Abstract
A considerable amount of people succumbs to lung adenocarcinoma (LUAD) due to its high incidence and mortality. This study attempted to reveal the impacts of GOLM1 on LUAD. This work analyzed GOLM1 expression in LUAD and normal tissue and studied its prognostic value utilizing data from The Cancer Genome Atlas. RNA and protein levels were, respectively, determined utilizing qRT-PCR and western blot. Cell-aggressive behaviors were assessed employing Cell Counting Kit-8, scratch healing, and Transwell assays. The targetting relationship between GOLM1 and miR-30a-3p was assayed by dual-luciferase method. GOLM1 up-regulation in LUAD was found in TCGA and it was also a negative factor for survival in patients. GOLM1 overexpression promoted cell progression in LUAD. Down-regulated miR-30a-3p in LUAD was an upstream regulatory miRNA of GOLM1 in terms of molecular mechanism. Further, rescue assays illustrated that miR-30a-3p overexpression attenuated the GOLM1 facilitating impacts on LUAD progression. Finally, we proved that miR-30a-3p/GOLM1 regulated progression of LUAD cells via JAK-STAT pathway. Collectively, the inhibitory impacts of miR-30a-3p on LUAD growth may be mediated by GOLM1/JAK-STAT, which may contribute to the diagnosis of LUAD therapy and the development of therapeutic tools.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
After years of research, lung cancer has been classified into several types, and non-small cell lung cancer (NSCLC) is the most frequent one [1]. Lung adenocarcinoma (LUAD) is the predominant classification of NSCLC [2]. LUAD development is a complex process modulated at genetic and epigenetic manners [3]. Inhibition of tumors is an effective measure for treatment of malignant tumors [4]. Platinum-based therapy is currently the main tumor-suppressant therapy, and it can repress the growth of LUAD cells [5]. However, its side-effects are concerned for body damage, which contribute to LUAD patients’ poor prognosis [6]. Hence, it is meaningful to clarify the mechanisms underlying LUAD progression and identify novel biomarkers and targets for LUAD.
Golgi membrane protein 1 (GOLM1) serves as a transmembrane glycoprotein of the Golgi body [7]. Since Golgi apparatus plays a vital role in protein modification and transport, aberrant expression of related proteins might lead to prominent changes in cellular processes and functions [8]. Many recent studies have manifested that GOLM1 exerts a tumor-promoting role in many cancers, like hepatocellular carcinoma [9], breast cancer [10], prostate cancer [11], and colorectal cancer [12]. GOLM1 overexpression pertains to bad prognostic outcomes of cancers, which might be a novel treatment target [13, 14]. GOLM1 can mediate metastasis of hepatocellular carcinoma (HCC) by modulating epidermal growth factor receptor/receptor tyrosine kinase (EGFR/RTK) cell surface recycling [15]. Moreover, GOLM1 up-regulation represses P53 tetramer formation to increase non-small cell lung cancer aggressiveness [16]. Although it was reported that overexpression of GOLM1 can predict patients’ survival [17], the biological function of GOLM1 in LUAD needs to be further researched.
miRNAs can suppress or promote cancer progression via mediating the posttranscriptional expression of target mRNAs [18]. Multiple miRNAs have been identified as the markers for cancer diagnosis and prognosis [19]. MiR-30a was demonstrated to play a part in cancer-related processes, like metastasis and proliferation [20]. It was less expressed in breast cancer and could suppress the progression of breast cancer via Wnt/β-catenin pathway [21]. The overexpression of miR-30a restrained colony formation and growth of lung cancer cells via targeting MEF2D [22]. Jing Sui et al. discovered that miR-30a-3p decreased LUAD cell proliferation and stimulated apoptosis via mediating AKT3 [23]. However, the exact relevance of miR-30a-3p in LUAD remains to be defined.
Here, GOLM1, miR-30a-3p and the downstream regulatory pathway were research objects. How these two genes and JAK-STAT signaling interacted to mediate tumor-relevant functions of LUAD was defined. Our results are conducive to developing novel strategies for LUAD therapy.
Materials and Methods
Microarray Analysis
Mature miRNA data (46 normal, 521 cancer) and mRNA data (59 normal, 535 cancer) of The Cancer Genome Atlas (TCGA)-LUAD were acquired from TCGA. Based on the download data, the “edgeR” package was utilized to introduce differential expression analysis on miRNAs (|logFC|> 2.0, padj < 0.01). To identify the upstream regulatory miRNAs of GOLM1, miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/) were used for prediction. Then, an overlapping set of the predicted and the significantly down-regulated miRNAs was made. Pearson correlation analysis was conducted between the obtained miRNAs and GOLM1, and the miRNA with a stronger negative correlation was selected for the study.
Cell Culture and Transfection
Human alveolar epithelial cell line HPAEpiC (BNCC337856), human LUAD cell lines A549 (BNCC337696), PC-9 (BNCC340767), and SPC-A-1 (BNCC100120) were all accessed from BeNa Culture Collection (BeNa, China). HPAEpiC was cultured in Roswell Park Memorial Institute-1640 (RPMI-1640, Gibco, USA) medium with 10% fetal bovine serum (FBS, Gibco, USA), A549 in F-12 K medium containing 10% FBS, SPC-A-1 in Dulbecco’s modified eagle medium–high glucose (DMEM-H, Gibco, USA) medium containing 10% FBS, and PC-9 in RPMI-1640 complete medium. Cells were incubated at 37 °C with 5% CO2. All mediums should be replaced every 1–2 days.
When the cell confluence reached 50%, they were transfected with miR-30a-3p mimics (miR-micmic) or negative control (miR-NC) with Lipofectamine 2000 (Invitrogen, USA). Cell harvesting was undertaken 48 h after transfection.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA isolation from cells was performed utilizing Trizol reagent (Invitrogen, USA). Then, reverse transcription was performed with the PrimeScript RT kit (Takara, Japan). The concentration of the extracted RNA was measured with a spectrophotometer. qRT-PCR was performed by SYBR Premix Ex Taq TM (Takara, Japan). The relative gene expression was calculated by the 2−ΔΔCt method. GAPDH and U6 were utilized as the endogenous controls for detection of GOLM1 and miR-30a-3p expression levels. The Primer sequences are displayed in Table 1.
Cell Counting Kit-8 (CCK-8) Assay
Cells (5 × 103 cells/well) were laid in 96-well plates and cultured in an incubator at 37 ℃ with 5% CO2. At 0, 24, 48, 72, and 96 h, 10 μL CCK-8 reagent (Dojindo, Japan) was, respectively, added to each well. The cells were then cultured for another 4 h, and the absorbance was detected at 450 nm using a Microplate Reader (Bio-Rad, USA).
Scratch Healing Assay
Cells (1 × 105 cells/well) in each group were seeded into 6-well plates. Once transfection completed, the monolayer cells were scraped with a sterile 200 μL micropipette. After three PBS washes, the cells were incubated in serum-free medium containing 5% CO2 at 37 ℃. The healing areas of the cells were then observed and photographed at 0 and 24 h. Wound healing area was calculated as: wound healing (%) = (W0–Wn) / W0 × 100, in which W0 denotes the initial wound healing distance and Wn denotes the wound healing distance after n h.
Transwell Assay
Cell invasion assay was done using 24-well Transwell chamber (Corning, USA). 50 μL Matrigel (BD Biosciences, USA) was applied to the upper chamber. The cells were starved for 12 h without serum, washed twice with PBS, and resuspended with serum-free bovine serum albumin (BSA, Invitrogen, USA). Next, 500 μl DMEM-H medium containing 20% FBS (Invitrogen, USA) was added in the lower chamber. After 24-h incubation, a cotton swab was employed to remove non-invasive cells. Cells that invaded the lower chamber were then fixed with 4% paraformaldehyde for 15 min, rinsed twice with PBS and then stained with 0.1% crystal violet (Thermo Scientific™, #R40052, USA). Finally, five random fields per filter were counted and imaged under a microscope.
Western Blot Assay
Cell lysis was performed with radioimmunoprecipitation assay buffer (Beyotime, China). Bicinchoninic acid protein assay kit (Beyotime, China) was used for quantify protein concentration. Protein extracts were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transferring the proteins to polyvinylidene fluoride membranes (Beyotime, China). Subsequently, the membranes were incubated with the primary antibodies rabbit anti-GOLM1 (1:5000, ab109628, Abcam, UK) and the rabbit anti-GAPDH (1:2500, ab9485, Abcam, UK) overnight, followed by the secondary antibody goat anti-rabbit IgG H&L (HRP) (1:10,000, ab6721, Abcam, UK). Measurements were made using a chemiluminescence system (Bio-Rad, USA) and Image Lab software (Bio-Rad, USA) was applied for analysis.
Dual-luciferase Detection
GOLM1 mutant (MUT) and wild-type 3′-untranslated regions (UTR) sequences were constructed by site mutation method. After annealing, all double-stranded segments were inserted into pmirGlO double-luciferase expression vectors (Promega, USA). Dual-luciferase assay was processed in A549 cells. MiR-30a-3p mimics or control mimics were co-transfected into cells using Lipofectamine 3000 (Invitrogen, USA). 48 h later, luciferase activity was assessed and standardized using the Reporter Assay System Kit (Promega, USA).
Statistical Analysis
Data were processed on GraphPad Prism 6.0 (GraphPad, USA) and SPSS 21.0 Software (SPSS, USA). The difference between two groups or among groups was determined by the Student’s t test or one-way analysis of variance, respectively. Survival was measured by Kaplan–Meier. Outcomes were displayed in a manner of as mean ± standard deviation (SD). All assays underwent three repetitions. p < 0.05 refers to a statistically prominent difference.
Results
GOLM1 Is Markedly Overexpressed in LUAD
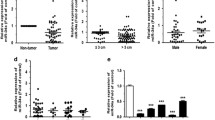
Comparison of GOLM1 level utilizing RNA-Seq data from TCGA uncovered an evidently increased GOLM1 expression in LUAD tissue over the normal control group (Fig. 1A). Combined with patient’s survival, we believed that the increased expression of GOLM1 was a negative factor for the survival of LUAD patients (Fig. 1B). Moreover, literature exhibited that the prominent activation of GOLM1 could deteriorate cancer development, and GOLM1 was expected to become a potential prognostic marker [14, 24, 25]. Hence, we took GOLM1 as the research target. qRT-PCR and western blot outcomes showed a remarkable up-regulation in mRNA and protein levels of GOLM1 in LUAD cell lines compared with HPAEpiC cells (Fig. 1C–D). In summary, we concluded that GOLM1 expression was dramatically increased in LUAD. A549 with the most significantly up-regulated expression of GOLM1 was selected for subsequent functional analyses.
Differential level of GOLM1 in LUAD. A Expression violin plot of GOLM1. Green: normal, red: tumor. B The survival rate of patients in the groups with different degrees of GOLM1 expression. C–D GOLM1 mRNA and protein expression in LUAD cell lines (A549, PC-9, and SPC-A-1) and human alveolar epithelial cells (HPAEpiC); *means p < 0.05
Overexpression of GOLM1 Accelerates the Aggressive Phenotypes of LUAD Cells
To seek for the way GOLM1 mediates LUAD cell phenotypes, we transfected oe-GOLM1 into A549 cells. The result of qRT-PCR manifested a remarkable up-regulation of GOLM1 in oe-GOLM1 group compared with control group (Fig. 2A). As demonstrated by CCK-8 method, cell-proliferative ability was markedly increased after overexpression of GOLM1 (Fig. 2B). The following scratch healing assay showed that overexpressed GOLM1 evidently increased the wound closure area of LUAD cells, indicating that overexpression of GOLM1 prominently strengthened cell migratory ability (Fig. 2C). The invasive ability after overexpressing GOLM1 was remarkably up-regulated (Fig. 2D). Combined with bioinformatics analysis and our experimental results, it was believed that GOLM1 had a promoting effect on the progression of LUAD cells.
MiR-30a-3p Is Notably Less Expressed in LUAD Cells
To gain insight into the possible reasons for the abnormal expression of GOLM1 in LUAD, we downloaded mature miRNA data (Fig. 3A). The differentially down-regulated miRNAs were overlapped with the miRNAs predicted by the database. As a result, miR-30a-3p and miR-143-3p were acquired (Fig. 3B). We further observed that miR-30a-3p had a higher negative correlation with GOLM1, and it was therefore regarded as the most likely upstream miRNA of GOLM1 (Fig. 3C). Subsequently, through TCGA-LUAD dataset, we found that miR-30a-3p was down-regulated in LUAD tissues, and the detection of miR-30a-3p at the cellular level further confirmed the result (Fig. 3D–E). Altogether, miR-30a-3p, less expressed in LUAD, was the most likely upstream regulatory miRNA of GOLM1.
miR-30a-3p is down-regulated in LUAD. A Volcano map of differential miRNAs. Red: high expression, green: low expression; B Venn diagram about miRNAs with binding sites with GOLM1 and down-regulated miRNAs; C Pearson correlation analysis between two miRNA and GOLM1; D expression violin plot of miR-30a-3p. Green: normal, red: tumor; E the mRNA expression of miR-30a-3p in LUAD cell lines and human alveolar epithelial cells; *means p < 0.05
MiR-30a-3p Targets GOLM1 in LUAD Cells
In the present study, the analysis using a bioinformatics prediction tool showed that miR-30a-3p directly bound to GOLM1 (Fig. 4A). To confirm it, we conducted dual-luciferase reporter detection and the result illustrated that miR-30a-3p mimics remarkably inhibited the luciferase activity of WT GOLM1, while the luciferase activity of MUT GOLM1 had no significant change (Fig. 4B). According to qRT-PCR and western blot, miR-30a-3p mimics treatments suppressed mRNA and protein expression of GOLM1 in A549 cells (Fig. 4C–D). Taken together, these data indicated that miR-30a-3p could directly target and modulate GOLM1 expression in LUAD.
MiR-30a-3p Restrains LUAD Development Through Mediating GOLM1
To further test for the role of miR-30a-3p on regulating GOLM1 expression, A549 cell line was used to construct transfection groups. First of all, qRT-PCR and western blot assays manifested that oe-GOLM1 treatment triggered the prominently fostered GOLM1 expression, which could be counteracted by further treatment with miR-mimics (Fig. 5A–B). As revealed by cell function experiments, cell proliferative, migratory, and invasive abilities in LUAD were significantly increased when GOLM1 was overexpressed alone, while these abilities were prominently weakened after the simultaneous overexpression of miR-30a-3p and GOLM1 (Fig. 5C–E). Altogether, miR-30a-3p could inhibit LUAD development through down-regulating GOLM1 expression.
MiR-30a-3p restrains the development of LUAD by targeting GOLM1. A–B The mRNA and protein expression of GOLM1 in different transfection groups; C cell proliferation level in different transfection groups; D the cell migration level of different transfection groups (40 ×); E the cell invasion in different transfection groups (100 ×); *means p < 0.05
MiR-30a-3p/GOLM1 Mediates JAK/STAT Signaling to Restrain LUAD Progression
We predicted the signaling pathways potentially affected by miR-30a-3p/GOLM1 axis in LUAD cells. Through GSEA enrichment analysis, the JAK/STAT signaling pathway was found to be closely related to miR-30a-3p/GOLM1 expression in LUAD (Supplementary Fig. 1). Toward that end, we assessed the protein levels of p-JAK2, JAK2, p-STAT3, and STAT3, in the differently transfected groups. The western blot experiment demonstrated that GOLM1 could enhance JAK2 and STAT3 phosphorylation, but miR-30a-3p overexpression reduced the abilities of GOLM1 to promote the activation of these proteins in A549 cells (Fig. 6A). We also observed that GOLM1 promoted epithelial–mesenchymal transition progression in LUAD cells, whereas miR-30a-3p overexpression was able to block this trend. On the above, miR-30a-3p/GOLM1 axis modulated JAK/STAT signaling as well as EMT progression in LUAD cells.
Discussion
With the increasing atmospheric pollution, the occurrence of LUAD has gradually attracted people’s attention. As a type of lung cancer, it is a non-small cell carcinoma originating from the mucus-producing glands of the lungs [26]. Its incidence is lower than that of squamous cell carcinoma and undifferentiated carcinoma, the age of onset age is younger, and it is more common in women [27]. It was uncovered that many mRNAs are involved in several important biological and pathological processes in LUAD. For instance, Hongxiang Feng et al. found PTEN could modulate the cell growth, migration, and invasion of LUAD [28]. Pengyu Jing et al. found KLF8 regulated LUAD metastasis by forming a signaling axis with miR-24-3p [29]. The overexpression of DAB2 can facilitate metastasis and drug resistance of stage I LUAD [30]. On this basis, we focused on finding new therapeutic targets and potential molecular mechanisms of LUAD.
Previous studies proved that GOLM1 throw a flood of light upon cancer therapies. For example, GOLM1 mediates EGFR/RTK to boost tumor metastasis in hepatocellular carcinoma [15]. GOLM1 accelerates human glioma progression by activating Akt [31]. In melanoma, GOLM1 aggravates tumor-associated behaviors by mediating PI3K/Akt [11]. Herein, we uncovered that GOLM1 was prominently overexpressed in LUAD, and it could facilitate the progression of LUAD cells.
Subsequently, we predicted potential upstream regulatory miRNAs of GOLM1. Combined with the predicted results and Pearson correlation analysis, miR-30a-3p was the most likely upstream mediator of GOLM1, and we speculated that miR-30a-3p modulated LUAD through mediating GOLM1. Previous work elaborated that miR-30a-3p could constrain tumor progression [32, 33]. For instance, inactivation of miR-30a-3p/5p accelerated cell proliferation in esophageal squamous cell carcinoma [34]. MiR-30a-3p repressed the development of helicobacter pylori gastric cancer by targeting COX-2 and BCL9 [35]. MiR-30a-3p restrained renal cell cancer cells through WNT2 mediation [36]. For the purpose of verifying our speculation, we attested miR-30a-3p binding GOLM1 and negatively mediating its expression. As illustrated by rescue assay, miR-30a-3p could debilitate the promotion of GOLM1 enforced expression on LUAD progression. Therefore, miR-30a-3p might be a key gene involved in LUAD tumorigenesis.
JAK/STAT pathway is pivotal in regulating cellular mechanisms, such as proliferation and survival, and also contributes to tumor growth [37]. C-S Zhang et al. demonstrated that miR-409 could suppress the progression of liver cancer via JAK-STAT pathway [38]. Zhongxing Bing et al. discovered that miR-153/PRDM2 modulated invasion and proliferation of NSCLC through JAK/STAT signaling [39]. This work determined that miR-30a-3p could inhibit JAK/STAT signaling via targeting GOLM1, thus suppressing the progression of LUAD.
Collectively, this study revealed the underlying molecular mechanisms of the tumor-relevant behaviors in LUAD. Through in vitro experiments, we confirmed that miR-30a-3p interacted with GOLM1 to mediate LUAD progression via JAK/STAT signaling pathway. Finally, our results indicated that miR-30a-3p/GOLM1 could be a potent candidate marker for clinical diagnosis and could be used as a therapeutic agent for LUAD. Nevertheless, the present work lacks evidence of miR-30a-3p regulating in vivo tumor metastasis, while other targets of miR-30a-3p may also influence LUAD cancerization, which prompts us to scrutinize the corresponding mechanism in LUAD progression.
Data Availability
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
References
Chen, Z., Fillmore, C. M., Hammerman, P. S., Kim, C. F., & Wong, K. K. (2014). Non-small-cell lung cancers: A heterogeneous set of diseases. Nature reviews. Cancer, 14, 535–546. https://doi.org/10.1038/nrc3775
Zhao, X., et al. (2018). LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Science, 109, 3068–3079. https://doi.org/10.1111/cas.13764
Zhang, R., et al. (2020). independent validation of early-stage non-small cell lung cancer prognostic scores incorporating epigenetic and transcriptional biomarkers with gene-gene interactions and main effects. Chest, 158, 808–819. https://doi.org/10.1016/j.chest.2020.01.048
Li, R., Lou, Y., Zhang, W., Dong, Q., & Han, B. (2014). Vitamin D inhibition of lung adenocarcinoma cell proliferation in vitro. Tumour Biology, 35, 10953–10958. https://doi.org/10.1007/s13277-014-1994-x
Qi, L., et al. (2016). An individualised signature for predicting response with concordant survival benefit for lung adenocarcinoma patients receiving platinum-based chemotherapy. British journal of cancer, 115, 1513–1519. https://doi.org/10.1038/bjc.2016.370
Qixing, M., et al. (2017). The expression levels of CYP3A4 and CYP3A5 serve as potential prognostic biomarkers in lung adenocarcinoma. Tumour Biology, 39, 1010428317698340. https://doi.org/10.1177/1010428317698340
Kladney, R. D., et al. (2000). GP73, a novel Golgi-localized protein upregulated by viral infection. Gene, 249, 53–65. https://doi.org/10.1016/s0378-1119(00)00136-0
Wei, S., Dunn, T. A., Isaacs, W. B., De Marzo, A. M., & Luo, J. (2008). GOLPH2 and MYO6: Putative prostate cancer markers localized to the Golgi apparatus. Prostate, 68, 1387–1395. https://doi.org/10.1002/pros.20806
Yan, J., Zhou, B., Li, H., Guo, L., & Ye, Q. (2020). Recent advances of GOLM1 in hepatocellular carcinoma. Hepat Oncol. https://doi.org/10.2217/hep-2020-0006
Cheng, L., Brzozowska-Wardecka, B., Lisowska, H., Wojcik, A., & Lundholm, L. (2019). Impact of ATM and DNA-PK inhibition on gene expression and individual response of human lymphocytes to mixed beams of alpha particles and X-rays. Cancers (Basel). https://doi.org/10.3390/cancers11122013
Chen, W. Y., Xu, Y. Y., & Zhang, X. Y. (2019). Targeting GOLM1 by microRNA-200a in melanoma suppresses cell proliferation, invasion and migration via regulating PI3K/Akt signaling pathway and epithelial-mesenchymal transition. European Review for Medical and Pharmacological Sciences, 23, 6997–7007. https://doi.org/10.26355/eurrev_201908_18740
Liewen, H., et al. (2019). Therapeutic targeting of golgi phosphoprotein 2 (GOLPH2) with armed antibodies: A preclinical study of anti-GOLPH2 antibody drug conjugates in lung and colorectal cancer models of Patient Derived Xenografts (PDX). Targeted Oncology, 14, 577–590. https://doi.org/10.1007/s11523-019-00667-z
Chen, M. H., et al. (2013). Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Annals of surgical oncology, 20(Suppl 3), S616-624. https://doi.org/10.1245/s10434-013-3101-8
Zhang, R., et al. (2019). Golgi membrane protein 1 (GOLM1) promotes growth and metastasis of breast cancer cells via regulating matrix metalloproteinase-13 (MMP13). Medical Science Monitor, 25, 847–855. https://doi.org/10.12659/msm.911667
Ye, Q. H., et al. (2016). GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell, 30, 444–458. https://doi.org/10.1016/j.ccell.2016.07.017
Song, Q., et al. (2021). The functional landscape of Golgi membrane protein 1 (GOLM1) phosphoproteome reveal GOLM1 regulating P53 that promotes malignancy. Cell death discovery, 7, 42. https://doi.org/10.1038/s41420-021-00422-2
Liu, X., Chen, L., & Zhang, T. (2018). Increased GOLM1 expression independently predicts unfavorable overall survival and recurrence-free survival in lung adenocarcinoma. Cancer Control, 25, 1073274818778001. https://doi.org/10.1177/1073274818778001
Di Leva, G., Garofalo, M., & Croce, C. M. (2014). MicroRNAs in cancer. Annual Review of Pathology, 9, 287–314. https://doi.org/10.1146/annurev-pathol-012513-104715
Sun, Z., et al. (2018). Effect of exosomal miRNA on cancer biology and clinical applications. Molecular Cancer, 17, 147. https://doi.org/10.1186/s12943-018-0897-7
Jiang, L. H., Zhang, H. D., & Tang, J. H. (2018). MiR-30a: A novel biomarker and potential therapeutic target for cancer. J Oncol, 2018, 5167829. https://doi.org/10.1155/2018/5167829
Miao, Y., et al. (2019). miR-30a inhibits breast cancer progression through the Wnt/beta-catenin pathway. International Journal of Clinical and Experimental Pathology, 12, 241–250.
Luan, N., Wang, Y., & Liu, X. (2018). Absent expression of miR-30a promotes the growth of lung cancer cells by targeting MEF2D. Oncology Letters, 16, 1173–1179. https://doi.org/10.3892/ol.2018.8719
Sui, J., et al. (2017). Comprehensive analysis of aberrantly expressed microRNA profiles reveals potential biomarkers of human lung adenocarcinoma progression. Oncology Reports, 38, 2453–2463. https://doi.org/10.3892/or.2017.5880
Yang, L., Luo, P., Song, Q., & Fei, X. (2018). DNMT1/miR-200a/GOLM1 signaling pathway regulates lung adenocarcinoma cells proliferation. Biomedicine & Pharmacotherapy, 99, 839–847. https://doi.org/10.1016/j.biopha.2018.01.161
Yan, G., et al. (2018). GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate, 78, 166–177. https://doi.org/10.1002/pros.23461
Wang, B., et al. (2021). ETV4 mediated lncRNA C2CD4D-AS1 overexpression contributes to the malignant phenotype of lung adenocarcinoma cells via miR-3681–3p/NEK2 axis. Cell Cycle, 20, 2607–2618. https://doi.org/10.1080/15384101.2021.2005273
Donner, I., et al. (2018). Germline mutations in young non-smoking women with lung adenocarcinoma. Lung Cancer, 122, 76–82. https://doi.org/10.1016/j.lungcan.2018.05.027
Feng, H., Zhang, Z., Qing, X., French, S. W., & Liu, D. (2019). miR-186-5p promotes cell growth, migration and invasion of lung adenocarcinoma by targeting PTEN. Experimental and Molecular Pathology, 108, 105–113. https://doi.org/10.1016/j.yexmp.2019.04.007
Jing, P., et al. (2020). miR-24-3p/KLF8 signaling axis contributes to LUAD metastasis by regulating EMT. Journal of Immunology Research, 2020, 4036047. https://doi.org/10.1155/2020/4036047
Zhang, L., Huang, P., Li, Q., Wang, D., & Xu, C. X. (2019). miR-134-5p promotes stage I lung adenocarcinoma metastasis and chemoresistance by targeting DAB2. Molecular Therapy Nucleic Acids, 18, 627–637. https://doi.org/10.1016/j.omtn.2019.09.025
Xu, R., et al. (2017). PDGFA/PDGFRalpha-regulated GOLM1 promotes human glioma progression through activation of AKT. Journal of Experimental & Clinical Cancer Research, 36, 193. https://doi.org/10.1186/s13046-017-0665-3
Chen, Q., et al. (2020). miR-30a-3p inhibits the proliferation of liver cancer cells by targeting DNMT3a through the PI3K/AKT signaling pathway. Oncology Letters, 19, 606–614. https://doi.org/10.3892/ol.2019.11179
Wang, W., et al. (2014). MicroRNA-30a-3p inhibits tumor proliferation, invasiveness and metastasis and is downregulated in hepatocellular carcinoma. European Journal of Surgical Oncology, 40, 1586–1594. https://doi.org/10.1016/j.ejso.2013.11.008
Qi, B., et al. (2017). Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World Journal of Gastroenterology, 23, 7965–7977. https://doi.org/10.3748/wjg.v23.i45.7965
Zhang, J. W., et al. (2020). MiR-30a-5p promotes cholangiocarcinoma cell proliferation through targeting SOCS3. Journal of Cancer, 11, 3604–3614. https://doi.org/10.7150/jca.41437
Liu, L., Chen, L., Wu, T., Qian, H., & Yang, S. (2019). MicroRNA-30a-3p functions as a tumor suppressor in renal cell carcinoma by targeting WNT2. Am J Transl Res, 11, 4976–4983.
Pencik, J., et al. (2016). JAK-STAT signaling in cancer: From cytokines to non-coding genome. Cytokine, 87, 26–36. https://doi.org/10.1016/j.cyto.2016.06.017
Zhang, C. S., et al. (2019). miR-409 down-regulates Jak-Stat pathway to inhibit progression of liver cancer. European Review for Medical and Pharmacological Sciences, 23, 146–154. https://doi.org/10.26355/eurrev_201901_16758
Bing, Z., Zheng, Z., Wang, Y., & Zhang, J. (2021). miR-153 targeting PRDM2 gene affects the proliferation and invasion of non-small cell lung cancer through JAK / STAT signaling pathway. Minerva Surgery. https://doi.org/10.23736/s2724-5691.21.09200-5
Acknowledgements
Not applicable.
Funding
None.
Author information
Authors and Affiliations
Contributions
DX contributed to the study design. HH conducted the literature search. YQ and XD acquired the data. KS wrote the article. WJ performed data analysis. JJ drafted the manuscript. LW and ZJ revised the article and gave the final approval of the version to be submitted. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no potential conflicts of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors consent to submit the manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, D., Zhang, Y., Zhang, X. et al. MiR-30a-3p Suppresses the Growth and Development of Lung Adenocarcinoma Cells Through Modulating GOLM1/JAK-STAT Signaling. Mol Biotechnol 64, 1143–1151 (2022). https://doi.org/10.1007/s12033-022-00497-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00497-x