Abstract
This work aimed to study the effect of NFE2 like bZIP transcription factor 3 (NFE2L3) on clear cell renal cell carcinoma (ccRCC) cells and whether NFE2L3 expression was mediated by DNA methylation. Twenty-one ccRCC patients were collected. The gene methylation and expression data of TCGA-KIRC were accessed from TCGA. Candidate methylation driver genes were identified by “MethylMix” package, and finally, NFE2L3 was selected as the target gene. The methylation of NFE2L3 was assayed by Ms PCR and QMSP. mRNA level of NFE2L3 was analyzed by qRT-PCR. Protein level of NFE2L3 was measured by Western blot. Demethylation was performed with methylation inhibitor 5-Aza-2’-deoxycytidine (5-Aza-CdR). Proliferative, migratory, and invasive abilities of ccRCC cells were assayed via cell colony formation assay, scratch healing assay, and transwell assay, respectively. Analysis of TCGA database presented that DNA hypomethylation occurred in the NFE2L3 promoter region in ccRCC tissues. NFE2L3 was significantly upregulated in ccRCC tissues and cells. Its expression in cells treated with 5-Aza-CdR was proportional to the concentration of methylation inhibitor. In cell function experiments, overexpressing NFE2L3 or demethylation could stimulate proliferation, migration, and invasion abilities of ccRCC and normal cells. 5-Aza-CdR treatment rescued repressive impact of knockdown NFE2L3 on malignant phenotypes of ccRCC and normal cells. DNA hypomethylation could induce high expression of NFE2L3 and facilitate malignant phenotypes of ccRCC cells. These results may generate insights into ccRCC therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In developed countries, kidney cancer is a relatively common cancer [1] with treatment difficulties and poor prognosis. More than 90% of kidney cancer is renal cell carcinoma (RCC) from renal tubular structure, and the rest less than 10% are transitional cell carcinoma originating from cells lining the renal pelvis and ureter [2, 3]. Clear cell RCC (ccRCC) is the most common subtype of RCC, which comprises about 75% of all RCCs. Since ccRCC is not sensitive to traditional chemotherapy, the research about targeted therapies for this cancer is attracting increasing attention from doctors and scholars. In order to further study the targeted therapies of ccRCC, we should have a deep understanding of the internal mechanism affecting its occurrence and development.
NFE2 like bZIP transcription factor 3 (NFE2L3) was first discovered in 1999 and reported to be involved in Wnt signaling pathway and oxidative stress [4]. NFE2L3 plays an imperative part in regulating expression of developmental-related genes [5,6,7] and participates in cell differentiation, inflammation, and carcinogenesis [8,9,10]. The function of NFE2L3 varies in the carcinogenesis process of different cancer types. For example, Chevillard et al. [10] found that deletion of NFE2L3 predisposed mice to lymphoma, indicating that NFE2L3 inhibited lymphomas. Chowdhury et al. [11] confirmed that NFE2L3 facilitated cell proliferation via upregulating UHMK1 in colon cancer. Therefore, NFE2L3 is a colon cancer-promoting factor. However, the effect of NFE2L3 on ccRCC has not been clearly studied, and it is worthy of further discussion.
Hypomethylation of oncogenes and hypermethylation of tumor suppressors have been extensively regarded as important regulatory factors in the occurrence and progression of cancer [12]. DNA methylation means adding a methyl group to 5’ cytosine of a CG dinucleotide, which is called CpG for short. Aberrant CpG island methylation occurs in gene promoter regions at different stages of tumor development, resulting in transcriptional silencing and aberrant levels of varying key genes [13]. In ccRCC, Shen et al. [14] proved that DNA hypomethylation in the 3’UTR region leads to high expression of BHLHE41, thereby enhancing the bioactivity of tumor cells. Kang H W et al. [15] also demonstrated that DNA methylation is associated with progression of ccRCC. It followed that DNA methylation could not only affect the development of ccRCC but also affect the prognosis of patients. NFE2L3 has been recently labeled as a new DNA methylation-driven gene in human ccRCC, exhibiting a hypomethylated state in the ccRCC cancer cohort [16]. However, what kind of biological function NFE2L3 plays in ccRCC and whether influence of NFE2L3 on biological function of ccRCC cells is related to its hypomethylation status are still underexplored.
This study attempts to explore influence of NFE2L3 on malignant progression of ccRCC cells and whether this process is mediated by hypomethylation, thus providing theoretical support for the development of new ccRCC treatment options.
Materials and Methods
Bioinformatics Methods
The methylation data (Illumina human methylation 450, Normal: 160, Tumor: 325) and expression data (Normal: 72, Tumor: 539) of TCGA-KIRC were downloaded from TCGA database on June 15th, 2020. The “limma” package was used to normalize the methylation data. The annotation files were downloaded from the GENECODE (https://www.gencodegenes.org/human/) database to annotate the ENsembl ID to separate mRNA. The average expression of duplicated genes was computed. Genes with an average expression of less than 1 were filtered out, and the ‘edgeR’ package was used for normalization and differential analysis. Genes with average methylation of 0 were filtered out, and the “limma” package was used to normalize the methylation data. The “MethylMix” was applied to select candidate methylation driver genes (log|FC|> 0.3, adjustP < 0.05, Cor < 0.3). Ultimately, NFE2L3 was determined to be the target gene. The tumor samples were separated into two groups according to median NFE2L3 methylation level in tumor tissues, and “survival” package was applied for survival analysis. Meanwhile, survival analysis was performed in combination with the expression level of NFE2L3 gene. Then, Chi-square test was employed to ascertain correlation between NFE2L3 methylation level and clinical practice.
Similarly, survival analysis of NFE2L3 was conducted, and the correlation between NFE2L3 and clinical practice was explored. Finally, 539 tumor samples were separated into two groups (high expression: 270, low expression: 269) according to median NFE2L3 level. Overall survival rates of patients in the two groups were compared. Survival rate was assayed using Kaplan–Meier method and log-rank test. The “c2.cp.kegg.v7.0. symbols.gmt” data were downloaded as a reference gene set from MSigDB (http://software.broadinstitute.org/gsea/msigdb /index.jsp). Gene Set Enrichment Analysis (GSEA) for single-gene was performed using GSEA 4.0.1 software.
Patients and Tissue Collection
From July 2021 to March 2022, 21 pairs of ccRCC tissue and adjacent normal tissue from patients who underwent ccRCC surgery in People’s Hospital of Lishui City were collected. All patients were confirmed by two or more pathologists and had not treated with any anticancer therapy before sampling. The collected samples were snap-frozen in liquid nitrogen for use in subsequent experiments. Written informed consent was obtained from every participant. This study was approved by the Ethics Committee of People’s Hospital of Lishui City.
Cell Line Culture
Human renal proximal tubular epithelial cell line RPTEC (BNCC340070) and four human ccRCC cell lines, LoMet-ccRCC (BNCC101803), Caki-1 (BNCC100682), Caki-2 (BNCC101647), and 769-P (BNCC100976) were offered by BeNa Culture Collection (BNCC, China). For cell biological function experiments, Caki-1 was maintained in McCoy’s 5A Media (BNCC338366) containing 10% FBS, while other cells were kept in Dulbecco’s modified Eagle’s medium (Gibco, USA) plus 10% fetal bovine serum (FBS, BNCC341830). The culture conditions were 37 °C and 5% CO2.
Cell Transfection
Cells in logarithmic phase were utilized for transfection. The oe-NFE2L3 (NFE2L3 overexpression system constructed by pcDNA3.1), oe-NC (control group of NFE2L3 overexpression system constructed by pcDNA3.1), sh-NC (control group of NFE2L3 knockdown system), and sh-NFE2L3 (knockdown system of NFE2L3) were all purchased from Sangon Biotech (China). Plasmids were diluted using serum-free McCoy’s 5A Media. 4 μL lipofectamine 2000 (Invitrogen, USA) reagent was supplemented to 50 μL of McCoy’s 5A Media. The diluted plasmid solution was mixed with the prepared lipofectamine solution, followed by shaking and 20 min of standing. After cells were rinsed twice with PBS, the above mixture was added to cells in each group. After 48 h of transfection, cells were harvested for the follow-up experiments.
DNA Extraction and Ms PCR
Genomic DNA was isolated by protease K method. The cells were digested with DNA digestion buffer (pH 8.0, 25 mM EDTA, 10 mM TrIS–Cl, 100 μg/mL protease K, 1% SDS) and isolated with phenol/chloroform. The EpiTect bisulfite kit (Qiagen, Germany) was used for DNA bisulfite conversion. Online software (MethPrimer) was adopted for primer design, and primer sequences are shown in Table 1. Methylation analysis of hydrogenated DNA was performed by PCR. The thermal cycling parameters were 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 40 s; and 72 °C for 5 min. Ms PCR products were separated by 2% agarose gel, stained with ethidium bromide, and analyzed by nucleic acid gel imaging system (BIO-RAD, USA).
TaqMan Quantitative Methylation-Specific PCR (QMSP)
The methylation primers for QMSP were the same as Ms PCR. The reaction volume was 10 μL (5 μL of 2 × Master Mix, 2.5 μL of BS-DNA (10 ng/μL), 0.3 μL of forward primer and 0.3 μL of reverse primer (10 μmol/L), 0.4 μL of probe (5 μmol/L), and 1.5 μL of ddH2O). Three parallel wells were set for each DNA sample. PCR detection was done using the ABI PRISM® 7900HT Sequence Detection System. QMSP cycling program was: 95 °C for 10 min, 50 cycles of 95 °C for 15 s, and 60 °C for 1 min. ACTB gene was utilized as a methylation reference gene. Relative methylation levels of NFE2L3 DNA were normalized to ACTB. NFE2L3 was considered hypomethylated when its methylation level relative to that of ACTB gene in ccRCC tissues was half that of paired adjacent normal tissue samples.
Total RNA Extraction and qRT-PCR
Total RNA was isolated from cells with TRIzol reagent (Sigma, USA). cDNA synthesis was performed using PrimeScript RT reagent Kit (Takara, Japan). qPCR was carried out using SYBR premixed Ex Taq (Takara, Japan) and ViiATM 7 real-time PCR system (Applied Biosystems, USA). PCR conditions were 95 °C for 30 s, 30 cycles of 95 °C for 5 s, 60 °C for 30 s and 95 °C for 15 s, 60 °C for 30 s and 95 °C for 15 s. Relative mRNA expression was calculated using 2−ΔΔCt method with GAPDH as reference. Primers for NFE2L3 and GAPDH were bought from Sangon Biotech (China). The primer sequences are presented in Table 2.
Western Blot
Total proteins were isolated with RIPA lysis buffer and extraction buffer (Thermo Fisher Scientific, USA). The protein concentration was assayed by BCA kit (Thermo Fisher Scientific, USA). The proteins were concentrated and divided via SDS-PAGE gel (Bio-Rad, USA), then blotted onto PVDF membrane, and blocked with 5% skim milk for 1 h. Afterward, membrane was incubated overnight with primary antibodies and cultured for 1 h with secondary antibody. Images were programmed using standard Western blotting techniques on Bio-Rad ChemiDoc MP imaging system (Hercules, USA). Primary antibodies were rabbit anti-NFE2L3 (1:1000, AP19864B, APCEPTA, USA) and rabbit anti-GAPDH (1: 2500, PA1-988, Thermo Fisher Scientific, USA). Secondary antibody was goat anti-rabbit IgG (0.25 μg/mL, A32731, Thermo Fisher Scientific, USA).
Demethylation Assay
ccRCC cells with low or deficient NFE2L3 expression and human renal proximal tubular epithelial cells (RPTECs) and ccRCC cells (Caki-1, Caki-2, 769-P, LoMet-ccRCC) were inoculated in a 6 cm petri dish at a concentration of 5 × 105 cells per dish. After 24 h, 5-Aza-2′-deoxycytidine (5-Aza-CdR, Selleckchem, USA) at doses of 0, 0.5, 1, 2.5, 5, and 10 μM was added for 72 h, then RNA and proteins were extracted.
Colony Formation Assay
ccRCC cells of different groups were plated into 6-well plates (1 × 103 cells/well). The culture was finished when the dilute cell colonies were visible to the naked eye. ccRCC cells were fixed with 75% ethanol for 30 min, and then dyed with 0.5% crystal violet solution for statistics. Five visual fields were randomly selected for counting. Each assay was repeated three times.
Scratch Healing Assay
ccRCC cells of different groups were plated into 6-well plates (1 × 105 cells/well) until they covered bottom of the plate. A 200 μL sterile pipette tip was utilized to create the wound on cell monolayer. Following rinsing with phosphate-buffered saline (PBS) three times, medium was cultured at 37 °C without FBS for 24 h. The cells were photographed at 0 and 24 h after wound creation. Cell migration distance was measured after image acquisition. Migration rate = (0 h scratch width − 24 h scratch width)/0 h scratch width. Five visual fields were randomly selected to count migrated cells. The experiment was repeated three times.
Cell Invasion Assay
Briefly, cell invasion assay means using Transwell Chambers (8 μm, BD Biosciences, USA) to study invasion. The chambers were placed in a 24-well plate, and 2 × 104 cells suspended in 100 μL serum-free medium were placed in the bottom chamber precoated with matrix. 600 μL medium containing 10% FBS was added to the well plate as the inducer. After incubation for 24 h, cells on the upper surface were removed, and those invading to the lower surface of the membrane were fixed with methanol, stained with 0.1% crystal violet at 37 °C for 30 min, and rinsed with PBS twice. Cells were photographed under a microscope in three separate areas and counted for staining. Five visual fields were randomly selected to count invading cells. The experiment was repeated three times.
Data Processing
All results were statistically analyzed by SPSS21.0. Quantitative data were presented as mean ± standard deviation. T-test was utilized for hypothesis test for inter-group comparison. Before hypothesis testing, normality analysis and homogeneity of variance test should be carried out for each group of data. Non-parametric test should be used for data that do not conform to normal distribution. When p < 0.05, there was a prominent statistical difference. When p < 0.01, there was an extremely significant statistical difference.
Results
NFE2L3 Shows Hypomethylation and High Expression in ccRCC
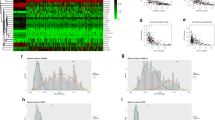
To investigate the influence of NFE2L3 methylation and expression on ccRCC, we firstly selected candidate methylation driver genes by MethylMix. The result showed that there was methylation modification of NFE2L3 in cancer tissues (Fig. 1A). Then, we used software (www.urogene.org/) to search the Human Genome Database for CpG islands in the NFE2L3 gene promoter and found that the CpG islands located in 1070 bp (875–1944) behind the promoter. This indicated that the expression of NFE2L3 was related to abnormal methylation (Fig. 1B). It was indicated that the DNA in the NFE2L3 promoter region was hypomethylated. Then, we predicted the methylation levels of NFE2L3. In TCGA database, methylation levels in the NFE2L3 promoter region were substantially decreased in tumor tissues in relevant to normal tissues (Fig. 1C). We collected tumor tissues and corresponding normal adjacent tissues from ccRCC patients for validation. We found that NFE2L3 was stimulated in tumor tissues in relevant to normal tissues (Fig. 1D). MethylMix analysis showed a substantial negative association between NFE2L3 methylation level and its gene expression (Fig. 1E). By bioinformation analysis and qRT-PCR, we manifested that NFE2L3 was highly expressed in ccRCC patient tissues (Fig. 1F). Combined with the methylation and expression levels of NFE2L3, we found that survival time of patients with hypermethylation and low expression of NFE2L3 was significantly longer than that of patients with hypomethylation and high expression of NFE2L3. In addition, separate survival analyses of NFE2L3 gene expression level and methylation level also showed that high expression or hypomethylation of NFE2L3 predicted a poor prognosis (Fig. 1G). It was indicated that the low expression of NFE2L3 mediated by DNA methylation was significantly conducive to the prognosis of patients. Hypomethylation of NFE2L3 in ccRCC was correlated with high NFE2L3 gene expression and a poor prognosis.
NFE2L3 is hypomethylated in ccRCC. A For the mixture model of NFE2L3 methylation, the abscissa represents the degree of methylation; the ordinate represents the number of methylated samples; the histogram represents the methylation distribution of the tumor group; the peak of the curve indicates the mean level of methylation distribution in the tumor group; the bold horizontal line represents the mean level of methylation distribution in the normal group; B Methylation status of NFE2L3 CpG sites in TCGA dataset; C Boxplot of NFE2L3 methylation level in normal group (blue) and tumor group (red); D The methylation level of NFE2L3 was measured by QMSP in 21 pairs of ccRCC patient tumor tissues and adjacent normal tissues; E Correlation between methylation level and expression level of NFE2L3; F. Boxplot of NFE2L3 expression level in normal group (blue) and tumor group (red), and the mRNA expression level of NFE2L3 in 21 pairs of ccRCC patent tumor tissues and adjacent normal tissues was measured by qRT-PCR; G Survival curve of patients with different NFE2L3 methylation and expression levels (red line for hypermethylation and low expression group, and blue line for hypomethylation and high expression group); Survival curve of patients with different NFE2L3 gene expression (red line for high expression group, and blue line for low expression group); Survival curve of patients with different NFE2L3 methylation levels (red line for hypermethylation group, blue line for hypomethylation group); H Ms PCR detection results of methylation in normal cells and ccRCC cells (U: unmethylated, M: methylated); I qRT-PCR detection result of mRNA expression of NFE2L3 in each cell line; J Western blot detection result in protein expression of NFE2L3 in each cell line. (* denotes p < 0.05)
The result of Ms PCR detection in RPTEC and LoMet-ccRCC, Caki-1, Caki-2 and 769-P cells were shown in Fig. 1H. When fluorescence was only displayed under U band, we believed that methylation did not occur. When fluorescence was displayed under M band or under both M and U bands, methylation or partial methylation was considered. Therefore, we found that the methylation degree of RPTEC cells was higher than that of ccRCC cells. Among all ccRCC cells, the most methylated cells were LoMet-ccRCC cells and the lowest were Caki-1 cells. NFE2L3 level in ccRCC cells and RPTEC cells were assessed. Notably, qRT-PCR detection illustrated that NFE2L3 mRNA level in all ccRCC cells was higher than in RPTEC, (Fig. 1I) and the same trend was observed in the result of Western blot (Fig. 1J). In conclusion, NFE2L3 presented hypomethylation and high expression in ccRCC cells.
Hypomethylation of NFE2L3 Promoter Region Mediates High Gene Expression
To further confirm that the expression of NFE2L3 in ccRCC was mediated by methylation, we then treated ccRCC cells and human renal proximal tubular epithelial cell line RPTEC with 5-Aza-CdR (demethylating drugs). The results presented that NFE2L3 mRNA expression in RPTEC, LoMet-ccRCC, Caki-2, and 769-P cell lines increased with the increase of 5-Aza-CdR concentration and did not increase after a certain concentration reached a plateau. Meanwhile, the expression level of NFE2L3 mRNA in RPTEC cell line was higher than that in ccRCC cell lines. In Caki-1 cell line, the impact of demethylation treatment on NFE2L3 mRNA expression was not obvious. This may be associated with methylation degree of NFE2L3 in different cell lines (Fig. 2A). As such, Western blot presented that NFE2L3 protein level in human renal proximal tubular epithelial cell line RPTEC and all ccRCC cells (Caki-1, Caki-2, 796-P, LoMet-ccRCC) increased with the increase of 5-Aza-CdR concentration (Fig. 2B). Thus, the degree of demethylation was proportional to expression of NFE2L3.
Hypomethylation of NFE2L3 promoter region mediates high gene expression. A qRT-PCR detected mRNA expression of NFE2L3 in ccRCC cells and RETPC cells treated with methylation inhibitor 5-Aza-CdR; B Western blot detected the protein expression of NFE2L3 in RPTEC cells and ccRCC cells treated with methylation inhibitor 5-Aza-CdR
NFE2L3 Overexpression Promotes Malignant Phenotypes of ccRCC Cells
Substantial evidence suggests that NFE2L3 serves as a modulator of proliferation and migration/invasion in a variety of cancers [17,18,19]. In the following step, we investigated the regulatory role of NFE2L3 in ccRCC cells and RPTEC cells. As the expression of NFE2L3 in Caki-1 cell line was the highest and that in LoMet-ccRCC cell line was the lowest among the four ccRCC cell lines, LoMet-ccRCC was selected for NEF2L3 overexpression treatment and Caki-1 for NFE2L3 knockdown treatment. First, NFE2L3 was overexpressed in the LoMet-ccRCC and RPTEC cells, which were divided into oe-NC group and oe-NFE2L3 group. Transfection efficiency was assayed via qRT-PCR and Western blot (Fig. 3A–B, Supplementary Fig. 2A). NFE2L3 mRNA and protein levels in ccRCC and RPTEC cells were noticeably increased after oe-NFE2L3 treatment, indicating the success of transfection. Then, we detected the change in methylation state of NFE2L3 gene in LoMet-ccRCC and RPTEC cells after transfection. The results showed that methylation state of NFE2L3 in oe-NFE2L3 group was lower than that in oe-NC group (Fig. 3C, Supplementary Fig. 2B). Meanwhile, we designed multiple cell function experiments to evaluate the malignant behaviors of cells. Colony formation assay illustrated that overexpression of NFE2L3 could notably enhance LoMet-ccRCC and RPTEC cell proliferative property (Fig. 3D, Supplementary Fig. 2C). Furthermore, tumor cells with overexpressed NFE2L3 were substantially enhanced in both cell migration and invasion, as revealed by wound healing assay (Fig. 3E, Supplementary Fig. 2D) and Transwell assay (Fig. 3F, Supplementary Fig. 2E). These suggested that NFE2L3 overexpression could facilitate the development of ccRCC cells.
Overexpression of NFE2L3 promotes malignant phenotypes of LoMet-ccRCC cells. A qRT-PCR detected transfection efficiency of LoMet-ccRCC cells; B Western blot detected the protein expression of NFE2L3 in transfected LoMet-ccRCC cells; C Change in methylation state of NFE2L3 gene in ccRCC cell line LoMet-ccRCC after transfection; D Cell colony formation assay detected proliferative ability of LoMet-ccRCC cells; E Scratch healing assay detected the migratory ability of LoMet-ccRCC cells (× 40); F Cell invasion assay detected the invasive ability of LoMet-ccRCC cells (× 100). (* denotes p < 0.05)
Repression of Methylation Attenuates the Tumor Suppressive Effect Caused by NFE2L3 Knockdown
Since high level of NFE2L3 was mediated by its hypomethylation, overexpression of NFE2L3 could facilitate the growth of ccRCC cells in vitro and thus, we used 5 μM/L 5-Aza-CdR to explore whether NFE2L3 regulates gene expression through demethylation and thereby affects malignant progression of ccRCC. In this respect, we first constructed two sh-NFE2L3 plasmids and examined the transfection efficiency by qRT-PCR (Supplementary Fig. 1). We selected sh-NFE2L3-1 with higher transfection efficiency for subsequent experiments and nominated it as sh-NFE2L3. Cells expressing sh-NC or sh-NFE2L3 were then treated with 5-Aza-CdR, respectively. qRT-PCR and Western blot denoted that NFE2L3 mRNA and protein levels were substantially decreased in the sh-NFE2L3 group and increased noticeably in 5-Aza-CdR group, and 5-Aza-CdR could restore NFE2L3 expression (Fig. 4A–B, Supplementary Fig. 3A–B). The results of cell colony formation assay presented that in comparison with the control, cell colony formation of Caki-1 and Caki-2 cells transfected with sh-NFE2L3 was markedly reduced, and 5-Aza-CdR treatment notably increased cell colony formation ability of Caki-1 and Caki-2 cells. Additionally, 5-Aza-CdR rescued repressive impact of NFE2L3 knockdown on growth of Caki-1 and Caki-2 cells (Fig. 4C, Supplementary Fig. 3C). As such, wound healing assay (Fig. 4D, Supplementary Fig. 3D) and Transwell (Fig. 4E, Supplementary Fig. 3E) assay assessed that knockdown of NFE2L3 hindered migration and invasion of Caki-1 and Caki-2 cells, while concurrent 5-Aza-CdR treatment attenuated this effect, and the level was recovered to the level of the sh-NC group. In summary, knockdown of NFE2L3 repressed malignant behaviors of ccRCC cells, and demethylation treatment substantially fostered malignant phenotypes of ccRCC cells by increasing NFE2L3 expression.
Repression of methylation attenuates the tumor suppressive effect caused by NFE2L3 knockdown in Caki-1 cells. A qRT-PCR detected Caki-1 cell transfection efficiency; B Western blot detected the protein expression of NFE2L3 in transfected Caki-1 cells; C. Cell colony formation assay detected proliferative ability of Caki-1 cells; D Scratch healing assay detected the migratory ability of Caki-1 cells (× 40); E Cell invasion assay detected the invasive ability of Caki-1 cells (× 100). (* denotes p < 0.05)
Discussion
As the subtype with the highest incidence of renal cancer, ccRCC is characterized by difficulty in early screening and poor prognosis after surgery. So far, specific drugs targeting ccRCC have not been developed, and the molecular mechanism controlling the malignant progression of ccRCC remains unclear. Here, we demonstrated the association between NFE2L3 and ccRCC and high expression of NFE2L3 in ccRCC. The hypomethylation of NFE2L3 regulated its high gene expression, thus promoting the proliferation, migration, and invasion of cancer cells.
We first proved that NFE2L3 could facilitate malignant phenotypes of ccRCC cells via bioinformatics analysis and a series of in vitro assays. Ralf Küppers et al. confirmed high expression of NFE2L3 in Hodgkin’s lymphoma by large-scale gene expression profiling and validation of gene expression in Hodgkin’s lymphoma cells [20]. In addition to hematopoietic malignancies, NFE2L3 mRNA level was also reported to be elevated in breast cancer cells [21]. These studies are in line with this study. In addition, NFE2L3 usually functions as a transcription factor to regulate tumor growth. It can regulate the expression of cell cycle-related genes to promote cell proliferation [11], which has been relatively well studied in colon cancer. For example, some scholars found that NFE2L3 transcription factor plays a key role in the pathway connecting NF-κB signal and CDK1 activity control, thus promoting the proliferation of colon cancer cells [17]. This may provide an idea for our follow-up study on the mechanism of NFE2L3 promoting the development of ccRCC. However, the roles of NFE2L3 in different cancers are different. NFE2L3 also functions as a repressor in other cancer types. For example, NFE2L3 can inhibit the development of lymphoma [10], inconsistent with our study. The specific molecular mechanism of NFE2L3 upregulation in ccRCC was studied in depth. Epigenetic changes are vital in tumor initiation and progression and can be used as tumor markers [22, 23]. Global methylation dysregulation and methylation phenotypes in CpG islands have also been widely observed in some cancer studies [24]. Bioinformatics prediction has found that NFE2L3 may serve as a new DNA methylation driver gene and prognostic marker in ccRCC [16], which was also confirmed in our study. NFE2L3 was hypomethylated in ccRCC patient tissues and cell lines. Although degree of methylation of NFE2L3 in the ccRCC cell line Caki-2 was noticeably lower compared to RPTEC, the methylated and unmethylated levels of NFE2L3 in Caki-2 cells were almost equal. It was not completely consistent with protein expression levels, possibly because NFE2L3 expression could be affected by factors other than methylation. For example, NFE2L3 is a target of miR-23b-3p in colon adenocarcinoma [25] and a target of miR-1246 in breast cancer [26]. Hence, NFE2L3 expression in ccRCC cell lines may also be regulated by multiple miRNAs, which needs future studies.
However, whether the upregulation of NFE2L3 expression is caused by dysregulation of methylation level has not been demonstrated experimentally. That is why we explore this issue further. We treated ccRCC cells with methylation inhibitor with gradient concentrations and then detected the expression level of NFE2L3. Our results proved that in LoMet-ccRCC, Caki-2 and 769-P cell lines, expression increased dependently on concentration of methylation inhibitor. Demethylation treatment had little effect on Caki-1 cells with extremely low NFE2L3 methylation. In other words, NFE2L3 level was closely associated with methylation degree in the gene promoter region. This is in line with earlier studies illustrating that activating genes with carcinogenic potential through DNA hypomethylation promotes the development of cancer [27]. We further unveiled that demethylation treatment substantially attenuated the repressive effect of NFE2L3 knockdown on cell malignant behavior in ccRCC. This suggested that the NFE2L3 upregulation promoting ccRCC development was activated by DNA hypomethylation.
At present, research on DNA methylation is not extensive, and scholars generally believe that DNA methylation will be a new target in cancer research and therapy. In the present study, regulation of hypomethylated NFE2L3 on ccRCC was studied. We hope we can provide new ideas for ccRCC therapy and therefore improve ccRCC patients’ prognoses.
Data Availability
The data used to support the findings of this study are included within the article.
References
Siegel, R. L., Miller, K. D., & Jemal, A. (2018). Cancer statistics, 2018. CA: A Cancer Journal for Clinicians, 68, 7–30. https://doi.org/10.3322/caac.21442
Cohen, H. T., & McGovern, F. J. (2005). Renal-cell carcinoma. New England Journal of Medicine, 353, 2477–2490. https://doi.org/10.1056/NEJMra043172
Shaw, G. (2016). The silent disease. Nature, 537, S98-99. https://doi.org/10.1038/537S98a
Chowdhury, I., Mo, Y., Gao, L., Kazi, A., Fisher, A. B., & Feinstein, S. I. (2009). Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radical Biology & Medicine, 46, 146–153. https://doi.org/10.1016/j.freeradbiomed.2008.09.027
Andrews, N. C. (1998). The NF-E2 transcription factor. International Journal of Biochemistry & Cell Biology, 30, 429–432. https://doi.org/10.1016/s1357-2725(97)00135-0
Chevillard, G., & Blank, V. (2011). NFE2L3 (NRF3): The Cinderella of the Cap’n’Collar transcription factors. Cellular and Molecular Life Sciences, 68, 3337–3348. https://doi.org/10.1007/s00018-011-0747-x
Derjuga, A., Gourley, T. S., Holm, T. M., Heng, H. H. Q., Shivdasani, R. A., Ahmed, R., Andrews, N. C., & Blank, V. (2004). Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Molecular and Cellular Biology, 24, 3286–3294. https://doi.org/10.1128/MCB.24.8.3286-3294.2004
Braun, S., Hanselmann, C., Gassmann, M. G., Keller, U. A. D., Born-Berclaz, C., Chan, K., Kan, Y. W., & Werner, S. (2002). Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Molecular and Cellular Biology, 22, 5492–5505. https://doi.org/10.1128/MCB.22.15.5492-5505.2002
Xiao, Q., Pepe, A. E., Wang, G., Luo, Z., Zhang, Li., Zeng, L., Zhang, Z., Yanhua, Hu., Ye, S., & Qingbo, Xu. (2012). Nrf3-Pla2g7 interaction plays an essential role in smooth muscle differentiation from stem cells. Arteriosclerosis, Thrombosis, and Vascular Biology, 32, 730–744. https://doi.org/10.1161/ATVBAHA.111.243188
Chevillard, G., Paquet, M., & Blank, V. (2011). Nfe2l3 (Nrf3) deficiency predisposes mice to T-cell lymphoblastic lymphoma. Blood, 117, 2005–2008. https://doi.org/10.1182/blood-2010-02-271460
Masudul Azad Chowdhury, A. M., Katoh, H., Hatanaka, A., Iwanari, H., Nakamura, N., Hamakubo, T., Natsume, T., Waku, T., & Kobayashi, A. (2017). Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Scientific Reports, 7, 12494. https://doi.org/10.1038/s41598-017-12675-y
Gonzalo, S. (2010). Epigenetic alterations in aging. Journal of Applied Physiology, 1985(109), 586–597. https://doi.org/10.1152/japplphysiol.00238.2010
Wang, Y. P., & Lei, Q. Y. (2018). Metabolic recoding of epigenetics in cancer. Cancer Communications (Lond), 38, 25. https://doi.org/10.1186/s40880-018-0302-3
Shen, Z., Zhu, L., Zhang, C., Cui, X., & Lu, J. (2019). Overexpression of BHLHE41, correlated with DNA hypomethylation in 3’UTR region, promotes the growth of human clear cell renal cell carcinoma. Oncology Reports, 41, 2137–2147. https://doi.org/10.3892/or.2019.7004
Kang, H. W., Park, H., Seo, S. P., Byun, Y. J., Piao, X. M., Kim, S. M., Kim, W. T., Yun, S. J., Jang, W., Shon, H. S., & Ryu, K. H. (2019). Methylation signature for prediction of progression free survival in surgically treated clear cell renal cell carcinoma. Journal of Korean Medical Science, 34, e144. https://doi.org/10.3346/jkms.2019.34.e144
Jie Wang, Hu., Zhao, H. D., Zhu, L., Wang, S., Wang, P., Ren, Q., Zhu, H., Chen, J., Lin, Z., Cheng, Y., Qian, B., Zhang, Yi., Jia, R., Weizheng, Wu., Jun, Lu., & Tan, J. (2019). LAT, HOXD3 and NFE2L3 identified as novel DNA methylation-driven genes and prognostic markers in human clear cell renal cell carcinoma by integrative bioinformatics approaches. Journal of Cancer, 10, 6726–6737. https://doi.org/10.7150/jca.35641
Bury, M., Le Calvé, B., Lessard, F., Dal Maso, T., Saliba, J., Michiels, C., Ferbeyre, G., & Blank, V. (2019). NFE2L3 controls colon cancer cell growth through regulation of DUX4, a CDK1 inhibitor. Cell Reports, 29(1469–1481), e1469. https://doi.org/10.1016/j.celrep.2019.09.087
Wang, X., Li, Y., Fang, Z., & Li, Y. (2021). Elevated expression of NFE2L3 promotes the development of gastric cancer through epithelial-mesenchymal transformation. Bioengineered, 12, 12204–12214. https://doi.org/10.1080/21655979.2021.2005915
Ren, Y., Wang, Y., Hao, S., Yang, Y., Wendong Xiong, Lu., & Qiu, J. T. (2020). Aifa Tang NFE2L3 promotes malignant behavior and EMT of human hepatocellular carcinoma (HepG2) cells via Wnt/betacatenin pathway. Journal of Cancer, 11, 6939–6949. https://doi.org/10.7150/jca.48100
Küppers, R., Klein, U., Schwering, I., Distler, V., Bräuninger, A., Cattoretti, G., Yuhai, Tu., Stolovitzky, G. A., Califano, A., Hansmann, M.-L., & Dalla-Favera, R. (2003). Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. Journal of Clinical Investigation, 111, 529–537. https://doi.org/10.1172/JCI16624
Rhee, D. K., Park, S. H., & Jang, Y. K. (2008). Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics, 92, 419–428. https://doi.org/10.1016/j.ygeno.2008.08.005
Shen, H., & Laird, P. W. (2013). Interplay between the cancer genome and epigenome. Cell, 153, 38–55. https://doi.org/10.1016/j.cell.2013.03.008
Ren, X., Yang, X., Cheng, B., Chen, X., Zhang, T., He, Q., Li, B., Li, Y., Tang, X., Wen, X., Zhong, Q., Kang, T., Zeng, M., Liu, Na., & Ma, J. (2017). HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nature Communications, 8, 14053. https://doi.org/10.1038/ncomms14053
Viren Amin, R., Harris, A., Onuchic, V., Jackson, A. R., Charnecki, T., Paithankar, S., Subramanian, S. L., Riehle, K., Coarfa, C., & Milosavljevic, A. (2015). Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nature Communications, 6, 6370. https://doi.org/10.1038/ncomms7370
Huang, G., Yang, Y., Lv, M., Huang, T., Zhan, X., & Yao, Y. (2021). Jianghou Hou miR-23b-3p Inhibits the oncogenicity of colon adenocarcinoma by directly targeting NFE2L3. Journal of Oncology, 2021, 8493225. https://doi.org/10.1155/2021/8493225
Dai, Y. C., Pan, Y., Quan, M. M., Chen, Q., Pan, Y., Ruan, Y. Y., & Sun, J. G. (2021). MicroRNA-1246 mediates drug resistance and metastasis in breast cancer by targeting NFE2L3. Frontiers in Oncology, 11, 677168. https://doi.org/10.3389/fonc.2021.677168
Feinberg, A. P., & Tycko, B. (2004). The history of cancer epigenetics. Nature Reviews Cancer, 4, 143–153. https://doi.org/10.1038/nrc1279
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors contributed to data analysis, drafting, and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Ethical Approval
The study was approved by the ethics committee of People’s Hospital of Lishui City.
Research Involving Human and Animals Rights
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2023_727_MOESM2_ESM.tif
Supplementary file2 (TIF 5885 KB)—Supplementary Figure 2 Overexpression of NFE2L3 promotes malignant phenotypes of RPTEC cells. A. qRT-PCR and Western blot detected the mRNA and protein expression of NFE2L3 in transfected RPTEC cells; B. Change in methylation state of NFE2L3 gene in RPTEC cells after transfection; C. Cell colony formation assay detected proliferative ability of RPTEC cells; D. Scratch healing assay detected the migratory ability of RPTEC cells (×40); E. Cell invasion assay detected the invasive ability of RPTEC cells (×100). (* denotes p<0.05)
12033_2023_727_MOESM3_ESM.tif
Supplementary file3 (TIF 7647 KB)—Supplementary Figure 3 Repression of methylation attenuates the tumor suppressive effect caused by NFE2L3 knockdown in Caki-2 cells. A. qRT-PCR detected Caki-2 cell transfection efficiency; B. Western blot detected the protein expression of NFE2L3 in transfected Caki-2 cells; C. Cell colony formation assay detected proliferative ability of Caki-2 cells; D. Scratch healing assay detected the migratory ability of Caki-2 cells (×40); E. Cell invasion assay detected the invasive ability of Caki-2 cells (×100). (* denotes p<0.05)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hou, X., Liao, Q., Wu, Y. et al. Hypomethylation-Mediated Upregulation of NFE2L3 Promotes Malignant Phenotypes of Clear Cell Renal Cell Carcinoma Cells. Mol Biotechnol 66, 198–207 (2024). https://doi.org/10.1007/s12033-023-00727-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-023-00727-w