Abstract
Objective
The methylation of DNA promoter region mediates the low expression of many tumor suppressor genes and plays an essential part in cancer progression. We investigated methylation and expression of ZNF582 in clear cell renal cell carcinoma (ccRCC), and to study the function of ZNF582 in ccRCC cells.
Methods
Methylation data and mRNA expression data of TCGA-KIRC were obtained from TCGA database to screen methylation-driven genes. Survival analysis and gene set enrichment analysis (GSEA) were done for the target gene. The methylation degree and mRNA level of ZNF582 in ccRCC cell line were detected by methylation-specific PCR (MSP) and qRT-PCR, respectively. Effects of overexpression of ZNF582 on ccRCC cells were assessed via CCK-8, flow cytometry, wound healing, Transwell, and cell adhesion assays.
Results
Eighteen methylation-driven genes were identified via bioinformatics methods. Among them, ZNF582 was noticeably hypermethylated and lowly expressed in tumor tissue, and ZNF582 methylation and expression levels were pronouncedly associated with prognosis and clinical stage. MSP also displayed that the ZNF582 DNA promoter region was hypermethylated in ccRCC cells, and the mRNA expression of ZNF582 was dramatically elevated after demethylation. In vitro cell experiments disclosed that overexpression of ZNF582 markedly hindered cell proliferation, invasion, migration, and fostered cell apoptosis and adhesion of ccRCC.
Conclusion
ZNF582 was hypermethylated in ccRCC, which mediated its low level. Overexpression of ZNF582 inhibited tumor cell proliferation, migration and invasion. This study generates novel ideas for ccRCC diagnosis and treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The occurrence of cancer involves changes in genome sequence and epigenetic modification [1]. Among them, epigenetics refers to the gene expression change caused by chemical modifications of DNA and related proteins without affecting DNA transcription sequence [2]. Compared with genetics, epigenetics determines the time, place and way of gene expression to a large extent, and is considered to be the perfect and complement of classical genetics [3]. Moreover, epigenetic aberrations are pharmacologically reversible [2]. Hence, the development of oncology drugs has increasingly emphasized the development of epigenome [1].
Clear cell RCC (ccRCC) is the most frequent subtype of renal cell carcinoma (RCC) [4, 5]. In the progression of RCC, gene silencing induced by promoter methylation plays a substantial role [6], and DNA methylation is also an important biomarker for the early detection of cancer, and prediction of prognosis and therapeutic response [7,8,9]. For example, Iris JH van Vlodrop et al. [10] comprehensively analyzed ccRCC methylation genes based on microarray-based RNA expression profiles and determined that 4 methylation markers (GREM1, NEURL, LAD1 and NEFH) are possible prognostic markers for ccRCC patients. Besides, another research team used a reduced centroid classification method on the basis of genome-wide CpG methylation profiling to identify CpG-based biomarkers that can distinguish ccRCC tumor tissues from adjacent tissues, which clinically provides a potential biomarker for ccRCC early detection [11]. In addition to its value as a biomarker, accumulating investigations pay attention to the effect of tumor suppressor gene silencing caused by methylation in DNA promoter region on biological functions of ccRCC cells, and these studies found that DNA methylation-mediated gene silencing affects proliferation, metastasis, and apoptosis of cancer cells [12,13,14]. This also supplies a novel approach to probe into the mechanism of ccRCC occurrence and progression.
ZNF582 protein is a Kruppel-associated box (KRAB) protein [15]. Methylation of ZNF582 promoter region is a likely biomarker for a variety of cancers, including oral cancer [16, 17], cervical cancer [18, 19], and esophageal squamous cell carcinoma [20], etc. However, there are relatively few studies on its methylation in ccRCC. We denoted that ZNF582 was highly methylated and lowly activated in ccRCC by analyzing TCGA-KIRC methylation and expression data, and verified through cell experiments that the methylation of ZNF582 promoter region mediated low gene expression, thereby causing changes in the biological functions of cancer cells. Our research explained the mechanism of ZNF582 in ccRCC and provided a novel direction for ccRCC targeted therapy.
Materials and methods
Bioinformatics analysis
The tumor tissue and adjacent tissue of patients with ccRCC were both obtained from TCGA-KIRC. Methylation data (Normal adjacent tissue: 160, ccRCC tumor tissue: 325) and expression data (Normal adjacent tissue: 72, ccRCC tumor tissue: 539) of TCGA-KIRC were acquired from TCGA database, along with their clinical data (download time: 2019/11/30). The “limma” package was utilized to standardize methylation data, and the “MethylMix” package was further taken to screen candidate methylation-driven genes with |logFC|> 0.5, adjust p < 0.05 and Cor < −0.3 as threshold. The “survival” package was utilized to evaluate the impact of methylation level and expression level of candidate methylation-driven gene on the prognosis of patients. GSEA software was implemented to evaluate the enriched GSEA pathway of the candidate methylation-driven gene.
Cell culture and transfection
The ccRCC cell lines Caki-1 (BNCC100682), 786-O (BNCC100681), Caki-2 (BNCC340136), A498 (BNCC350808) and human renal tubular epithelial cell line HKC (BNCC338628) were all accessed from BeNa Culture Collection (BNCC). All cells were cultivated according to the instructions in a saturated humidity incubator at 37 °C, with 5% CO2.
The pcDNA3.1 vector subcloned with the full sequence of ZNF582 (pcDNA3.1-ZNF582) was transfected into Caki-2 cells to construct a ZNF582 overexpressed cell line. The transfection process was done with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA isolation from each group of ccRCC cells and HKC cells was done by Trizol (Invitrogen, CA, USA). Complementary DNA (cDNA) was synthesized with the reverse transcription system kit (Invitrogen, CA, USA). qRT-PCR was completed on the ABI 7900HT instrument (Applied Biosystems, USA). The miScript SYBR Green PCR Kit (Qiagen, Germany) was taken to do quantitative. GAPDH was utilized as an internal reference to normalize the data, while the 2−ΔΔCt value was utilized to compare relative level of target gene mRNA. The primer sequences were detailed in Supplementary Table 1.
Methylation-specific PCR (MSP) and DNA demethylation
Genomic DNA from Caki-1, 786-O, Caki-2, A498, and HKC cells was extracted by QIAmap DNA Mini Kit (50) (Qiagen 51304, Germany). The DNA was treated with bisulfite via the EZ DNA Methylation-GoldTM Kit (Zymo Research, CA, USA). The genomic DNA treated with bisulfite was selected as the template for MSP. Methylated (IVMD) and unmethylated (IVUD) control DNAs (Qiagen, Duesseldorf, Germany) were used as reaction controls in MSP. MSP primers were shown in Supplementary Table 1.
In the demethylation experiment, cell lines were treated with 2.5 μmol/L of the demethylation drug 5-aza-2′-deoxycytidine (5-aza-dC) for 6 days. Then, the cells were harvested. Total RNA was isolated to determine ZNF582 level.
Cell counting kit-8 (CCK-8) assay
Cell viability was detected by CCK-8 (Beyotime Biotechnology). Caki-2 cells (3 × 103 cells/well) of each group were seeded in 96-well plates. At 0, 24, 48, 72, and 96 h, optical density (OD) values at 450 nm were recorded under a microplate reader, respectively.
Flow cytometry measurement of apoptosis
At 48 h after cell transfection, 3 × 105 cells were digested with trypsin (without EDTA), harvested and then resuspended in PBS at 4 °C. Cells were centrifuged at 1000 rpm, 4 °C. After PBS was removed, binding buffer (1 ×) was supplemented to resuspend cells. Next, the annexin V-FITC supplied by Annexin V-FITC apoptosis detection kit (Biovision, K101) and PI were added to the stain cell suspension. The suspension was allowed to react for 15 min in the dark. Finally, the apoptotic ratio in cells was assessed by flow cytometry (BD Biosciences).
Transwell
A 24-well Transwell chamber with 8 μm pore size (BD Biosciences) was handled for the Transwell invasion assay. About 2 × 104 Caki-2 cells were placed to the upper chamber, previously coated with Matrigel (Corning, NY). DMEM containing 10% FBS was added to the lower chamber. After incubation at 37 °C for 24 h, the cells that remained on the upper side of membrane were discarded, while cells under the membrane were stained with 3% crystal violet. 4 fields were chosen under a microscope (100 ×) randomly to count cell number.
Wound healing assay
Different groups of Caki-2 cells were inoculated into different wells of a 6-well plate and maintained to compete for fusion. After removing the medium, the cell layer was scraped with a 200 μL pipette tip to form a wound. Scraped cells were rinsed off with PBS, and then remained cells were kept with serum-free medium under routine conditions. Wound healing distance was recorded with a digital camera (40 ×). The wound healing degree was measured, and the wound healing rate was calculated.
Cell adhesion assay
Fibronectin (10 mg/mL) was spread on a 96-well plate overnight at 4 °C, and sealed with 1% BSA for 1 h at 37 °C. Next, 3 × 104 Caki-2 cells were seeded on a 96-well plate and cultured in DMEM without FBS. After cultured for 2 h, cells were rinsed 3 times with PBS and non-adherent cells were gently discarded. Afterwards, the attached cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet (Sangon Biotech). Crystals were dissolved with sodium lauryl sulfate (Amresco, Solon, OH, USA). At last, the OD value at 570 nm was measured with a microplate reader to calculate relative cell adhesion activity.
Statistical analysis
Data from three repeats of experiments were processed by SPSS21.0 statistical software (SPSS, USA). Measurement data were presented as mean ± SD. Comparison between two groups was performed by t-test. The overall survival curves of patients were calculated by Kaplan–Meier, while survival differences of patients were analyzed via log-rank test. p < 0.05 means difference was statistically significant. Cell experiments were conducted three times.
Results
ZNF582 is hypermethylated in ccRCC tissue and is prominently related to prognosis as indicated by bioinformatics analysis
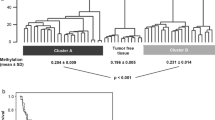
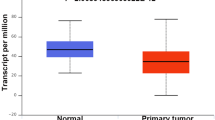
MethylMix analysis screened out 18 candidate methylation-driven genes (Fig. 1A), wherein ZNF582 was dramatically hypermethylated in ccRCC patients’ tumor tissue (Fig. 1B). Meanwhile, ZNF582 methylation level was remarkably negatively correlated with the expression level (Fig. 1C). ZNF582 expression level in ccRCC patients’ tumor tissue was markedly down-regulated (Fig. 1D). Moreover, the survival analysis revealed that the methylation level of ZNF582 influenced ccRCC patient’s prognosis (Fig. 1E). Combined with the expression level, it was also disclosed that survival time of ccRCC patients in ZNF582 hypermethylation and low expression group was conspicuously shorter than ZNF582 hypomethylation and high expression group (Fig. 1E). These suggested that DNA methylation might mediate the low expression of ZNF582, which was pronouncedly detrimental to ccRCC patient’s prognosis. Afterwards, the correlation between the ZNF582 expression level and clinical staging was further analyzed. The results clarified that ZNF582 expression was significantly associated with tumor grade, disease stage, and TNM stage, and the later the staging, the lower the expression level (Fig. 1F). Overall, hypermethylation of ZNF582 was likely to regulate the low expression of ZNF582, thereby affecting the progression of ccRCC.
ZNF582 is hypermethylated in ccRCC tissue, and low expression of ZNF582 will promote the progression of ccRCC. A Heat map of methylation-driven genes related to ccRCC, color from green to a red indicating trend of methylation level from low to high (blue: normal adjacent tissue; pink: ccRCC tumor tissue); B Methylation mixture model of ZNF582 (histogram shows the distribution of methylation in ccRCC samples); C Correlation between methylation level and expression level of ZNF582 in ccRCC tumor tissue; D Violin plot of ZNF582 level in normal group and ccRCC tumor group in TCGA (blue: normal adjacent tissue; red: ccRCC tumor tissue); E The survival curves depicted influence of ZNF582 methylation and expression levels on ccRCC patient’s prognosis (the red line shows the hypermethylation (low expression) group, and the blue line shows the hypomethylation (high expression) group); F Violin plot of the expression level of ZNF582 in different grade, disease stage, T, M, and N stage of ccRCC. ****p < 0.001
DNA methylation in the promoter region mediates low expression of ZNF582 in ccRCC cells
ZNF582 expression in the renal epithelial cell line HKC and ccRCC cell lines Caki-1, Caki-2, 786-O, and A498 was detected by qRT-PCR. The results were congruous with the previous bioinformatics analysis that the expression of ZNF582 was dramatically low in all ccRCC cell lines (Fig. 2A). To further verify that methylation of ZNF582 promoter region regulated the low expression of ZNF582, the MethPrimer website was utilized to predict the distribution of CpG islands of the ZNF582 promoter region (Fig. 2B), and then MSP primers were designed to detect the methylation degree of ZNF582 in each cell line. Our results disclosed that compared with the normal cell line HKC, the methylation degree of the ZNF582 gene in all ccRCC cell lines was higher (Fig. 2C). After treating ccRCC cell lines with 5-aza-dC, ZNF582 mRNA level was remarkably increased (Fig. 2D). The above results confirmed that DNA methylation in the promoter region mediated low expression of ZNF582 in ccRCC cells.
Methylation of ZNF582 promoter region mediates the down-regulation of its expression level. A The expression of ZNF582 in the renal epithelial cell line HKC and ccRCC cell lines Caki-1, Caki-2, 786-O, and A498 was assayed by qRT-PCR; B The distribution of CpG islands in the ZNF582 promoter region was predicted on MethPrimer website; C Methylation degree of ZNF582 promoter in each cell line was analyzed via MSP. (U) unmethylated alleles, (M) methylated alleles; D The expression of ZNF582 restored in ccRCC cell lines using 5-aza-dC. *p < 0.05
Overexpression of ZNF582 restrains cell proliferation and facilitates apoptosis of ccRCC
To further investigate the function of ZNF582 in ccRCC, GSEA pathway enrichment analysis was performed on ZNF582 and found that ZNF582 was prominently enriched in the TIGHT JUNCTION (TJ) pathway (Fig. 3A). Interestingly, relative literature has shown that the TJ pathway is related to cancer cell proliferation, metastasis, and adhesion [21]. Therefore, we restored the expression of ZNF582 in Caki-2 cells (Fig. 3B) to observe its effect on cell proliferation and apoptosis. Compared with the control group, forced ZNF582 expression notably suppressed cell viability (Fig. 3C). Besides, the results of flow cytometry pointed out that the proportion of apoptosis in the oe-ZNF582 group was appreciably increased (Fig. 3D). Forced ZNF582 expression could inhibit cell proliferation and promote cell apoptosis of ccRCC.
Forced ZNF582 expression restrained cell proliferation and promotes apoptosis of ccRCC. A The results of GSEA pathway enrichment analysis of ZNF582; B Overexpression efficiency of ZNF582 was detected by qRT-PCR; C Cell viability of each group was measured via CCK-8; D The level of apoptosis in each group was measured by flow cytometry. *p < 0.05
Overexpression of ZNF582 suppresses cell migration, invasion, and enhances cell adhesion of ccRCC
In the other aspect, the impact of forced ZNF582 expression on cell migration, invasion, and adhesion was tested. The results of the wound healing assay displayed that is relevant to oe-NC group, cell migratory ability was conspicuously lessened in oe-ZNF582 group (Fig. 4A), while the results of the Transwell assay suggested that overexpression of ZNF582 reduced the number of invading cells (Fig. 4B). Cell adhesive ability is often closely related to tumor metastasis. Cell adhesion assay disclosed that forced ZNF582 expression enhanced the adhesive property of Caki-2 cells (Fig. 4C). The above findings manifested that ZNF582 was likely to repress migration and invasion of ccRCC cells by enhancing cell adhesion.
Overexpression of ZNF582 suppresses cell migration, invasion, and enhances cell adhesion of ccRCC. A Cell migratory ability of each group was measured via wound healing assay; B Cell invasive property of each group was assessed via transwell assay; C Cell adhesive ability of each group was detected through cell adhesion assay. *p < 0.05
Discussion
In recent years, research on the molecular biological characteristics of RCC by TCGA and ICGC has greatly promoted the development of targeted therapy. These research advancements increased median survival time of patients with advanced disease from less than 10 months before 2004 to 30 months in 2011 [22]. But about 30% of patients with regional ccRCC show recurrence or metastasis after tumor resection, which severely reduces the survival time and life quality of patients. Hence, it is vital to evaluate the possible molecular mechanisms of cancer progression and discover new diagnostic and therapeutic markers. DNA methylation is a major epigenetic change associated with cancer that may lead to transcriptional silencing of tumor suppressor genes [23]. Reversal of gene suppression by the inhibition of DNA methyltransferases has been successful in the treatment of benign and malignant cells [24, 25]. For example, 5-aza-dC has been demonstrated to reverse the inhibition of many tumor suppressor genes in human tumor cell lines [26, 27]. At present, numerous studies have revealed that DNA methylation plays a substantial role in the occurrence and progression of ccRCC [28,29,30]. For instance, Gooskens SL et al. [14] displayed that in the ccRCC cell line 786-O, the promoter region of the transcription factor TCF21 gene is hypermethylated, which leads to the low expression of TCF21, while up-regulating TCF21 will reduce cell proliferation and migration. In this study, it was disclosed through bioinformatics analysis that the ZNF582 gene was hypermethylated in ccRCC, and the methylation degree was pronouncedly negatively correlated with ZNF582 expression level. Patients with hypermethylation were often accompanied by low expression of ZNF582.
The KRAB-ZNF family may be related to various physiological processes associated with the repair of DNA damage, cell cycle control, and tumor transformation, and ZNF582 is a member of the KRAB-ZNF family [31, 32]. Most of the present studies on the methylation of ZNF582 have focused on its value as a cancer biomarker [19, 20, 33], and its methylation degree has been noted to indicate different treatment responses and clinical outcomes in different cancers. For example, in cervical cancer, high protein level of ZNF582 is implicated in negative methylation and increases the resistance of Hela cells to radiotherapy and chemotherapy [34]. In oral cancer, some scholars found that the average methylation (M) index of ZNF582 gene is noticeably increased, and increased M index of methylated ZNF582 (ZNF582 M) is related to more advanced clinical stage, which indicates that ZNF582 M is a factor for dismal prognosis of oral cancer [16]. These findings demonstrate that ZNF582 methylation may play a prominent role in cancer progression. In this study, bioinformatics analysis denoted that patients with high methylation and low expression of ZNF582 often had a poor prognosis. Besides, the decrease in the expression level of ZNF582 had a remarkable relation to the staging of clinical cases, and the later the stage, the lower the expression level.
The correlation between the methylation of ZNF582 DNA promoter region and its expression in the ccRCC cell lines was further verified. Methylation level of ZNF582 in cancer cells was markedly higher than that in normal cells, and the ZNF582 mRNA expression in cells was restored after demethylation treatment, which confirmed that methylation mediated low gene expression. Subsequently, the role of ZNF582 in ccRCC cells was studied. The GSEA pathway enrichment analysis illustrated that ZNF582 was dramatically enriched in the TJ pathway. TJ defines the extremes of cells by dividing upper and lower areas of cells, thereby giving the cell polarity. TJ proteins participate in modulation of cell proliferation, differentiation, migration, and other important functions [21]. TJ abnormalities caused by inflammation, mutations or abnormal signaling mechanisms will disrupt normal cell functions, leading to cancer and other diseases [35, 36]. Thus, it was judged that the abnormal expression of ZNF582 is likely to affect the phenotype progression of ccRCC cells via modulating TJ signaling pathway. To confirm this conjecture, a cell line with stably overexpressed ZNF582 was constructed. Through a series of cell experiments, it was discovered that forced ZNF582 expression prominently restrained cell proliferation, migration, invasion, and promoted cell apoptosis and adhesion of ccRCC. These results revealed that ZNF582 served as a repressor in ccRCC cells.
Over the past decade, targeted therapies for ccRCC have made significant advances. For example, the tyrosine kinase inhibitor sunitinib, as well as everolimus and temsirolimus that inhibit mTOR complex 1, have received Food and Drug Administration approval [37,38,39]. The administration of these drugs for ccRCC treatment usually results in tumor regression, but treatment resistance often develops within 1 year, which greatly limits their efficacy [40, 41]. Epigenetic drugs are emerging as new therapies in the drug combination that may help prevent or overcome drug resistance [42]. In this study, we found that ZNF582 was a tumor suppressor in ccRCC, which was hypermethylated in ccRCC cells and was an important epigenetic regulator in cancer. These findings provide reliable theoretical support for ZNF582 as a new epigenetic drug for ccRCC treatment. In the future, ZNF582 may be used in combination with targeted agents as a promising and effective treatment strategy for advanced-stage ccRCC.
Viewed in total, our research found that ZNF582 was hypermethylated in ccRCC, which mediated low ZNF582 expression. The overexpression of ZNF582 restrained tumor phenotype progression. But the specific regulatory mechanism of ZNF582 is still unknown, which will be our research direction in the future. This study generates a novel target for ccRCC diagnosis and treatment.
Availability of data and materials
The datasets generated during analyzed are not publicly available but are available from the corresponding author on reasonable request.
References
Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. https://doi.org/10.1038/nrg.2016.93.
Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. https://doi.org/10.1007/978-981-15-3449-2_1.
Yang Q, et al. Epigenetics in ovarian cancer: premise, properties, and perspectives. Mol Cancer. 2018;17:109. https://doi.org/10.1186/s12943-018-0855-4.
Siegel RL, Miller KD. Jemal A (2018) Cancer statistics. CA Cancer J clin. 2018;68:7–30. https://doi.org/10.3322/caac.21442.
Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. 2016;70:93–105. https://doi.org/10.1016/j.eururo.2016.02.029.
Morris MR, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30:1390–401. https://doi.org/10.1038/onc.2010.525.
Shridhar K, et al. DNA methylation markers for oral pre-cancer progression: a critical review. Oral Oncol. 2016;53:1–9. https://doi.org/10.1016/j.oraloncology.2015.11.012.
Ma K, Cao B, Guo M. The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin Epigenetics. 2016;8:43. https://doi.org/10.1186/s13148-016-0210-9.
Schulz WA, Goering W. DNA methylation in urothelial carcinoma. Epigenomics. 2016;8:1415–28. https://doi.org/10.2217/epi-2016-0064.
van Vlodrop IJH, et al. A four-gene promoter methylation marker panel consisting of GREM1, NEURL, LAD1, and NEFH predicts survival of clear cell renal cell cancer patients. Clin Cancer Res. 2017;23:2006–18. https://doi.org/10.1158/1078-0432.CCR-16-1236.
Wei JH, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun. 2015;6:8699. https://doi.org/10.1038/ncomms9699.
Kulkarni P, et al. Elevated miR-182-5p associates with renal cancer cell mitotic arrest through diminished MALAT-1 expression. Mol Cancer Res. 2018;16:1750–60. https://doi.org/10.1158/1541-7786.MCR-17-0762.
Beuselinck B, et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin Cancer Res. 2015;21:1329–39. https://doi.org/10.1158/1078-0432.CCR-14-1128.
Gooskens SL, et al. TCF21 hypermethylation regulates renal tumor cell clonogenic proliferation and migration. Mol Oncol. 2018;12:166–79. https://doi.org/10.1002/1878-0261.12149.
Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–77. https://doi.org/10.1101/gr.4842106.
Cheng SJ, et al. Hypermethylated ZNF582 and PAX1 genes in oral scrapings collected from cancer-adjacent normal oral mucosal sites are associated with aggressive progression and poor prognosis of oral cancer. Oral Oncol. 2017;75:169–77. https://doi.org/10.1016/j.oraloncology.2017.11.013.
Cheng SJ, et al. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck. 2018;40:355–68. https://doi.org/10.1002/hed.24958.
Shen-Gunther J, et al. Molecular Pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology. Clin Epigenetics. 2016;8:96. https://doi.org/10.1186/s13148-016-0263-9.
Liou YL, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics. 2015;7:50. https://doi.org/10.1186/s13148-015-0084-2.
Tang L, et al. Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;120:109488. https://doi.org/10.1016/j.biopha.2019.109488.
Bhat AA, et al. Tight junction proteins and signaling pathways in cancer and inflammation: a functional crosstalk. Front Physiol. 2018;9:1942. https://doi.org/10.3389/fphys.2018.01942.
Li QK, Pavlovich CP, Zhang H, Kinsinger CR, Chan DW. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): a critical step towards the personalized care of renal cancers. Semin Cancer Biol. 2019;55:8–15. https://doi.org/10.1016/j.semcancer.2018.06.004.
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–50. https://doi.org/10.1101/gad.276568.115.
Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40. https://doi.org/10.1200/jco.2002.04.117.
Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–46. https://doi.org/10.1158/1078-0432.Ccr-08-2783.
Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–73. https://doi.org/10.1016/j.cell.2015.07.056.
Chuang JC, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2’-deoxycytidine. Mol Cancer Ther. 2005;4:1515–20. https://doi.org/10.1158/1535-7163.Mct-05-0172.
Fu RJ, et al. DNMT1-maintained hypermethylation of Kruppel-like factor 5 involves in the progression of clear cell renal cell carcinoma. Cell Death Dis. 2017;8:e2952. https://doi.org/10.1038/cddis.2017.323.
Li Y, Su J, Sun W, Cai L, Deng Z. AMP-activated protein kinase stimulates osteoblast differentiation and mineralization through autophagy induction. Int J Mol Med. 2018;41:2535–44. https://doi.org/10.3892/ijmm.2018.3498.
Lin YL, Wang YL, Fu XL, Ma JG. Aberrant methylation of PCDH8 is a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Med Sci Monit. 2014;20:2380–5. https://doi.org/10.12659/MSM.892433.
Huang RL, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS ONE. 2012;7:e41060. https://doi.org/10.1371/journal.pone.0041060.
Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. https://doi.org/10.1186/gb-2003-4-10-231.
Cheng SJ, et al. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncol. 2016;62:34–43. https://doi.org/10.1016/j.oraloncology.2016.09.007.
Wu NY, et al. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann Transl Med. 2019;7:328. https://doi.org/10.21037/atm.2019.06.15.
Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE. Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Mod Pathol. 2005;18:511–8. https://doi.org/10.1038/modpathol.3800301.
Runkle EA, Mu D. Tight junction proteins: from barrier to tumorigenesis. Cancer Lett. 2013;337:41–8. https://doi.org/10.1016/j.canlet.2013.05.038.
Motzer RJ, et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006;295:2516–24. https://doi.org/10.1001/jama.295.21.2516.
Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–13. https://doi.org/10.1056/NEJMoa1510665.
Zanardi E, et al. Clinical experience with temsirolimus in the treatment of advanced renal cell carcinoma. Ther Adv Urol. 2015;7:152–61. https://doi.org/10.1177/1756287215574457.
Bielecka ZF, Czarnecka AM, Solarek W, Kornakiewicz A, Szczylik C. Mechanisms of acquired resistance to tyrosine kinase inhibitors in clear—cell renal cell carcinoma (ccRCC). Curr Signal Transduct Ther. 2014;8:218–28. https://doi.org/10.2174/1574362409666140206223014.
Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. https://doi.org/10.1016/s1470-2045(09)70240-2.
Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–27. https://doi.org/10.1016/j.molcel.2014.05.015.
Funding
Shaoxing City Medical and Health Science and Technology Plan (2020A13061).
Author information
Authors and Affiliations
Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work. MD and GX contributed to the study design. QW conducted the literature search. QW and WZ acquired the data. MD wrote the article. JC and HL performed data analysis and drafted. GX gave the final approval of the version to be submitted.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Ding, M., Wang, Q., Zhu, W. et al. DNA methylation-mediated low expression of ZNF582 promotes the proliferation, migration, and invasion of clear cell renal cell carcinoma. Clin Exp Nephrol 27, 24–31 (2023). https://doi.org/10.1007/s10157-022-02275-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10157-022-02275-0