal lncRNA FAM99B in HCC progression.
Stem cells are characterized by multi-directional differentiation and self-renewal abilities. As a subtype of stem cells, mesenchymal stem cells (MSCs) are stromal stem cells isolated from many adult tissues and can differentiate into cells of the mesodermal lineage [12]. HucMSCs are isolated from the Wharton’s jelly of the umbilical cord and are easy to collect because they are rich in sources [13,14,15]. HucMSCs have been validated to increase the apoptotic ratio of osteosarcoma cells and ovarian cancer cells [16]. HucMSCs can also suppress malignant biological phenotypes of HCC cells and lung cancer cells via inactivation of the Wnt signaling and induction of cell apoptosis [17]. Moreover, the inhibitory impact of hucMSC-Exo on HCC malignant cellular behaviour has been previously confirmed [18].
Exosomes are a subset of tiny extracellular vesicles secreted by almost all cells, with a density of 1.13–1.19 g/ml and a diameter of 30–150 nm. Exosomes contain a variety of nucleic acids, including miRNAs, mRNAs, and lncRNAs [19, 20]. Since lncRNAs can be transferred by exosomes to play regulatory roles in many human diseases, the personalized treatment might be possibly enabled by incorporating therapeutic lncRNAs into cells producing exosomes [21]. It is well-known that MSCs can produce exosomes among cell types [22]. Moreover, MSC-derived exosomal lncRNA c5orf66-AS1 was reported to hamper HCC malignant cell activities by regulating downstream factors miR-127-3p and DUSP1 [23]. HucMSC-derived exosomal miR-451a inhibits the epithelial-mesenchymal transition in HCC by regulating a downstream target gene ADAM10 [18].
In the current study, the effects of Exo-lncRNA FAM99B on HCC cell beahviour and tumorigenesis of xenografts were investigated. The study might extend the understanding of lncRNA FAM99B in cancer development and attract attentions to the functions of exosome-derived RNA in cancer treatment.
Materials and Methods
Cell Culture
Human HCC cell lines HepG2, MHCC97H, and MHCC97L were purchased from Otwo Biotech Inc. (Shenzhen, China). HCC cell line Hep3B was obtained from Yitabio (Beijing, China) and human normal liver LO2 cells were purchased from Fuyubio (Shanghai, China). These cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified atmosphere of 37℃ with 5% carbon dioxide (CO2).
HucMSC Preparation
The human umbilical cords were collected from full-term cesarean section surgery in Nanjing Tongren Hospital (Nanjing, China). The protocol of this study was approved by the Institutional Review Board of Nanjing Tongren Hospital and was in accordance with the guidelines of the Declaration of Helsinki. All patients had been priorly informed and had consented to the donation. Then, the umbilical cord was rinsed thrice with phosphate buffered saline (PBS; pH = 7.4) supplemented with 3% penicillin/streptomycin. The cord was cut into small fragments (3 cm). After the vein, artery, and membrane were removed, the Wharton’s jelly was peel off, cut into 1 mm3 using a scissor and then put into culture flasks. Afterwards, the samples were cultured in DMEM/F12 (#41420ES76, Yeasen, Shanghai, China) containing 10% FBS at 37℃ with 5% CO2. The culture medium was replaced with fresh medium every three days. After ten days, upon 80% cell confluence, the fibrous cells that grew out from the tissues were detached and passaged using 0.25% trypsin. The well-grown cells from the third generation to the sixth generation were selected for the following experiments.
Identification of HucMSCs
A microscope (Olympus, Tokyo, Japan) was used to observe the adherence and morphology of hucMSCs. Surface markers of hucMSCs were detected using a flow cytometer (Becton, Dickinson and Company, NJ, USA). Briefly, hucMSCs (2 × 105 cells/tube) in the third passage were subpackaged into flow tubes and incubated with antibodies of CD105-fluorescein isothiocyanate (FITC), CD29-FITC, CD44-FITC, HLA-DR-fluorescein isothiocyanate (FITC), CD14-FITC and CD31-FITC in darkness for 30 min at 4℃. IgG-FITC served as an isotype control. All these antibodies were purchased from Abcam (Cambridge, MA, USA).
Identification of Exosomes
Western blotting was used to measure protein levels of exosomal markers. The protein was extracted from hucMSCs or hucMSC-Exo using radioimmunoprecipitation lysis buffer (#HY-K1001, Medchem Express, Monmouth Junction, NJ, USA). Protein concentration was determined using the bicinchoninic acid method. Then, 20 µg protein samples were subjected to SDS-polyacrylamide gel electrophoresis, and then the samples were transferred onto PVDF membranes. After the membranes were blocked in 5% non-fat milk powder for 1 h, primary antibodies (Abcam) at the dilution of 1:1,000 including anti-TSG101 (ab125011), anti-CD63 (ab134045), and anti-Alix (ab275377) were incubated with the membranes at 4℃ overnight. The next day, the membranes were washed five times with PBS + tween (PBST) for five minutes each time and then incubated at 37℃ with goat anti-Rabbit IgG H&L secondary antibody (#ab96899, Abcam) for 1 h. After PBST washing again, enhanced chemiluminescent solution (#P0018A, Beyotime, Beijing, China) was added for visualization of the bands, and the signal intensity was quantified with ImageJ software (National Institutes of Health, USA).
Nanoparticle Tracking Analysis (NTA)
The suspension of exosomes was resuspended and diluted in PBS at the ratio of 1:10 at 4℃. Then, 0.1 ml exosome suspension was plated in a cuvette and then was placed into a particle size analyzer (Microtrac, USA) to measure the particle size of the exosomes.
Exosome Uptake Assay
MHCC97L and MHCC97H cells were plated onto culture plates (24-well, 3 × 104 cells/well), and then hucMSC-Exo pre-stained with 4 µl PKH67 were added into the plates [24, 25]. During co-culture of HCC cells and hucMSC-Exo, cell slides were fixed with 4% paraformaldehyde for 20 min at the timepoint of 0 and 48 h, followed by staining with 4ʹ,6-diamidino-2-phenylindole 2hci (DAPI) for 1 h. The images were captured using a laser confocal microscope (Olympus, Tokyo, Japan).
Co-culture of hucMSC-Exo with MHCC97L and MHCC97H Cells
To evaluate the effects of hucMSC-Exo and Exo-lncRNA FAM99B interference on HCC cell activities, MHCC97L and MHCC97H cells were divided into different groups as follows and then subjected to hucMSC-Exo co-culture for 48 h. For Blank group, no treatment was done to HCC cells. For Exo-vector group, Exo-FAM99B group, Exo-siRNA-negative control (Exo-si-NC) group, and Exo-si-FAM99B group, the hucMSC-Exo (200 µg) were first transfected with empty pcDNA vectors, pcDNA-FAM99B vectors, si-NC and si-FAM99B for 48 h, respectively; then hucMSC-Exo with indicated transfection were co-cultured with MHCC97L and MHCC97H cells.
The above-mentioned plasmids were all synthesized by Genepharma (Shanghai, China). The pcDNA-FAM99B vectors contain the full sequence of FAM99B and were used to overexpress FAM99B, with empty pcDNA vectors as the control. The si-FAM99B refers to small interfering RNA targeting FAM99B that can effectively knock down FAM99B expression.
Annexin V-FITC/Propidium Iodide (PI) Double Staining
After reaching 80% cell confluence, HCC cells were detached using trypsin and then plated to culture plates (6-well, 4 × 104 cells/well) for 72 h. After that, cells were centrifuged at 200 ×g for 5 min and then washed twice with pre-cooled PBS. Next, cells were suspended in binding buffer (5,000 µl; 1% bovine serum albumin), and then Annexin V-FITC (5 µl) and PI (5 µl) were added for 10 min of cell incubation in the dark. Cell apoptosis was measured by a flow cytometer (Becton, Dickinson and Company, USA).
Analysis of Cell Cycle Progression
The harvested cells were digested and fixed with 75% ethanol at 4℃ for 4 h, and then the supernatant was disposed. Next, the cells were incubated with RNase and PI for 10 min before rinsed thrice with PBS, and cell cycle progression was analyzed using a flow cytometry (Becton, Dickinson and Company, USA).
Methyl Thiazolyl Tetrazolium (MTT) Assay
HCC cells were detached by trypsin (0.25%) upon reaching 80% cell confluence. After HCC cells were seeded to culture plates (24-well, 5 × 103 cells/well) and incubated for 24 h, 50 ml of MTT solution (1 mg/ml; M8180-1, Solarbio, Beijing, China) diluted in PBS was added for another 3 h of cell incubation. After supplementation of dimethyl sulphoxide (150 ml), the formed formazan was solubilized, and the absorbance was measured at 570 nm using a microplate spectrophotometer (Thermo Fisher Scientific, USA).
Transwell Assay
Transwell chambers (Millipore, MA, USA) pre-coated with (for cell invasion assay) or without (for cell migration assay) matrigel were put into 24-well plates. MHCC97L and MHCC97H cell suspension (3 × 105 cells/ml) was appended into the upper chamber at 200 µl/well, and the bottom chamber was added with culture medium supplemented with 10% FBS at 600 µl/well. After 48 h of incubation at 37℃ with 5% CO2, 4% paraformaldehyde was used to fix the chambers, and the migrated or invaded cells were stained with crystal violet. The images of cells were captured and the number of migrated or invaded cells was quantified using ImageJ software (National Institutes of Health, USA).
Establishment of Xenograft Tumor Models
All animal experiments were approved by the Animal Ethics Committee of Southeast University (20,210,701,004) and were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MA, USA). Animals were kept in standard cages (3 mice/cage), under controlled light (a 12 h/12 h light/dark cycle) in a constant temperature (24℃ ± 1℃) and humidity (40% ± 5%). All mice were housed with ad libitum access to food and water.
Eighteen BALB/c nude mice (male, 6-week-old, 20–23 g) that purchased from Vital River Laboratory Animal Technology (Beijing, China) were divided into three groups (n = 6/group): Blank group, Exo + vector group, and Exo + FAM99B group. MHCC97L cells in the last two groups were treated with indicated plasmids and then co-cultured with hucMSC-Exo as mentioned above. Then, indicated MHCC97L cell suspension was mixed with Matrigel and adjusted to 1 × 104 cells/200 µl. After that, the mixed solution was subcutaneously injected to the ventral side of the nude mice. Tumor volume was measured every week according to the formula: volume = length×width2/2. Four weeks later, all mice were euthanized after injected with an overdose of pentobarbital sodium (200 mg/kg). After euthanasia, xenograft tumor tissues were collected for the following Ki67 immunofluorescence staining.
RT-qPCR Analysis
TRIzol reagent (Invitrogen) was used for the extraction of total RNA from HCC cells. The cDNA was synthesized by reverse transcription of 200 ng extracted RNA using ReverTra Ace qPCR RT Kit (Toyobo, Japan). GAPDH was used as the internal control of lncRNA FAM99B. PCR was performed using SYBR® qPCR Mix (Toyobo, Japan) on a LightCycler 480 Real-Time PCR system (Roche, Shanghai, China). Relative expression of lncRNA FAM99B was calculated using the 2−ΔΔCt method. Primer sequences are shown as follows: lncRNA FAM99B (Forward: 5′-ACAGTGACCGCCGAGACA-3′, Reverse: 5′- AGATTTGGGATTTAGGGAAGG-3′); GAPDH (Forward: 5′-TCAAGGCTGAGAACGGGAAG-3′, Reverse: 5′-TGGACTCCACGACGTACTCA-3′).
Immunofluorescence Staining for the Proliferation Marker Ki67
The collected xenograft tumor tissues were fixed with 4% paraformaldehyde at 4℃ overnight. Then, the samples were embedded in paraffin and cut into 4 μm sections. After the sections were dewaxed, antigen retrieval was achieved using citric acid in high temperature (98℃) for 5 min. Next, the sections were blocked using 5% normal goat serum for 1 h at room temperature followed by incubation with Ki67 antibody (#ab92742, 1:1000) at 4℃ overnight. The samples were subsequently washed in PBS and then treated with anti-Rabbit IgG (#ab150081, 1:200). The nuclei were stained with DAPI. A microscope (Leica, Germany) was used to capture fluorescent images. The percentage of positive Ki67 cells was determined by the ratio of positive cells and total cells.
Statistical Analysis
GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA) was used for statistical analysis. All data are shown as the mean ± standard deviation. The comparison between two groups was evaluated using Student’s t-test, and multiple group comparison was performed using one-way analysis of variance followed by Bonferroni test. A p value less than 0.05 was deemed as the threshold for statistical significance.
Results
Low Expression of lncRNA FAM99B is Associated with Poor Outcomes in Patients with HCC
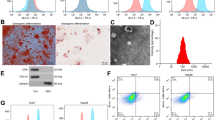
According to GEPIA database, lncRNA FAM99B is significantly downregulated in liver hepatocellular carcinoma (LIHC) tissues compared to that in healthy tissues (Fig. 1A). Bioinformatics tools, GEPIA and Kaplan-Meier, were used to analyze the correlation between lncRNA FAM99B expression and the survival of patients diagnosed with HCC. Bioinformatics results illustrated that low expression of lncRNA FAM99B is correlated to a poor overall survival rate in patients (Fig. 1B C) and is also associated with a low relapse-free survival probability in patients with HCC (Fig. 1D). Then, we examined the expression of lncRNA FAM99B in HCC cell lines (HepG2, MHCC97H, MHCC97L, and Hep3B) and human normal liver LO2 cells. RT-qPCR analysis revealed that lncRNA FAM99B expression was markedly decreased in HCC cells, especially in MHCC97H and MHCC97L cell lines (Fig. 1E). These results suggested that lncRNA FAM99B is downregulated in HCC cells and is correlated with poor survivals in patients with HCC.
Low expression of lncRNA FAM99B is associated with poor outcomes in patients with HCC. (A) LncRNA FAM99B expression in LIHC tissues and corresponding normal samples was analyzed using the bioinformatics tool GEPIA (http://gepia.cancer-pku.cn/). *p<0.05. (B-C) The correlation analysis of lncRNA FAM99B expression and the overall survival of patients diagnosed with HCC was performed using GEPIA and Kaplan-Meier plotter (http://kmplot.com/analysis/). (D) Kaplan-Meier analysis was also performed to explore the relationship between lncRNA FAM99B expression and the relapse-free survival possibility of patients with HCC. (E) RT-qPCR analysis was conducted to measure lncRNA FAM99B expression in HCC cell lines and normal human hepatocyte LO2 cells. *p<0.05 versus LO2 group
Identification of hucMSCs and Exosomes
To identify the extracted hucMSCs, 10% Exo-free DMEM/F12 supplemented with FBS was used for hucMSC incubation, and then cells were observed using a microscope. After adherence, the cells were all in good morphology. Cells in even long fusiform showed the shape of fibroblast and grew in whirl manner with fewer impurity, clear nuclei and abundant cytoplasm. Additionally, cells in other morphology were not found (Fig. 2A). Surface markers of hucMSCs were detected using a flow cytometer, and the results manifested that CD105, CD29, and CD44 were positively expressed while CD31, CD14, and HLA-DR were negatively expressed (Fig. 2B). Protein levels of exosomal markers in hucMSC-Exo and hucMSC were quantified using western blot analysis, and the positively expressed Alix, CD63 and TSG101 were discovered (Fig. 2C). Moreover, according to nanoparticle tracking analysis, the diameter of exosomes was ranged from 80 to 150 nm and was mainly concentrated at 94 nm (Fig. 2D). Moreover, Exo treatment upregulated FAM99B expression in HCC cells compared to that in the Blank group, and Exo-pcDNA-FAM99B vectors further amplified FAM99B level while Exo-si-FAM99B successfully reduced FAM99B expression compared to that in control groups (Fig. 2E).
Identification of hucMSCs and exosomes. (A) The morphology of hucMSCs was observed using a microscope. (B) Flow cytometry was used to detect the surface markers of hucMSCs. (C) The protein levels of exosomal markers in hucMSC-Exo were quantified by western blotting. (D) Nanoparticle tracking analysis was performed to measure the particle size of hucMSC-Exo. (E) RT-qPCR analysis of FAM99B expression in MHCC97L and MHCC97H cells of Blank group, Exo-control groups (Exo, Exo-vector, Exo-si-NC), Exo-FAM99B group, and Exo-si-FAM99B group. **p<0.01 versus Blank group, ##p < 0.01 versus Exo-vector group, &&p < 0.01 versus Exo-si-NC group
Exosome-derived lncRNA FAM99B Induces Cell Cycle Arrest and Cell Apoptosis
To validate the entry of hucMSC-Exo into MHCC97L and MHCC97H cells, 4 µl PKH67 was used to pre-stain hucMSC-Exo, and then hucMSC-Exo were co-cultured with MHCC97L and MHCC97H cells for 24 h. According to images captured by a fluorescence microscope, the entering of exosomes into MHCC97L and MHCC97H cells was not found at 0 h compared to cell morphology in the control group (HCC cells without Exo treatment). At 24 h, exosomes entered HCC cells and gathered along the nuclear membranes as evidenced by green fluorescence around the nuclei (Fig. 3A). MTT assays were performed to evaluate the effects of Exo treatment and Exo-lncRNA FAM99B on the viability of MHCC97L and MHCC97H cells. Compared with the Blank group, HCC cell viability was reduced by Exo-vector treatment and was further decreased in Exo-lncRNA FAM99B group (Fig. 3B C). Additionally, there was a significant increase in cell proportion at G0/G1 phases and a decrease in cell percentage at S and G2/M phases in Exo groups, especially in Exo-lncRNA FAM99B group (Fig. 3D and E). The finding suggested that Exo-lncRNA FAM99B induced HCC cell cycle arrest at the G0/G1 stage. Moreover, Exo-vector and Exo-lncRNA FAM99B induced significant increases in HCC cell apoptosis rates (Fig. 3F and G). Overall, exosome-derived lncRNA FAM99B contributes to cell cycle arrest and cell apoptosis in HCC.
Exo-lncRNA FAM99B promotes cell cycle arrest and cell apoptosis. (A) The uptake of hucMSC-Exo in MHCC97L and MHCC97H cells was imaged at 0 and 24 h, and the control group was defined as HCC cells without Exo treatment. (B-C) MTT assays were performed to measure the viability of MHCC97L and MHCC97H cells in blank group, Exo-vector group, and Exo-lncRNA FAM99B group. (D-E) The effects of Exo-vector and Exo-lncRNA FAM99B on HCC cell cycle progression was evaluated using flow cytometry. (F-G) HCC cell apoptosis in Blank, Exo-vector and Exo-lncRNA FAM99B groups was measured by flow cytometry analysis. *p<0.05 versus Blank group; #p<0.05 versus Exo-vector group
Exo-lncRNA FAM99B Inhibits HCC Cell Migration and Invasion
HCC cell migration and invasion were measured by Transwell assays. Our results manifested that Exo-vector significantly reduced the number of migrated cells, and the cell number was further decreased by overexpression of Exo-FAM99B (Fig. 4A C). Moreover, cell invasive ability was suppressed in Exo groups, and the suppressive effect was more prominent in Exo-lncRNA FAM99B group, as evidenced by the least number of invaded cells after Exo-lncRNA FAM99B treatment (Fig. 4D F). These findings suggested that the migratory and invasive capacities of HCC cells are suppressed by Exo-lncRNA FAM99B.
Exo-lncRNA FAM99B restrains cell migratory and invasive abilities in HCC. (A-C) HCC cell migration in Blank, Exo-vector and Exo-lncRNA FAM99B groups was measured by Transwell assays. (D-F) The number of invaded MHCC97L or MHCC97H cells in response to Exo treatment and Exo-lncRNA FAM99B overexpression was measured by Transwell assays. *p<0.05 versus Blank group; #p<0.05 versus Exo-vector group
Exosome Carrying si-FAM99B Inhibits HCC Cell Cycle Arrest and Apoptosis
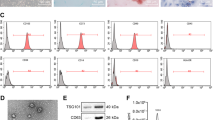
Subsequently, the influences of Exo-lncRNA FAM99B depletion on cell cycle progression and apoptosis were explored. As shown by Fig. 5A-B, cell viability was reduced in HCC cells co-cultured with hucMSC-Exo-si-NC, and the alteration was reversed by Exo-lncRNA FAM99B deficiency. In addition, HCC cell cycle arrest and cell apoptosis enhanced by Exo treatment was partially countervailed by Exo-si-FAM99B (Fig. 5C-F). These findings further confirmed the promoting effect of Exo-lncRNA FAM99B on cell cycle arrest and cell apoptosis.
Exosome carrying si-FAM99B inhibits HCC cell cycle arrest and apoptosis. (A-B) MTT assays were conducted to measure HCC cell viability in Blank group, Exo-si-NC group, and Exo-si-FAM99B group. (C-D) Flow cytometry analysis was performed to detect HCC cell cycle progression in response to Exo treatment and Exo-lncRNA FAM99B knockdown. (E-F) HCC cell apoptosis in three groups was measured using flow cytometry analysis. *p<0.05 versus Blank group; #p<0.05 versus Exo-si-NC group
Exosome lncRNA FAM99B Suppresses the Tumorigenesis of HCC
Finally, the role of Exo-lncRNA FAM99B in vivo was investigated. After indicated treatments, MHCC97L cell suspension were injected into nude mice for exploration of tumorigenesis. Tumor volume was measured weekly. It was found that Exo treatment decreased tumor growth rate and tumor weight, and overexpression of Exo-lncRNA FAM99B enhanced the anti-tumor influence (Fig. 6A and B). The xenograft tumor tissues were subjected to Ki67 immunofluorescence staining, and the results showed that the percentage of Ki67-positive cells was reduced by Exo treatment, and the change was amplified by Exo-lncRNA FAM99B overexpression (Fig. 6C and D). The schematic of the in vitro cell treatment and animal experiments is shown in Fig. 6E. In summary, the animal experiments confirmed the anti-tumor role of Exo-lncRNA FAM99B in HCC.
Exosome lncRNA FAM99B suppresses the tumorigenesis of HCC in vivo. (A) The volume of xenografts was measured and recorded weekly. (B) The weight of xenografts was measured on Week 4. (C-D) Immunofluorescence staining was performed to measure the number of Ki67-positive cells in xenograft tumor tissues. (E) A schematic of in vitro cell treatment and animal experiments. n = 6/group. *p<0.05 versus Blank group; #p<0.05 versus Exo-vector group
Discussion
Substantial evidence shows that the aberrant expression of lncRNA is implicated with the tumorigenesis and progression of human cancers [26]. The present study revealed that lncRNA FAM99B is poorly expressed in HCC cell lines, and its low expression is correlated with poor outcomes in patients diagnosed with HCC. HucMSC-Exo treatment upregulated FAM99B expression and suppressed HCC cellular process. Moreover, overexpression of Exo-lncRNA FAM99B further impaired the viability, migration, and invasion of HCC cells while enhancing cell cycle arrest and cell apoptosis. Knockdown of Exo-lncRNA FAM99B exerted the opposite effects on HCC cell activities.
LncRNA has been demonstrated to participate in tumor recurrence, metastasis, and growth in HCC [27]. For example, lncRNA ewing sarcoma associated transcript 1 (EWSAT1, also known as LINC00277) shows a high level in HCC tissues and contributes to HCC cell metastasis and proliferation by activating the Src-YAP signaling [28]. LINC00071 shows a high level in HCC cells, and its silencing represses HCC cell migration and invasion by targeting the miR-129-5p/ETV1 axis [29]. In this study, we examined lncRNA FAM99B expression in HCC cell lines HepG2, MHCC97H, MHCC97L. A reduction of lncRNA FAM99B expression was observed in HCC cells, which was consistent with its expression analysis in LIHC tissues using bioinformatics tools. From the pathological aspect, the aberrantly expressed lncRNA has the potential to be a prognostic maker [30]. Bioinformatics analysis revealed that downregulation of lncRNA FAM99B predicts poor survival outcomes in patients with HCC, suggesting that lncRNA FAM99B is involved in the regulation of HCC progression.
Recent studies have revealed that functional lncRNAs can be transferred by exosomes [31]. RNA and protein can be easily degraded outside the cells where exosomes can protect biological compounds from degradation [32]. A study validates that exosomes can protect cells from radiation injury via ingestion of survival proteins, thereby promoting cell proliferation and metastatic potential [33]. Previously, lncRNA FAM138B was reported to be found in the exosomes of HCC and show low expression in exosomes and HCC tissues [34]. This study revealed that lncRNA FAM99B can be upregulated by hucMSC-Exo treatment. In addition, FAM99B can be overexpressed in hucMSC-Exo and then transferred into HCC cells. Functional experiments verified that Exo-lncRNA FAM99B hampers the viability, migration, and invasion of HCC cells while promoting cell cycle arrest and cell apoptosis. Currently, exosomes are used as targeted drug delivery methods in clinical trials. The study not only confirms the inhibitory role of FAM99B in regulating HCC cellular process but also highlights the functions of Exo-lncRNA FAM99B in HCC cell progression and tumorigenesis.
Conclusion
Our study reveals that lncRNA FAM99B derived from hucMSC-Exo inhibits HCC cellular process and tumorigenesis. After Exo-lncRNA FAM99B overexpression, cell cycle arrest and apoptosis are enhanced while cell viability, migration, invasion, and tumor growth are inhibited. The present study may provide a new biomarker for investigation of hucMSC-derived exosomes in cancer treatment.
Limitation
Because of the complexity of molecular mechanisms, the upstream transcription factors regulating lncRNA FAM99B or downstream genes regulated by lncRNA FAM99B are not further explored in the present study, which could be directions for future works.
References
Shimada, S., Mogushi, K., Akiyama, Y., Furuyama, T., Watanabe, S., Ogura, T., Ogawa, K., Ono, H., Mitsunori, Y., Ban, D., Kudo, A., Arii, S., Tanabe, M., Wands, J. R., & Tanaka, S. (2019). Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine, 40, 457–470.
Li, Y., Chen, B., Yang, X., Zhang, C., Jiao, Y., Li, P., Liu, Y., Li, Z., Qiao, B., Bond Lau, W., Ma, X. L., & Du, J. (2019). S100a8/a9 signaling causes mitochondrial dysfunction and cardiomyocyte death in response to Ischemic/Reperfusion Injury. Circulation, 140, 751–764.
Liu, Y., Sun, L., Gao, F., Yang, X., Li, Y., Zhang, Q., Zhu, B., Niu, S., Huang, Y., Hu, Y., Feng, Y., Jiang, Y., & Wang, X. (2018). A new scoring model predicting macroscopic vascular invasion of early-intermediate hepatocellular carcinoma. Medicine (Baltimore).97,e13536.
Cheng, S., Chen, M., Cai, J., Sun, J., Guo, R., Bi, X., Lau, W. Y., & Wu, M. (2020). Chinese Expert Consensus on Multidisciplinary diagnosis and treatment of Hepatocellular Carcinoma with Portal Vein Tumor Thrombus (2018 Edition). Liver Cancer, 9, 28–40.
Pan, W., Li, W., Zhao, J., Huang, Z., Zhao, J., Chen, S., Wang, C., Xue, Y., Huang, F., Fang, Q., Wang, J., Brand, D., & Zheng, S. G. (2019). lncRNA-PDPK2P promotes hepatocellular carcinoma progression through the PDK1/AKT/Caspase 3 pathway. Molecular Oncology, 13, 2246–2258.
Marchese, F. P., & Huarte, M. (2017). A long noncoding RNA in DNA replication and chromosome dynamics. Cell Cycle, 16, 151–152.
Sheng, J. Q., Wang, M. R., Fang, D., Liu, L., Huang, W. J., Tian, D. A., He, X. X., & Li, P. Y. (2021). LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomedicine & Pharmacotherapy, 133, 111023.
Chen, Y. T., Xiang, D., Zhao, X. Y., & Chu, X. Y. (2021). Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m(6)a methylation promotes disease progression and sorafenib resistance. Human Cell, 34, 1800–1811.
Xu, K., Xia, P., Gongye, X., Zhang, X., Ma, S., Chen, Z., Zhang, H., Liu, J., Liu, Y., Guo, Y., Yao, Y., Gao, M., Chen, Y., Zhang, Z., & Yuan, Y. (2022). A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol Metab, 63, 101540.
Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., Habuka, M., Tahmasebpoor, S., Danielsson, A., Edlund, K., Asplund, A., Sjöstedt, E., Lundberg, E., Szigyarto, C. A., Skogs, M., Takanen, J. O., Berling, H., Tegel, H., Mulder, J., Nilsson, P., Schwenk, J. M., Lindskog, C., Danielsson, F., Mardinoglu, A., Sivertsson, A., von Feilitzen, K., Forsberg, M., Zwahlen, M., Olsson, I., Navani, S., Huss, M., Nielsen, J., Ponten, F., & Uhlén, M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular And Cellular Proteomics, 13, 397–406.
Mo, M., Liu, S., Ma, X., Tan, C., Wei, L., Sheng, Y., Song, Y., Zeng, X., Huang, D., & Qiu, X. (2019). A liver-specific lncRNA, FAM99B, suppresses hepatocellular carcinoma progression through inhibition of cell proliferation, migration, and invasion. Journal Of Cancer Research And Clinical Oncology, 145, 2027–2038.
Uccelli, A., Moretta, L., & Pistoia, V. (2006). Immunoregulatory function of mesenchymal stem cells. European Journal Of Immunology, 36, 2566–2573.
Zeddou, M., Briquet, A., Relic, B., Josse, C., Malaise, M. G., Gothot, A., Lechanteur, C., & Beguin, Y. (2010). The umbilical cord matrix is a better source of mesenchymal stem cells (MSC) than the umbilical cord blood. Cell Biology International, 34, 693–701.
Raio, L., Cromi, A., Ghezzi, F., Passi, A., Karousou, E., Viola, M., Vigetti, D., De Luca, G., & Bolis, P. (2005). Hyaluronan content of Wharton’s jelly in healthy and down syndrome fetuses. Matrix Biology, 24, 166–174.
Prasanna, S. J., Gopalakrishnan, D., Shankar, S. R., & Vasandan, A. B. (2010). Pro-inflammatory cytokines, IFNgamma and TNFalpha, influence immune properties of human bone marrow and Wharton jelly mesenchymal stem cells differentially. PLoS One, 5, e9016.
Gauthaman, K., Yee, F. C., Cheyyatraivendran, S., Biswas, A., Choolani, M., & Bongso, A. (2012). Human umbilical cord Wharton’s jelly stem cell (hWJSC) extracts inhibit cancer cell growth in vitro. Journal Of Cellular Biochemistry, 113, 2027–2039.
Yuan, Y., Zhou, C., Chen, X., Tao, C., Cheng, H., & Lu, X. (2018). Suppression of tumor cell proliferation and migration by human umbilical cord mesenchymal stem cells: A possible role for apoptosis and wnt signaling. Oncol Lett, 15, 8536–8544.
Xu, Y., Lai, Y., Cao, L., Li, Y., Chen, G., Chen, L., Weng, H., Chen, T., Wang, L., & Ye, Y. (2021). Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. Rna Biology, 18, 1408–1423.
Zhang, Z. G., Buller, B., & Chopp, M. (2019). Exosomes - beyond stem cells for restorative therapy in stroke and neurological injury. Nat Rev Neurol, 15, 193–203.
Xu, R., Rai, A., Chen, M., Suwakulsiri, W., Greening, D. W., & Simpson, R. J. (2018). Extracellular vesicles in cancer - implications for future improvements in cancer care. Nature Reviews. Clinical Oncology, 15, 617–638.
Chen, C., Luo, Y., He, W., Zhao, Y., Kong, Y., Liu, H., Zhong, G., Li, Y., Li, J., Huang, J., Chen, R., & Lin, T. (2020). Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest, 130, 404–421.
Yeo, R. W., Lai, R. C., Zhang, B., Tan, S. S., Yin, Y., Teh, B. J., & Lim, S. K. (2013). Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Advanced Drug Delivery Reviews, 65, 336–341.
Gu, H., Yan, C., Wan, H., Wu, L., Liu, J., Zhu, Z., & Gao, D. (2021). Mesenchymal stem cell-derived exosomes block malignant behaviors of hepatocellular carcinoma stem cells through a lncRNA C5orf66-AS1/microRNA-127-3p/DUSP1/ERK axis. Human Cell, 34, 1812–1829.
Chen, L., Wang, Y., Li, S., Zuo, B., Zhang, X., Wang, F., & Sun, D. (2020). Exosomes derived from GDNF-modified human adipose mesenchymal stem cells ameliorate peritubular capillary loss in tubulointerstitial fibrosis by activating the SIRT1/eNOS signaling pathway. Theranostics, 10, 9425–9442.
Wang, G., Jin, S., Huang, W., Li, Y., Wang, J., Ling, X., Huang, Y., Hu, Y., Li, C., Meng, Y., & Li, X. (2021). LPS-induced macrophage HMGB1-loaded extracellular vesicles trigger hepatocyte pyroptosis by activating the NLRP3 inflammasome. Cell Death Discov, 7, 337.
Yan, C., Wei, S., Han, D., Wu, L., Tan, L., Wang, H., Dong, Y., Hua, J., & Yang, W. (2020). LncRNA HULC shRNA disinhibits mir-377-5p to suppress the growth and invasion of hepatocellular carcinoma in vitro and hepatocarcinogenesis in vivo. Ann Transl Med, 8, 1294.
Hong, H., Sui, C., Qian, T., Xu, X., Zhu, X., Fei, Q., Yang, J., & Xu, M. (2020). Long noncoding RNA LINC00460 conduces to tumor growth and metastasis of hepatocellular carcinoma through miR-342-3p-dependent AGR2 up-regulation. Aging (Albany NY), 12, 10544–10555.
He, X., Chen, J., Zhou, J., Mao, A., Xu, W., Zhu, H., Pan, Q., Zhao, Y., Zhang, N., Wang, L., Wang, M., Liu, Z., Zhu, W., & Wang, L. (2022). LncRNA-EWSAT1 promotes hepatocellular carcinoma metastasis via activation of the Src-YAP signaling axis. Faseb j.36,e22663.
Chen, J., Yuan, D., Hao, Q., Zhu, D., & Chen, Z. (2021). LncRNA PCGEM1 mediates oxaliplatin resistance in hepatocellular carcinoma via miR-129-5p/ETV1 axis in vitro. Adv Clin Exp Med, 30, 831–838.
Tian, J., & Hu, D. (2021). LncRNA SLC16A1-AS1 is upregulated in hepatocellular carcinoma and predicts poor survival. Clin Res Hepatol Gastroenterol, 45, 101490.
Hewson, C., & Morris, K. V. (2016). Form and function of Exosome-Associated Long non-coding RNAs in Cancer. Current Topics In Microbiology And Immunology, 394, 41–56.
Konečná, B., Tóthová, Ľ., & Repiská, G. (2019). Exosomes-Associated DNA-New marker in pregnancy complications? Int J Mol Sci 20.
Han, F., Huang, D., Huang, X., Wang, W., Yang, S., & Chen, S. (2020). Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells. Journal Of Cellular And Molecular Medicine, 24, 7730–7742.
Zhuo, C., Yi, T., Pu, J., Cen, X., Zhou, Y., Feng, S., Wei, C., Chen, P., Wang, W., Bao, C., Wang, J., & Tang, Q. (2020). Exosomal linc-FAM138B from cancer cells alleviates hepatocellular carcinoma progression via regulating miR-765. Aging (Albany NY).12,26236–26247.
Acknowledgements
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, G., Ban, K., Mu, H. et al. Human Umbilical Cord Mesenchymal Stem Cells-derived Exosomal lncRNA FAM99B Represses Hepatocellular Carcinoma Cell Malignancy. Mol Biotechnol 66, 1389–1401 (2024). https://doi.org/10.1007/s12033-023-00795-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00795-y