Abstract
Lung adenocarcinoma (LUAD) is the most frequent histological subtype of non-small cell lung cancer. Cyclin B1 (CCNB1) is the vital initiator and controller of mitosis. Studies have indicated that CCNB1 overexpression is closely associated with cell proliferation and tumorigenesis in many cancers. Thus, discovery of molecular mechanism of CCNB1 in LUAD is conducive to developing new diagnostic or therapeutic targets for LUAD. We acquired mature miRNA and mRNA expression information of LUAD from TCGA database, as well as related clinical data. CCNB1 expression in normal and LUAD tissue was analyzed. Relationship between CCNB1 and patient’s survival and clinical stage was analyzed. Upstream regulatory gene miRNA of CCNB1 was predicted. qRT-PCR and western blot examined expression levels of CCNB1 and miR-139-5p in cells. CCK-8 tested cell proliferation. Scratch healing and Transwell determined cell migration and invasion. Flow cytometry analyzed the cell cycle. Dual-luciferase verified targeting relationship between the two genes. Compared to controls, CCNB1 expression was prominently high in LUAD patient samples, and associated with advanced tumor stages and shorter overall survival. MiR-139-5p expressed an evidently negative correlation with CCNB1 and was predicted to target CCNB1. MiR-139-5p mimics reduced CCNB1 mRNA and protein expression, and suppressed luciferase activity in a target-specific manner, as confirmed by a control construct with a mutated miR-139-5p binding site. CCNB1 overexpression fostered progression of LUAD cells. Mechanistically, miR-139-5p might negatively regulate CCNB1 in LUAD, thereby suppressing cell proliferation, migration, invasion and cell cycle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer accounts for more than 25% of all cancer deaths [1]. The 5-year overall survival rate of non-small cell lung cancer (NSCLC) is only 11% [2]. Lung adenocarcinoma (LUAD) is the most frequent subtype of NSCLC [3]. In recent years, development of molecular targeted therapy [4] and immunotherapy [5] has increased survival rate of LUAD. However, 5-year overall survival and prognosis of LUAD is still not optimistic [6]. The poor prognosis may result from the high rates of tumor metastasis and recurrence. As a result, a deeper understanding of the molecular mechanisms that control LUAD development helps to improve LUAD diagnosis and treatment.
Cyclins are proteins required to stimulate specific cyclin-dependent kinases (CDKs) in cell cycle [7]. Cyclin B1 (CCNB1) is the vital initiator and controller of mitosis. CCNB1 can regulate cyclin-dependent kinase 1 (CDK1) and combine with CDK1 to form a complex, thereby facilitating rapid passage of the cell cycle through the G2 phase of mitosis [8, 9]. Studies have shown that CCNB1, as an oncogenic factor, can participate in modulation of the progression of various cancers. For example, FOXM1 activates CCNB1 through transcription and then promotes the proliferation of human hepatocellular carcinoma (HCC) [10]. Chk1 induces overexpressed CCNB1 and fosters tumor growth of colorectal cancer cells [11]. Although some studies have expressed aberrant expression of CCNB1 in LUAD [12], the regulatory mechanism of CCNB1 in LUAD has not been studied.
MicroRNAs (miRNAs) are involved in mRNA degradation and translational repression by targeting the 3’-UTR of mRNA, and they can function as tumor suppressors or oncogenes [13, 14]. MiR-139-5p is a normal mature miRNA produced by the precursor miR-139 [15, 16]. MiR-139-5p is proven to be repressed in many tumors, like glioblastoma multiforme [17], bladder cancer [18], HCC [19], and breast cancer [20]. MiR-139-5p is aberrantly expressed in LUAD as well [21]. Nonetheless, specific mechanism and function of miR-139-5p in LUAD are still unknown.
Bioinformatics results showed that CCNB1 was markedly fostered in LUAD, and it was targeted by miR-139-5p. Hence, this study aimed to study association between miR-139-5p and CCNB1, and effect of miR-139-5p regulating CCNB1 on progression of LUAD cells. The analysis of mechanism of miR-139-5p/CCNB1 axis modulating LUAD growth can help us develop new targets for LUAD.
Materials and Methods
Microarray Analysis
The mature miRNA data and mRNA data of LUAD were provided by TCGA database. Differential expression of CCNB1 in normal and cancer tissue was tested by t-test based on the downloaded data. “Survival” package was employed for survival analysis [22]. One-way analysis of variance was applied to evaluate the correlation between CCNB1 and clinical features of LUAD. The differential analysis of miRNA data was carried out using the “EdgeR” package [23] with |logFC| > 1.5 and padj < 0.01 as thresholds. StarBase database was utilized to analyze upstream regulatory miRNAs of CCNB1, and then the results were intersected with differentially downregulated miRNAs. Pearson correlation analysis was conducted for acquired miRNA and CCNB1, and t-test was employed to further clarify the differential expression of miRNAs in TCGA.
Cell Cultivation and Transfection
Human bronchial epithelial cell line BEAS-2B (BNCC101727), LUAD cell lines A549 (BNCC100215), PC9 (BNCC340767), H1299 (BNCC100268) and H1975 (BNCC340345) were offered by BeNa Culture Collection (BNCC, China). Cell lines were incubated strictly complying with the operating instructions provided by BeNa Culture Collection. The following constructs were designed by GenePharma (Suzhou): miR-139-5p mimic (miR-mimic), pcDNA3.1-CCNB1 plasmids (oe-CCNB1) and si-CCNB1, as well as their control mimic NC (miR-NC), pcDNA3.1 plasmids (oe-NC) and si-NC. Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was applied for transfection.
qRT-PCR
For PCR analysis, miScript II RT kit (Qiagen, Germany) was employed to reversely transcribe extracted miR-139-5p into cDNA, and PrimeScript RT Master Mix (Takara, P.R. Japan) was used to reversely transcribe CCNB1 mRNA into cDNA. Step One Plus real-time PCR system (Applied Biosystems Inc., Foster City, CA, USA) and SYBR Green real-time PCR Master Mix (Cat.# QPK-212, Toyobo) were used to conduct qRT-PCR assay in line with the manufacturer’s specifications. MiR-139-5p took U6 as its internal reference, and CCNB1 took GAPDH as its internal reference. Primer sequences were expressed in Table 1. Relative expression of miR-139-5p and CCNB1 mRNA in each group was compared by 2−ΔΔCt value.
CCK-8 Assay
Cells (2 × 103 cells/well) were inoculated into 96-well plates after 24-h transfection. Cell viability was measured by CCK-8 assay (Dojindo, Kumamoto, Japan) at 0, 24, 48, 72, and 96 h after cultivation. The specific operation was carried out in accordance with the operating instructions. Finally, a plate reader from Molecular Devices (San Jose, CA, USA) was applied to count optical density at 450 nm.
Scratch Healing Assay
Cells were cultivated at 37 ℃ for 24 h and scraped with 10 µl pipette when cells reached 90% confluence. Later, phosphate buffer saline (PBS) was adopted for cells wash and then detached cells were removed. Remaining cells were cultivated at 37 ℃ in a medium plus 1% FBS for another 24 h and the motion of cells was analyzed by an optical microscope (Olympus Corporation, × 40). The relative mobility was calculated as: (d1-d2) / d1 × 100%, and d1 is the scratch width at 0 h, d2 is the scratch width at 24 h.
Transwell Invasion Assay
Matrigel (356234, BD, USA) was dissolved at 4 ℃ overnight, diluted with serum-free medium and added to Transwell (8.0 μm, 24 wells, Corning Incorporated, Corning, NY, USA) upper chamber. Thereafter, 3 × 105 cells were put in the upper chamber and 500 μl culture medium plus 10% FBS was placed to the lower chamber. Cells passing through the membrane were treated by 4% formaldehyde solution and 0.25% crystal violet solution at room temperature for 30 min. Then an inverted microscope (Olympus Corporation, Tokyo, Japan) was utilized to observe cells. The number of invading cells was counted in 4 random microscope fields.
Western Blot
RIPA buffer (Beyotime, Beijing, China) was applied to extract proteins. Samples (30 μg) were separated on 10% SDS-PAGE and then transferred to the nitrocellulose (NC, Millipore, Billerica, MA) membrane. After 1-h blocking with 5% skim milk, the membrane was cultivated with primary antibodies and secondary antibody in sequence. Protein bands were examined by an enhanced chemiluminescence detection system (Pierce, Billerica, MA).
Dual-Luciferase Assay
Mutant (MUT) and wild-type (WT) DNA sequences of CCNB1(GeneChem Co., Ltd., Shanghai, China) were inserted into pGL3-control vectors (Ambion, Austin, TX, USA). Lipofectamine 3000 (Invitrogen) was used to co-transfect 100 ng of the above pGL3-CCNB1 WT/MUT plasmids and 50 nm miR-139-5p mimic or mimic NC into A549 cells. Luciferase activity was tested 48 h later.
Cell Cycle Detection
Cell cycle analysis kits (Beyotime Institute of Biotechnology) were used for cell cycle measurement. Cells were collected and fixed in 70% ethanol at 4 ℃ overnight. Prodium Iodide was added to dye cells and then the cells were analyzed with flow cytometry. Finally, FlowJo software (FlowJo LLC) was introduced to analyze cell cycle.
Data Analysis
Data analysis was done by using GraphPad Prism 6 Software (GraphPad Software, Inc., La Jolla, USA). Student’s t-test was adopted to examine statistical significance of the difference among groups. Pearson correlation analysis was employed to clarify the correlation between miR-139-5p and CCNB1 expression. Data were acquired from at least 3 independent assays and in the form of mean ± SD. P < 0.05 indicates significant differences.
Results
CCNB1 Expression is Increased in LUAD Cells and Correlates with Tumor Stage of Patients with LUAD
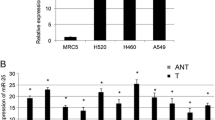
Literature shows that CCNB1 is relevant with adverse prognosis and can be a novel therapeutic target for cancer [24,25,26]. Therefore, CCNB1 was chosen as the research object. Analysis of TCGA-LUAD data found that CCNB1 expression was prominently elevated in LUAD tissue compared with normal tissue (Fig. 1A), and patients with LUAD with high CCNB1 expression level had an evidently shorter survival time (Fig. 1B). Besides, CCNB1 expression was positively associated with staging of LUAD (Fig. 1C). CCNB1 expression in 4 LUAD cell lines (A549, PC9, H1299 and H1975) and one normal bronchial epithelial cell line (BEAS-2B) was also verified. It was found that CCNB1 was also upregulated in these LUAD cell lines in comparison with that in normal cell line (Fig. 1D, E). Together, these findings suggested that CCNB1 was elevated in LUAD and indicated an adverse prognosis in patients with LUAD. A549 cell line with the greatest difference in CCNB1 expression was selected to further explore the specific role of CCNB1 in LUAD and its molecular mechanism.
Expression of CCNB1 increases in LUAD cells and correlates with tumor stage of patients with LUAD. A CCNB1 expression levels in LUAD tumor tissue (red) and adjacent normal tissue (green); B Overall survival analysis based on CCNB1 in LUAD patients; C Expression level of CCNB1 in LUAD at different stages; D and E CCNB1 mRNA and protein levels in bronchial epithelial cell line BEAS-2B and LUAD cell lines A549, PC9, H1299 and H1975; *p < 0.05
CCNB1 Promotes Progression LUAD Cells
To investigate effect of CCNB1 in LUAD cells. oe-CCNB1, si-CCNB1 and their negative controls were transfected into A549 cells. As expected, CCNB1 expression was remarkably increased in A549 cells transfected with oe-CCNB1 and decreased in the one transfected with si-CCNB1. (Fig. 2A). Subsequently, as result of CCK-8 manifested, the activity of A549 cells was significantly potentiated by transfection with oe-CCNB1 in contrast with the cells with oe-NC. The viablity of A549 cells was markedly weakened by transfection with si-CCNB1 in contrast with the cells treated with si-NC (Fig. 2B). Transwell (Fig. 2C) and scratch healing (Fig. 2D) assays observed that compared with oe-NC group, the invasion and mobility of A549 cells treated with oe-CCNB1 were remarkably increased. Compared with si-NC group, invasion and mobility of A549 cells treated with si-CCNB1 were evidently declined. Cell cycle progression was analyzed via flow cytometry to further evaluate growth-enhancing effect of CCNB1 on A549 cells in LUAD. As results displayed, the overexpression of CCNB1 led to decrease of cells in G0/G1 phase, and silencing CCNB1 triggered the increase of cells in G0/G1 phase (Fig. 2E). To further confirm the conclusion, H1299 was also transfected with si-CCNB1 and si-NC. The results implied that the conclusion drawn from A549 cells were also applicable to H1299 cells (Supplementary Fig. 1). All above results unveiled that CCNB1 was helpful to promote the cell cycle progression in LUAD and to accelerate growth of LUAD cells. Inhibition of CCNB1 expression repressed the malignant progression of LUAD.
CCNB1 elevates cellular viability and hastens progression of LUAD cells. A CCNB1 level in A549 cells was determined; B LUAD cell proliferation; C Invasive capacity of transfected LUAD cells (×100); D Migratory capacity of transfected LUAD cells (×40). E Cell cycle analysis of transfected LUAD cells; The cell grouping settings were as follows: oe-CCNB1, si-CCNB1 and corresponding negative controls were transfected into A549 cells. *p < 0.05
MiR-139-5p is Less Expressed in LUAD
To study mechanism of CCNB1 in LUAD progression, a total of 184 differential miRNAs were obtained through differential analysis (Fig. 3A). The potential upstream miRNAs of CCNB1 were obtained using the starBase database, and were intersected with 39 differentially downregulated miRNAs. MiR-139-5p and miR-144-3p were predicted to have binding sites on CCNB1 (Fig. 3B). MiR-139-5p and CCNB1 showed a highly evident negative correlation as revealed by Pearson correlation analysis (Fig. 3C). In addition, miR-139-5p level in LUAD tissue was prominently lower than that in normal tissue (Fig. 3D). Hence, we chose miR-139-5p as the study object. Subsequently, it was found by qRT-PCR that miR-139-5p in LUAD cells was notably declined in comparison with normal lung cells (Fig. 3E). Subsequently, it was unveiled that miR-139-5p and CCNB1was negatively correlated (R2 = 0.974, p < 0.01), which further verified the results of the above-mentioned bioinformatics analysis (Figure F). Overall, results above confirmed that miR-139-5p level was low in LUAD and might target CCNB1 in LUAD.
MiR-139-5p is repressed in LUAD cells. A Differential miRNAs in LUAD data set of TCGA (red dots: differentially upregulated miRNAs, green dots: differentially downregulated miRNAs); B Candidate differential miRNAs with binding sites on CCNB1; C CCNB1 and miR-139-5p expression manifested a highly evidently-negative Pearson correlation; D MiR-139-5p level in LUAD tissue (red) and adjacent tissue (green); E: MiR-139-5p level in BEAS-2B, A549, PC9, H1299 and H1975; F CCNB1 and miR-139-5p showed a prominently highly negative correlation in 4 LUAD cells. *p < 0.05
CCNB1 is Targeted by miR-139-5p
Further analyses were carried out to dig whether miR-139-5p can target and even regulate CCNB1. CCNB1 was predicted by starBase database as a target of miR-139-5p (Fig. 4A). Firstly, qRT-PCR tested the transfection efficiency of miR-139-5p in the experimental group, uncovering that miR-139-5p expression in miR-mimic group was prominently boosted compared to that in miR-NC group (Fig. 4B). Luciferase activity analysis displayed that, miR-139-5p remarkably hampered luciferase activity of CCNB1-WT 3’-UTR group in comparison with miR-NC group, but did not influence luciferase activity of the CCNB1-MUT 3’-UTR group (Fig. 4C). Moreover, increased miR-139-5p in A549 cells notably reduced mRNA (Fig. 4D) and protein (Fig. 4E) levels of CCNB1. All the above results proved that CCNB1 was targeted and modulated by miR-139-5p.
CCNB1 is targeted by miR-139-5p. A Targeted binding sites of miR-139-5p and CCNB1; B: MiR-139-5p level in each transfection group; C Luciferase activity of A549 cells co-transfected with miR-139-5p mimic or miR-NC, CCNB1-WT or CCNB1-MUT; D and E The mRNA and protein levels of CCNB1 in LUAD A549 cells; *p < 0.05
MiR-139-5p Suppresses Progression of LUAD Cells Through Targeting CCNB1
To dig whether miR-139-5p participates in affecting LUAD cell function through targeting CCNB1, normal cell line (miR-NC + oe-NC), CCNB1-overexpressed cell line (miR-NC + oe-CCNB1), miR-139-5p and CCNB1-overexpressed cell line (miR mimic + oe-CCNB1) were constructed. Firstly, we found through qRT-PCR and western blot that CCNB1 mRNA and protein levels in miR-NC + oe-CCNB1 group was facilitated evidently, however, when miR-139-5p and CCNB1 both were accelerated, CCNB1 mRNA and protein in the cells were evidently reduced compared with those of miR-NC + oe-CCNB1 group (Fig. 5A, B). Additionally, overexpressed miR-139-5p eliminated the promotion influence of LUAD cells by overexpressed CCNB1 on proliferation (Fig. 5C), invasion and migration (Fig. 5D, E) and cell cycle progression (Fig. 5F). As suggested by these findings, miR-139-5p played an inhibitory part in LUAD by inhibiting CCNB1.
MiR-139-5p inhibits progression of LUAD cells through targeting CCNB1. A and B The expression levels of mRNA and protein of CCNB1 in A549 cells was determined; C Proliferation of A549 cells; D Invasion of A549 cells (×100); E Migration of A549 cells (×40); F Flow cytometry analyzed the cell cycle of A549 cells; *p < 0.05
Discussion
So far, several mRNAs that regulate LUAD progression have been identified as biomarkers for LUAD. For example, SGK1 [27], AKT1 [28], ZEB2 [29], SATB1 [30] are identified as biomarkers of LUAD. In our paper, expression, biological function, and possible mechanisms of CCNB1 in LUAD progression were analyzed. The results suggested that increased CCNB1 could function as a therapeutic target for LUAD.
Previous literature shows that the hastened CCNB1 is relevant with poor prognosis of a variety of cancers and can be a novel target for cancer [24, 31]. For instance, CCNB1 targeted by miR-718 plays an oncogenic role in NSCLC [25]. PKMYT1 targets and regulates CCNB1 and CCNE1 to affect the growth of prostate cancer cells [32]. FOXM1 activates CCNB1 to promote proliferation of human hepatocellular carcinoma cells [10]. In this study, analysis of TCGA-LUAD data and cell experiment results uncovered high expression of CCNB1 in LUAD. As expected, CCNB1 fostered progression of LUAD cells in vitro. In addition, si-CCNB1 inhibited LUAD cell activity and cell cycle progression. These results suggested that CCNB1 could modulate the development of LUAD as an oncogenic factor.
In the process of screening and verifying the upstream miRNAs of CCNB1, we unveiled that miR-139-5p had binding sites and was highly correlated with CCNB1. Hence, it was speculated that miR-139-5p modulated CCNB1 in LUAD and even affected the progression of LUAD. Other works found that miR-139-5p is suppressed in a variety of cancers and can be a cancer suppressor [33,34,35,36]. For instance, a study demonstrated that miR-139-5p is an inhibitor for endometrial cancer [34]. Forced expression of miR-139-5p in breast cancer alleviates radiation resistance by inhibiting multiple genes associated with ROS defense and DNA repair [37]. Paradoxically, however, miR-139-5p level in the blood of prostate cancer patients is higher than that of healthy controls [38]. Reduced miR-139-5p level facilitates prostate cancer progression via modulating SOX5 [39]. These conclusions indicate that miR-139-5p may play different roles in varying species and tissues. In this article, analysis of TCGA-LUAD data and results of cell assays manifested that miR-139-5p was repressed in LUAD. CCNB1 was targeted by miR-139-5p in LUAD cells. Hastened miR-139-5p eliminated the promoting effect of overexpression of CCNB1 on LUAD cells. Consistent with our research, miR-139-5p affects invasion and proliferation of LUAD cells by reducing SLITRK4 expression [40]. These results demonstrated that miR-139-5p might suppress LUAD tumor growth by inhibiting CCNB1. In addition, a study by Jian Wu et al. demonstrated that the lncRNA SNHG3/miR-139-5p axis plays an imperative role in cell progression of hepatocellular carcinoma [41]. Based on previous studies and current experimental results, it is speculated that SNHG3 may affect the malignant phenotypes of LUAD cells through miR-139-5p/CCNB1 signaling pathway. This hypothesis has not been tested, but it will be the direction of future research.
In summary, in this study, CCNB1 was identified for the first time as a downstream target of miR-139-5p in LUAD. MiR-139-5p restrained LUAD cell proliferation, migration, invasion and cell cycle progression through targeting CCNB1. These outcomes uncover a novel mechanism for miRNA dysregulation and offer promising potential biomarkers for LUAD.
Data Availability
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
References
Siegel, R. L., Miller, K. D., & Jemal, A. (2019). Cancer statistics, 2019. CA: A Cancer Journal for Clinicians, 69, 7–34. https://doi.org/10.3322/caac.21551
Thomson, C. S., & Forman, D. (2009). Cancer survival in England and the influence of early diagnosis: What can we learn from recent EUROCARE results? British Journal of Cancer, 101(Suppl 2), S102-109. https://doi.org/10.1038/sj.bjc.6605399
Jemal, A., Siegel, R., Xu, J., & Ward, E. (2010). Cancer statistics, 2010. CA: A Cancer Journal for Clinicians, 60, 277–300. https://doi.org/10.3322/caac.20073
Zhou, C., & Yao, L. D. (2016). Strategies to improve outcomes of patients with EGRF-mutant non-small cell lung cancer: Review of the literature. Journal of Thoracic Oncology, 11, 174–186. https://doi.org/10.1016/j.jtho.2015.10.002
Villanueva, N., & Bazhenova, L. (2018). New strategies in immunotherapy for lung cancer: Beyond PD-1/PD-L1. Therapeutic Advances in Respiratory Disease, 12, 1753466618794133. https://doi.org/10.1177/1753466618794133
Riaz, S. P., et al. (2012). Trends in incidence of small cell lung cancer and all lung cancer. Lung Cancer, 75, 280–284. https://doi.org/10.1016/j.lungcan.2011.08.004
Shin, J. U., et al. (2012). Prognostic significance of ATM and cyclin B1 in pancreatic neuroendocrine tumor. Tumour Biology, 33, 1645–1651. https://doi.org/10.1007/s13277-012-0420-5
Krek, W., & Nigg, E. A. (1991). Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: Identification of major phosphorylation sites. EMBO Journal, 10, 305–316.
Morgan, D. O. (1995). Principles of CDK regulation. Nature, 374, 131–134. https://doi.org/10.1038/374131a0
Chai, N., et al. (2018). FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochemical and Biophysical Research Communications, 500, 924–929. https://doi.org/10.1016/j.bbrc.2018.04.201
Fang, Y., Yu, H., Liang, X., Xu, J., & Cai, X. (2014). Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biology & Therapy, 15, 1268–1279. https://doi.org/10.4161/cbt.29691
Li, S., et al. (2018). Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res, 10, 3383–3392. https://doi.org/10.2147/CMAR.S173941
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136, 215–233. https://doi.org/10.1016/j.cell.2009.01.002
Huang, L. L., et al. (2017). Potential role of miR-139-5p in cancer diagnosis, prognosis and therapy. Oncology Letters, 14, 1215–1222. https://doi.org/10.3892/ol.2017.6351
Krishnan, K., et al. (2013). miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA, 19, 1767–1780. https://doi.org/10.1261/rna.042143.113
Bushati, N., & Cohen, S. M. (2007). microRNA functions. Annual Review of Cell and Developmental Biology, 23, 175–205. https://doi.org/10.1146/annurev.cellbio.23.090506.123406
Adam, L., et al. (2009). miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clinical Cancer Research, 15, 5060–5072. https://doi.org/10.1158/1078-0432.CCR-08-2245
Qiu, G., Lin, Y., Zhang, H., & Wu, D. (2015). miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochemical and Biophysical Research Communications, 463, 315–321. https://doi.org/10.1016/j.bbrc.2015.05.062
Asangani, I. A., et al. (2008). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene, 27, 2128–2136. https://doi.org/10.1038/sj.onc.1210856
Shao, Y., Liang, B., Long, F., & Jiang, S. J. (2017). Diagnostic MicroRNA biomarker discovery for non-small-cell lung cancer adenocarcinoma by integrative bioinformatics analysis. BioMed Research International, 2017, 2563085. https://doi.org/10.1155/2017/2563085
Survival Analysis v. 2.37–7 (2014).
Robinson, M. D., McCarthy, D. J., & Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England), 26, 139–140. https://doi.org/10.1093/bioinformatics/btp616
Zhuang, L., Yang, Z., & Meng, Z. (2018). Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted worse overall survival and disease-free survival in hepatocellular carcinoma patients. BioMed research international, 2018, 7897346. https://doi.org/10.1155/2018/7897346
Wang, S., Sun, H., Zhan, X., & Wang, Q. (2020). MicroRNA718 serves a tumorsuppressive role in nonsmall cell lung cancer by directly targeting CCNB1. International Journal of Molecular Medicine, 45, 33–44. https://doi.org/10.3892/ijmm.2019.4396
Choudhury, T. R. Is Hepatoglobin therapy justified? J Assoc Physicians India 39, 724; author reply 724–725 (1991).
Greenawalt, E. J., et al. (2019). Targeting of SGK1 by miR-576-3p inhibits lung adenocarcinoma migration and invasion. Molecular Cancer Research, 17, 289–298. https://doi.org/10.1158/1541-7786.MCR-18-0364
Pan, Z. H., Guo, X. Q., Shan, J., & Luo, S. X. (2018). LINC00324 exerts tumor-promoting functions in lung adenocarcinoma via targeting miR-615-5p/AKT1 axis. European Review for Medical and Pharmacological Sciences, 22, 8333–8342. https://doi.org/10.26355/eurrev_201812_16531
Zhu, D., et al. (2019). MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathology, Research and Practice, 215, 861–872. https://doi.org/10.1016/j.prp.2019.01.029
Zhou, L. Y., Zhang, F. W., Tong, J., & Liu, F. (2020). MiR-191-5p inhibits lung adenocarcinoma by repressing SATB1 to inhibit Wnt pathway. Molecular Genetics & Genomic Medicine, 8, e1043. https://doi.org/10.1002/mgg3.1043
Wang, F., Chen, X., Yu, X., & Lin, Q. (2019). Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination promotes cell cycle progression and proliferation of non-small cell lung cancer cells. Am J Transl Res, 11, 7166–7185.
Wang, J., et al. (2020). PKMYT1 is associated with prostate cancer malignancy and may serve as a therapeutic target. Gene, 744, 144608. https://doi.org/10.1016/j.gene.2020.144608
Li, P., Xiao, Z., Luo, J., Zhang, Y., & Lin, L. (2019). MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. Journal of Cellular and Molecular Medicine, 23, 2475–2488. https://doi.org/10.1111/jcmm.14121
Liu, J., et al. (2018). Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell International, 18, 51. https://doi.org/10.1186/s12935-018-0545-8
Yong-Hao, Y., Xian-Guo, W., Ming, X., & Jin-Ping, Z. (2019). Expression and clinical significance of miR-139-5p in non-small cell lung cancer. Journal of International Medical Research, 47, 867–874. https://doi.org/10.1177/0300060518815379
Wang, K., Jin, J., Ma, T., & Zhai, H. (2017). MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. Journal of Cellular and Molecular Medicine, 21, 3730–3740. https://doi.org/10.1111/jcmm.13282
Pajic, M., et al. (2018). miR-139-5p modulates radiotherapy resistance in breast cancer by repressing multiple gene networks of DNA repair and ROS defense. Cancer Research, 78, 501–515. https://doi.org/10.1158/0008-5472.Can-16-3105
Pang, C., et al. (2016). MiR-139-5p is increased in the peripheral blood of patients with prostate cancer. Cellular physiology and biochemistry: International journal of experimental cellular physiology, biochemistry, and pharmacology, 39, 1111–1117. https://doi.org/10.1159/000447819
Yang, B., et al. (2019). Downregulation of miR-139-5p promotes prostate cancer progression through regulation of SOX5. Biomedicine & Pharmacotherapy, 109, 2128–2135. https://doi.org/10.1016/j.biopha.2018.09.029
Wu, J., Zhang, T., Chen, Y., & Ha, S. (2020). MiR-139-5p influences hepatocellular carcinoma cell invasion and proliferation capacities via decreasing SLITRK4 expression. Bioscience Reports. https://doi.org/10.1042/BSR20193295
Wu, J., et al. (2019). LncSNHG3/miR-139-5p/BMI1 axis regulates proliferation, migration, and invasion in hepatocellular carcinoma. OncoTargets and Therapy, 12, 6623–6638. https://doi.org/10.2147/ott.S196630
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no potential conflicts of interest.
Ethical Approval
Not applicable.
Consent for Publication
All authors consent to submit the manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12033_2022_465_MOESM1_ESM.tif
Supplementary Fig. 1 LUAD malignant progression was suppressed by silencing CCNB1. A: CCNB1 level in H1299 cells; B: The proliferation ability of H1299 cells; C: The invasion of H1299 cells (100×); D: The migration ability of H1299 cells (40×); E: The cell cycle of H1299. The cell grouping settings were as follows: H1299 cells with si-NC and si-CCNB1. * denotes p<0.05 (TIF 3302 kb)
Rights and permissions
About this article
Cite this article
Bao, B., Yu, X. & Zheng, W. MiR-139-5p Targeting CCNB1 Modulates Proliferation, Migration, Invasion and Cell Cycle in Lung Adenocarcinoma. Mol Biotechnol 64, 852–860 (2022). https://doi.org/10.1007/s12033-022-00465-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00465-5