Abstract
The sericulture industry faces substantial economic losses due to severe pathogenic infections caused by fungi, viruses, and bacteria. The development of transgenic silkworms against specific pathogens has been shown to enhance disease resistance against a particular infection. A single gene or its products that can confer protection against multiple pathogens is required. In an attempt to develop silkworms with enhanced immunity against multiple pathogens, we generated transgenic silkworm lines with an overexpressed NF-kB transcription factor, Relish 1, under two different promoters. Separately, a potent anti-fungal gene, Drosomycin, was also expressed in transgenic silkworms. Both Relish 1 and Drosomycin transgenic silkworms had single copy genomic integration, and their mRNA expression levels were highly increased after infection with silkworm pathogens. The overexpression of the Relish 1 in transgenic silkworms resulted in the upregulation of several defense-related genes, Cecropin B, Attacin, and Lebocin, and showed enhanced resistance to Nosema bombycis (microsporidian fungus), Nucleopolyhedrovirus (BmNPV), and bacteria. The Drosomycin expressing transgenic silkworms showed elevated resistance to N. bombycis and bacteria. These findings demonstrate the role of Relish 1 in long-lasting protection against multiple pathogens in silkworms. Further, the successful introduction of a foreign gene, Drosomycin, also led to improved disease resistance in silkworms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sericulture is an agro-based industry practiced in China, India, Brazil, and other developing countries. The majority of commercial silk produced in the world comes from the silkworm, Bombyx mori. It and other silkworms are susceptible to various infectious diseases caused by pathogens. The major pathogens infecting silkworms are microsporidia, including Nosema bombycis, nucleopolyhedrovirus (NPV), densovirus (DNV), infectious flacherie virus (IFV), cytoplasmic polyhedrovirus (CPV), bacteria, and fungi, causing substantial crop loss in the sericulture industry [1, 2]. Therefore, the use of silkworm breeds resistant to diseases is a prerequisite for effective disease prevention and increased productivity. Conventional breeding strategies are labor-intensive, time-consuming and imprecise, whereas, through transgenesis, desired traits can be introduced into an organism with higher accuracy.
The use of transgenic technology to enhance disease resistance in silkworms is a promising strategy [3,4,5]. The development of individual transgenic silkworm lines for every pathogen to be controlled is challenging and not practical in sericulture. Hence, the need for a single gene or its products that can be used to develop transgenic silkworms conferring resistance to multiple pathogens is desirable. Several transgenic silkworms conferring protection against a particular pathogen such as BmNPV, BmCPV, N. bombycis, and bacteria by overexpressing endogenous/exogenous or by targeting pathogen-specific genes have been reported [6,7,8,9,10,11,12,13]. The Sprouty (Spry) protein is reported to regulate the activation of ERK and inhibit replications of different classes of silkworm viruses, NPV, CPV, and BDNV [14, 15]. To our knowledge, there are no reports on transgenic silkworms expressing a single gene to enhance immunity against multiple categories of pathogens (Fungi, viruses, and bacteria). Here, we show that overexpression of a single gene, Relish 1, can confer enhanced protection against multiple pathogens of silkworms. Analogously, we intended to look into whether a foreign gene and its promoter borrowed from Drosophila melanogaster would be functional in silkworms. Thus, transgenic silkworm expressing Drosomycin, a potent anti-fungal gene, was successfully developed and functionally tested.
Relish is a transcription factor that is an important part of the IMD (immunodeficient) signaling pathway. It is a key component in the induction of the humoral immune response in Drosophila and other insects, inducing anti-bacterial, anti-fungal, and anti-plasmodium factors [16,17,18]. The antimicrobial peptides (AMPs) under the regulation of Relish include Cecropin, Diptericin, Attacin, Defensin, and Metchnikowin [19]. Relish 1 and Relish 2 genes have been identified and characterized in insects, and Relish 1 is found to be more active than Relish 2 in silkworm, B. mori [20, 21]. Relish 1 consists of Rel homology domain (RHD), nuclear localization sequence (NLS), acidic and hydrophobic amino acid (AHAA)-rich region, ankyrin repeats, and death domain [20]. Relish 1 is present as an inactive form in the cytoplasm. It becomes active by endoproteolytic cleavage of ankyrin repeats and the death domain. In addition, Relish 1 lacking the ankyrin repeats (ANK) at the C-terminal region has been shown to strongly activate the promoters of antimicrobial genes in insect mbn-2 cells [20]. Hence, we chose Relish 1 as a candidate gene to test whether transgenic silkworm overexpressing the same can enhance immunity against different pathogens. The Drosomycin was isolated from bacteria- infected flies [22], effective against fungal infection but inactive against bacteria. Its expression is regulated by the Toll pathway in the fat body of D. melanogaster [23]. It is a 44-residue peptide composed of one α-helix and a twisted three-stranded β-sheet [24].
We have selected a productive and commercial breed of the silkworm, B. mori, CSR2, which is used in the sericulture industry in India, but susceptible to diseases. We generated transgenic silkworm lines expressing Relish 1 or Drosomycin under the control of constitutive (CMV) and inducible (Cecropin A or Drosomycin) promoters. Next, we compared their expression levels and resistance against protozoan/fungus (microsporidian, N. bombycis), virus (BmNPV), and gram-negative and positive bacteria, the major pathogens infecting silkworms.
Materials and Methods
Silkworm
The silkworm B. mori breed, CSR2, was reared on fresh mulberry leaves at 25 ± 1 °C with 75 ± 5% relative humidity. Rearing and grainage activities were carried out as per standard protocols. All transgenic procedures and maintaining of transgenic silkworms were followed as per the Institute Biosafety Committee procedures (Approval No. CSB/SBRL/IBSC/2017-18-Project Code AIT 3540). Silkworm pathogens used in the study were as reported by us earlier [1].
Vector Constructions
The piggyBac (PB) transposon vector (System Biosciences, USA) consists of CMV promoter for transgene expression and green fluorescent protein (GFP) reporter gene under the elongation factor 1α (EF1α) promoter, was used for making transgenes. The active form of Relish 1 lacking ankyrin repeats and death domain was used separately under CMV and Cecropin A promoters. An 804 bp DNA fragment of Cecropin A promoter (Accession No. D84395.1, from nt 3 to 806) was amplified from genomic DNA using primer 1F & 1R with an additional NFĸB site incorporated in the forward primer (Table 1). An 1881 bp Relish 1 gene (Accession No. AB298441.1, from nt 324 to 2205) was amplified from fat body cDNA of B. mori larvae using primers, 2aF/2bF & 2R. For Drosomycin promoter, a 1168 bp DNA fragment, 5’ upstream regulatory region of Drosomycin gene was amplified from genomic DNA of D. melanogaster using the primers 3F & 3R. A 213 bp Drosomycin gene (Accession No. NM_079177.4 from nt 1 to 213) was amplified from fat body cDNA of D. melanogaster using primers, 4aF & 4aR or 4bF & 4bR. The GFP and SV40 poly A signal was individually amplified from PB plasmid employing primers 5F & 5R and 6F & 6R. The PCR amplification was set with an initial denaturation of 95 °C for 2 min, followed by 30 cycles at 95 °C for 30 s, 68 °C for 30 s and 72 °C for 1 min/kb using Platinum Taq DNA polymerase (Thermo Fisher Scientific, USA). For SV40 amplification, the annealing temperature was 52 °C. The amplified Cecropin A or Drosomycin promoter, Relish 1 or Drosomycin, SV40 poly A signal and GFP after restriction digestions were ligated in-frame to PB plasmid separately to generate the vector constructs: PBCMV-Relish1-SV40, PBCecropinA-Reishl1-SV40, PBCecropinA-GFP-SV40, PBCMV-Drosomycin-SV40, PBDrosomycin-Drosomycin-SV40 and PBDrosomycin-GFP-SV40], transformed in E. coli (DH5α) and plasmids were purified. The plasmids were sequenced to confirm their integrity and correctness of the frame. Finally, six transgenic PB vectors were constructed and designated as PBCMVPRelish1, PBCecAPRelish1, PBCecAPGFP, PBCMVPDrs, PBDrsPDrs, and PBDrsPGFP. The list of primers and their product sizes which include the primer sequences are given in Table 1. Restriction enzymes (New England BioLabs, USA) and other reagents were purchased from Thermo Fisher Scientific, USA.
Electroporation and Microinjection
Female moths were allowed to lay eggs post 4 h of copulation. Eggs were disinfected with 2% formalin for 5 min and then washed with water. To prevent the embryo from entering into a diapause state, eggs were treated with HCl (specific gravity of 1.110) at 25 °C for 90 min and washed under running water and air-dried [25]. Transgenic silkworms were essentially generated using previously described methods [9, 26]. Eggs were collected in a 0.4 cm chilled electroporation cuvette containing 10:1 of vector and helper vector (System Biosciences, USA) in deionized water. Electroporation was performed using Gene Pulser (BioRad, USA) at constant resistance of 200 Ω, the capacitance of 25 μF giving single pulse at 1700 V. For microinjection, eggs were arranged on a glass slide and microinjected with vector and helper (5:1) in injection buffer (0.5 mM Na3PO4 pH 7 and 5 mM KCl) using a microinjector (FemtoJet, Eppendorf, Germany). Electroporated/microinjected eggs were incubated at 25 °C and 75% relative humidity to allow hatching.
Screening and Selection of Transgenic Silkworm
The hatched G0 larvae were reared on fresh mulberry leaves under standard conditions, and each surviving moth was mated with a parental CSR2 moth to generate G1. Transgenic eggs, larvae, and pupae were screened for GFP at 480 nm under a fluorescent microscope (Euromex, Holland). The positive broods were inbred or backcrossed in subsequent generations. Apart from fluorescent screening, at each generation, the presence of GFP, Relish 1, and Drosomycin transgenes was confirmed by PCR using gene/vector-specific primers 5F & 5R, 7F & 7R, and 4aF & 4aR or 4bF & 4bR, respectively, using genomic DNA of moths.
Quantitative Real-Time PCR Analyses
Total RNA was extracted from the fat body of 5th instar (day 1) infected and non-infected larvae using RNeasy Kit (Qiagen, Germany). Contaminating DNA was removed using RNase-free DNase I (Thermo Fisher Scientific, USA). First-strand cDNA was synthesized from 2 μg of RNA using RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, USA) employing oligo(dT)18 primers. Resultant cDNA was used as a template for qPCR. The qPCR analysis was carried out on an ABI StepOne Plus real-time PCR (Applied Biosystems, USA) with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, USA), according to the manufacturer's instructions. All the reactions were carried out using three biological replicates. Expression of the Relish 1, Lebocin 4 (Accession No. AB003036.1), Cecropin B (Accession No. D11113) and Attacin (Accession No. NM_001043541.1) was quantified using primers 8F & 8R, 9F & 9R, 10F & 10R and 11F & 11R, respectively (Table1). Drosomycin was quantified using primers 12F & 12R. Relative quantification using a modified comparative Ct method [27] was employed for gene expression analysis. The relative expression levels of respective genes were calculated by normalization to GAPDH (Accession: NM_001043921.1) using primers 13F & 13R.
Inverse PCR
Genomic DNA was extracted from transgenic silkworms and digested with KpnI and NdeI, and self-ligated. The products were then used as templates for inverse PCR with primers specific for the transposon, pBacL (14F & 14R) and pBacR (15F & 15R) as reported [7, 28]. The PCR products were cloned into the pJET vector (Thermo Fisher Scientific, USA) or directly sequenced, then analyzed using the silkworm genome database, silkDB (https://silkdb.bioinfotoolkits.net/).
Mortality Analyses
B. mori Nucleopolyhedrovirus (BmNPV, 3000 polyhedra/larva), N. bombycis (3000 spores/larva) or bacteria—Escherichia coli (JM 109) or Staphylococcus aureus (ATCC 25923) (104 CFU/ml/larva) were injected into the hemocoel of transgenic and control larvae (5th instar, day 1) at G7. Infected and non-infected non-transgenic larvae were used as controls. Surviving larvae were collected at 0, 6 and 12 h or 0, 4, and 8 h for gene expression analyses. The mortality rate was calculated for each group till moth emergence. The experiment was carried out in three groups with 100 larvae in each group. Microscopic examination was performed in the moth stage to determine the presence of injected pathogens. Comparative expression levels of transgenes were obtained by quantitative real-time PCR as above.
Economical Characters
The economical characters of control and transgenic silkworms were recorded in G7. Pupation rate was determined from the percentage of healthy pupae to the numbers of larvae at 5th instar, day 1, from 5 random batches. Fecundity was obtained by counting the number of eggs laid by 15 moths, and the hatching percentage was calculated by considering the number of larvae hatched from the total number of eggs laid. Cocoon and shell weight of 15 randomly selected cocoons were obtained.
Statistical Analyses
All statistical values are represented as the mean ± SE and analyzed by Student’s t-test at *p < 0.05 and **p < 0.01 significance levels. The experiments were performed in triplicates unless otherwise stated.
Results
Generation and Screening of Transgenic Silkworms Overexpressing Relish 1 and Expressing Drosomycin
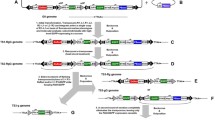
The transgenic vectors, PBCMVPRelish1, PBCecAPRelish1, PBCecAPGFP, PBCMVPDrs, PBDrsPDrs, and PBDrsPGFP, were generated for overexpression/expression of the Relish 1 (active form) or Drosomycin under the control of constitutive and inducible promoters (Fig. 1). GFP under EF1α promoter was used as a reporter to screen transgenic silkworms (Fig. 1). Each vector was mixed with transposase encoding a helper vector and was introduced into silkworm embryos of CSR2 breed by electroporation or microinjection. The G0 moth (s) was crossed with wild moth (s) to produce heterozygous G1 broods. The success percentage was determined from the number of G1 broods having GFP-positive larvae to the number of G0 moths backcrossed. A difference in efficiency was observed in the transformation between the two gene delivery methods. In G1 broods, 16.6% and 9.5% of silkworms were found to be expressing GFP by electroporation and microinjection methods, respectively (Table 2). As shown in Fig. 2, GFP expression was observed in eggs (b’), 3rd instar larva (d’), pupae (f’), and cocoon (h’) of transgenic silkworms compared with that of controls (a’, c’, e’, and g’). The GFP expression in the fat body of CecAGFP transgenic larvae confirms that Cecropin A promoter (including an extra NFκB binding site) activated GFP transcription in the fat body (Fig. 2j’). The vector construction also included a GFP under the control of the Drosomycin promoter (Fig. 1f), which showed the same results. The G1 transgenic silkworms were inbred or backcrossed for generations to obtain stable transgenic lines. The transgenic lines showing good fitness, economical characteristics and lifespan equivalent to controls were selected from each generation and continued breeding for further generations.
Generation of transgenic lines of B. mori. Schematic representation of overexpression/expression constructs in piggyBac (PB). a PBCMVPRelish1: Relish1 without C-terminal domain under the control of CMV (Cytomegalovirus) promoter; b PBCecAPRelish1: Relish1 without C-terminal domain under the control of Cecropin A promoter; c PBCecAPGFP: GFP under Cecropin A promoter; d PBCMVPDrs and e PBDrsPDrs: Drosomycin under the control of CMV and Drosomycin promoters; f PBDrsPGFP: GFP under Drosomycin promoter; SV40: polyadenylation signal; pBacR and pBacL: right and left terminal inverted repeats of piggyBac vector; CecA P: Cecropin A promoter; Drs P: Drosomycin promoter; Drs: Drosomycin; Ef1α P: Elongation factor-1alpha promoter. Restriction enzyme sites are indicated
GFP expression at different developmental stages of transgenic silkworms. Representative photographs were taken under white (a–j) or GFP (480 nm) (a’–j’) in G4. GFP fluorescence was observed in egg (b’), 3rd instar larval (d’), pupal (f’) stages, cocoon (h’) and larval fat body (j’). No GFP fluorescence was observed in egg (a’), 3rd instar larval (c’), pupal (e’) stages, cocoon (g’) and larval fat body (i’). Control silkworms: (a, a’), (c, c’), (e, e’), (g, g’), and (i, i’); Transgenic silkworms: (b, b’), (d, d’), (f, f’), (h, h’), and (j, j’). Scale bars—a, a’, b, b’: 0.2 mm; c, c’, d, d’: 2 mm; e, e’, f, f’: 3 mm; g, g’, h, h’: 5 mm; i, i’, j, j’: 4 mm
Apart from screening GFP expression microscopically, PCR amplification was carried out by GFP-specific primers using genomic DNA as templates. As shown in Fig. 3a–d, amplification of GFP resulted in a single band of 765 bp from CMVPRelish1, CecAPRelish1, CecAPGFP, and DrsPDrs transgenic silkworms, respectively. Further, to identify the Relish 1 transgene, genomic DNA extracted from CMVPRelish1 and CecAPRelish1 and control silkworms were subjected to PCR using a combination of gene and vector-specific primers which gave a discrete band of 1935 bp only in the former two (Fig. 3e, f). Likewise, a single band of 233 or 231 bp was observed when genomic DNA from CMVPDrs and DrsPDrs were subjected to PCR using Drosomycin primers (Fig. 3g, h). All the PCR products were sequenced to confirm their authenticity (Data not shown).
Screening of transgene by PCR amplification using gDNA in G2. a–d GFP detection showing a band of 765 bp in CMVPRelish1, CecAPRelish1, CecAPGFP, and DrsPDrs, respectively; e and f A band of 1935 bp representing Relish1 in CMVPRelish1 and CecAPRelish1, respectively; g and h Drosomycin detection yielded a band at 233 or 231 bp in CMVPDrs and DrsPDrs, respectively. Transgenic PB plasmids were used as positive control (C1) and gDNA from control moths (C2) was used as negative control. 1–4: Each lane shows the presence or absence of the respective transgene PCR product from random selection of moths; i Inverse PCR analysis of genomic insertion sites of CMVPRelish1, CecAPRelish1, CecAPGFP, CMVPDrs, DrsPDrs, and DrsPGFP
Analyses of Insertion Sites of the Transgenes
Inverse PCR was performed to detect and confirm the insertion sites of transgenes in silkworm lines. A single band was observed in PCR amplifications employing pBacL and pBacR primer pairs, indicating that transgenic insertion occurred as a single copy (Supplementary Fig. S1). Sequencing analysis showed that all the transgenic insertions were in intergenic regions of the silkworm genome. CMVPRelish1 was inserted on chromosome 14, and the nearest left and right genes identified were BGIBMGA009544 and BGIBMGA009543, which were 45.85 and 1.14 kb away from the insertion sites, respectively. The insertion site of CecAPRelish1 was found on chromosome 18, while the nearest genes on the left and right ends were BGIBMGA008250 and BGIBMGA008264, which were 2.8 and 4.5 kb away from the insertion site, respectively. The CecAPGFP insertion was observed on chromosome 11, and the nearest genes identified were BGIBMGA012123 and BGIBMGA012124, on the left and right sides that were 45.43 and 80.04 kb away from the insertion site, respectively. The CMVPDrs was inserted on chromosome 21, while the nearest gene on the left was BGIBMGA012446, and the gene on the right was BGIBMGA012445 which were 45 and 37 kb away from the insertion site, respectively. The insertion of DrsPDrs was found on chromosome 10, and the nearest genes to the left and right were BGIBMGA006565 and BGIBMGA013103, which were 1.2 and 8.5 kb away from the insertion site, respectively. The insertion of DrsPGFP occurred on chromosome 3, while the nearest genes were BGIBMGA007425 and BGIBMGA007424, which were 8.2 and 3.4 kb away from the insertion site on the left and right side, respectively (Fig. 3i).
Transgenes Show Increased Expression and Enhanced AMP Production
The expression level of Relish 1 transcripts in 5th instar larvae was determined post-infection (p.i.) with N. bombycis, BmNPV, and bacteria (E. coli and S. aureus). Without the immune challenge, the expression levels of Relish 1 transcripts were low in the larval fat body at 0 h of infection (Fig. 4). However, Relish 1 mRNA was significantly increased in transgenic lines (p < 0.01) compared to controls in response to N. bombycis, BmNPV, and bacterial challenges (Fig. 4a–d). In response to N. bombycis infection, elevated levels of Relish 1 transcripts were observed at 6 h followed by a slight decline at 12 h p.i. (Fig. 4a). Further its expression was elevated by 5.28, 8.78, and 7.75-fold at 6 h p.i. and declined by 3.90, 6.20, and 6.00-fold at 12 h p.i. in control, CMVPRelish1, and CecAPRelish1, respectively (Fig. 4a). Interestingly, the level of expression of Relish 1 mRNA was better manifested (p < 0.05) in CMV promoter than that of CecA promoter at 6 h p.i. However, no significant level of its expression was found between CMV and CecA promoters at 12 h p.i. (Fig. 4a). Similar expression patterns were observed in the case of BmNPV infection (Fig. 4b). The transcript levels were elevated by 5.13, 9.28, and 8.50-fold at 6 h p.i. and declined by 3.76, 7.03, and 6.82-fold at 12 h p.i. in control, CMVPRelish1, and CecAPRelish1, respectively (Fig. 4b). Notably, the transcript levels in CMVPRelish1 and CecAPRelish1 transgenic silkworms were higher against E. coli than S. aureus 4 h and declined 8 h p.i. when compared to control silkworms (Fig. 4c, d). The transcripts of Relish 1 were elevated by 42.56, 84.51, and 80.32-fold at 4 h p.i and then declined to 33.36, 65.97, and 63.24-fold at 8 h p.i. in control, CMVPRelish1 and CecAPRelish1, respectively (Fig. 4c). The same in response to S. aureus was 9.76, 15.38, and 14.1-fold at 4 h p.i. and 5.98, 10.24, and 8.97-fold at 8 h p.i. (Fig. 4d). No significant difference in expression was observed between CMV and CecARelish1 groups. Further, to confirm whether the overexpression of Relish 1 activated the IMD pathway, the expression levels of AMP genes were quantified post 4 h of E. coli challenge. As shown in Fig. 4e and f, the transcript levels of AMPs-Cecropin B, Attacin, and Lebocin 4 were significantly elevated in transgenic silkworms compared to controls. Expression of these AMPs manifested better in CMVPRelish1 than in CecAPRelish1.
Quantitative expression analyses of Relish 1, AMPs and Drosomycin in transgenic silkworms. a–d expression levels of Relish 1 against N. bombycis, BmNPV, E. coli and S. aureus, respectively. *p ˂ 0.05 and **p ˂ 0.01 with respect to control. e–f expression levels of Cecropin B, Attacin and Lebocin against E. coli. *p ˂ 0.05 and **p ˂ 0.01 with respect to control. g–j expression levels of Drosomycin against N. bombycis, BmNPV, E. coli, and S. aureus, respectively. *p ˂ 0.05 and **p ˂ 0.01 with respect to CMVPDrs. All data belong to G7
Drosomycin was found to be expressed in transgenic silkworms. The abundance of Drosomycin transcript was determined in response to N. bombycis and bacterial (E. coli and S. aureus) infections but not to BmNPV in CMVPDrs and DrsPDrs transgenic lines (Fig. 4g–j). Differing from Relish1, Drosomycin under its promoter significantly (p < 0.01) exhibited a higher expression level than under the CMV promoter. No significant difference was observed in the expression of Drosomycin between CMVPDrs and DrsPDrs at 4 or 6 h p.i. The expression levels of Drosomycin were not significantly altered against BmNPV infection (Fig. 4h). Transcript levels were significantly (p < 0.01) elevated from 6 to 12 h in response to N. bombycis (Fig. 4g) and from 4 to 8 h in response to bacterial infection in DrsPDrs (Fig. 4i, j). Expression level of Drosomycin was elevated from 1.59 at 6 h p.i. to 6.36-fold at 12 h p.i. in DrsPDrs against N. bombycis (Fig. 4g). It was increased from 1.36-fold at 4 h p.i. to 4.82-fold at 8 h p.i. in response to E. coli (Fig. 4i) and from 2.06 at 4 h p.i. to 8.36-fold at 8 h p.i. in response to S. aureus (Fig. 4j). However, no significant difference in hike of expression was observed from 6 to 12 h in response to N. bombycis and from 4 to 8 h in response to bacterial infections in CMVPDrs. Remarkably, the expression of Drosomycin was found to be more in DrsPDrs at 8 h against S. aureus infection (Fig. 4j). Together, these results indicated that the overexpression and expression of Relish 1 and Drosomycin, respectively, were successful and led to an increased immune response in transgenic silkworms.
Transgenic Silkworms Show Increased Resistance Against Pathogens and Thus Survivability
To determine the anti-pathogenic capacity of the transgenic silkworm, the mortality rate of transgenic and non-transgenic larvae was analyzed against N. bombycis, BmNPV, and bacterial infections. The fungal, viral or bacterial susceptibility of transgenic CMVPRelish1 and CecAPRelish1 silkworms were compared with non-transgenic silkworms, which showed higher mortality. The percentage of mortality of CMVPRelish1, CecAPRelish1 transgenic, and control silkworms was 42, 45, and 98 against N. bombycis (Fig. 5a), 51, 53, and 96 against BmNPV (Fig. 5b), 40, 43, and 80 against E. coli (Fig. 5c), and 46, 43, and 85 against S. aureus (Fig. 5d), respectively. Further, CMVPRelish1 showed a higher survival rate than CecAPRelish1 and was consistent with the high transcript levels observed. The Drosomycin was introduced into silkworms successfully and transgenic silkworms were challenged with N. bombycis, BmNPV, and bacteria. The transgenic lines CMVPDrs, and DrsPDrs showed a mortality rate of 51 and 45% against N. bombycis compared to 97% in controls (Fig. 5e), whereas in BmNPV infection, mortality rates were high in transgenic lines (82 and 80%) similar to control (83%) (Fig. 5f). Mortality rates were 56, 49, and 80% against E. coli (Fig. 5g), and 50, 39, and 85% against S. aureus (Fig. 5h) for transgenic lines and controls, respectively. These results indicate significant protection of transgenic silkworms expressing Drosomycin against N. bombycis and bacterial infections.
Mortality analyses of transgenic silkworms. Non-Inf and Inf-C: control without and with infection, respectively. a–d Transgenic Relish 1 and e–h Drosomycin silkworms. Transgenic silkworm lines show reduced mortality upon infection with various pathogens as indicated (G7). **p ˂ 0.01 with respect to infected control
Commercial Traits are not Affected in Transgenic Silkworms
To evaluate whether the overexpression/expression of Relish 1 or Drosomycin affected the economical characteristics of transgenic lines, we analyzed the shell rate, fecundity, pupation rate, and hatching in G7. The results are presented in Table 3. The transgenic silkworms were found weak in the initial few generations compared to control silkworms. After recurrent selective breeding, most economic traits were almost at par with control silkworms at G7. The pupation rate, fecundity, and hatching were slightly higher in control silkworms than that of transgenic ones. Our results show that the transgenic silkworms have commercial traits almost similar to that of non-transgenic lines.
Discussion
The silkworm, CSR2 breed is a bivoltine hybrid that is commercially used for silk production in India but is highly susceptible to infections. We used transgenic technology to confer enhanced immunity to the silkworm by overexpressing its Relish 1 gene. We designed transgenic silkworms with the BmRelish 1 gene lacking the C-terminal domain [20], an active form of Relish 1, under a constitutive and an inducible promoter, CMV and Cecropin A, respectively. Our results demonstrate that both promoters can drive the overexpression of Relish 1 and the reporter gene, GFP. In the transgene delivery methods, the percentages of GFP-positive G1 broods produced from electroporation and microinjection are 16.6 and 9.5, respectively. The efficiency of introducing a transgene into silkworm eggs was voltage-dependent and showed a high difference between polyvoltine and bivoltine silkworms. Further, the acid treatment of the electroporated 54A embryos may be responsible for the low efficiency [29]. In the present study, we obtained an efficiency of 16.6% at 1700 V for the bivoltine CSR2 breed.
The overexpression of the active form of Relish 1 in transgenic silkworms enhanced their anti-pathogenic activity to major pathogens of silkworms, microsporidian (N. bombycis), BmNPV, and gram-negative and positive bacteria. High levels of Relish 1 mRNA are expressed in transgenic silkworms compared to the controls in response to infections. Injections of pathogens to silkworms result in quick immune responses as evident by the Relish 1 expression at the early stages of infection. Our results are consistent with the expression patterns of Relish 1 and other immune-related genes in response to infections with bacteria, BmNPV, and N. bombycis in silkworms [20, 30,31,32]. The Relish 1 was able to induce the downstream expression of AMP genes, Cecropin B, Attacin, and Lebocin 4 at higher levels than in control silkworms. Interestingly, both BmNPV infection and cGAMP stimulation resulted in the activation of the IMD pathway and further, BmSTING promoted the processing of Relish to activate an antiviral immune response in silkworms [32]. The Relish is also reported to be expressed in response to N. bombycis infection in silkworms [33]. Further, BmSTING regulates the infection of N. bombycis in an LC3 (microtubule-associated protein) -dependent manner in the early stage of infection and then activates Relish as a protective response to survive N. bombycis infection [34].
Mainly Toll pathway is activated by gram-positive bacteria, and the IMD pathway responds to gram-negative bacterial infection. The Relish 1 expression in silkworm is upregulated stronger against E. coli than S. aureus [20]. The transgenic mosquitoes overexpressing IMD-Rel2 (a homologue of Drosophila Relish) show greater resistance to protozoan Plasmodium and gram-negative and positive bacteria [18, 35]. Further, they showed Rel2 overexpression resulted in transcriptional activation of Defensins, Cecropins, and other AMPs. Hence, most of the AMP genes can be regulated by either pathway [36, 37]. In Tenebrio molitor, knockdown of IMD transcript significantly reduced host resistance to E. coli and fungus Candida albicans [38]. Our results show that the overexpression of Relish 1 is effective against major pathogens of the silkworm, a protozoan/fungus (N. bombycis), a virus (BmNPV), and both gram-negative and positive bacteria showing its potential in combating multiple pathogens. Though both promoters effectively drive the expression of Relish 1, the CMV promoter performed comparatively better than Cecropin A promoter. The reasons for this difference require further study.
The effect of transgenic modification of Relish 1 on silkworm immunity is evaluated by a survival test. The overexpression of Relish1 in transgenic lines results in reducing the mortality rate significantly in response to infections by major pathogens. On average, we observed the resistance of 56.5, and 48% against N. bombycis and BmNPV, respectively, and 58.5 and 52.5% to gram-negative and positive bacteria, respectively, in transgenic silkworms. Previous reports have shown enhanced resistance due to overexpression/expression of endogenous/exogenous genes and anti-pathogen genes in transgenic silkworms. Resistance to BmNPV is achieved by overexpression of Bmlipase and expression of exogenous hycu-ep32 genes [7, 8], and knocking down of BmNPV genes using transgenic RNAi [6, 39]. In addition, RNAi-based downregulation of a nuclear receptor, BmNHR96, which promotes BmNPV entry into the silkworm cells, effectively enhanced survivability of silkworm against BmNPV infection [9]. The overexpression of the BmPGRP-S2 gene resulted in the activation of IMD, Relish, and AMP genes Attacin 2, Gloverin 2, and Moricin, and has shown resistance against BmCPV (cytoplasmic polyhedrosis virus) [10]. Knockdown of BmCPV and BmDNV (Densovirus) genes enhanced the antiviral capacity of transgenic silkworms [40, 41]. Further, BmSpry is involved in resistance against three silkworm viruses [14, 15]. Congruent with these findings, in our study, overexpression of Relish 1 resulted in increased resistance of transgenic silkworms to multiple pathogens. To our knowledge, this is the first study demonstrating overexpression of a single gene can confer greater resistance to multiple pathogens of different categories in silkworms. The active Relish 1 led to the upregulation of effector AMPs that protect against invading pathogens, and it may also be due to the production of other factors that are controlled by Relish 1. Further, our study shows it is a good strategy to improve disease resistance by developing transgenic silkworms overexpressing a transcription factor.
Drosomycin is a key antimicrobial peptide involved in Toll signaling and innate immune responses primarily against fungal infections in Drosophila [42]. Recombinant Drosomycin is shown to have an inhibitory effect on several fungi, yeast, and a protozoan parasite, Plasmodium berghei [43]. Microsporidia, including N. bombycis, which causes the destructive “pebrine” disease in silkworms, were earlier classified under protozoa but now reported to be closely related to fungi [44]. The successful expression of Drosomycin from D. melanogaster in transgenic silkworm effectively reduced the N. bombycis and bacterial infections. The level of resistance of immune-enhanced transgenic silkworm lines, CMVPDrs, and DrsPDrs to N. bombycis infection is 49 and 55%, respectively. The expression of Drosomycin in transgenic silkworms may also help to combat other fungi, such as Beauveria bassiana which causes “muscardine” disease in silkworms. Drosomycin has been highly expressed in response to B. bassiana infection in D. melanogaster [45]. It may also help in combatting Aspergillus spp., which causes aspergillosis in young silkworm larvae. Enhanced resistance to A. fumigatus was observed in transgenic D. melanogaster expressing Drosomycin peptide constitutively [46]. The transcript level of Drosomycin was higher in response to S. aureus, and hence it could be speculated that the transgenic silkworms may survive against flacherie disease caused by gram-positive bacteria. Toll pathway activation in response to gram-positive bacteria has been shown to control the expression of the Drosomycin gene [23, 47]. The AMPs including Gloverins, Lebocins, and Moricins are strongly upregulated against N. bombycis infection in silkworms and followed the Toll pathway [48]. However, Drosomycin is not found to be effective against BmNPV infection in silkworms. It has been shown that unlike in fungal and bacterial infections, the expression of a single AMP provides no resistance to viral infections in Drosophila [49]. Our results demonstrate the enhancement of the silkworm innate immune system through the successful introduction of a foreign recombinant gene driven by its promoter.
The commercial traits such as cocoon and shell weight, silk yield, fecundity, and hatching in transgenic CSR2 lines were at par with control CSR2, after continuous selection and breeding. This process is continuing in our laboratory further to enhance the level of resistance among transgenic silkworms. Cross-breeding of transgenic lines will also help improve resistance. Overall, we have developed transgenic silkworms with enhanced immunity to major pathogens. Our findings also provide a strategy for enhancing disease resistance against multiple pathogens by the overexpression of a single gene.
References
Ravikumar, G., Raje Urs, S., Vijaya Prakash, N. B., Rao, C. G., & Vardhana, K. V. (2011). Development of a multiplex polymerase chain reaction for the simultaneous detection of microsporidians, nucleopolyhedrovirus, and densovirus infecting silkworms. Journal of Invertebrate Pathology, 107, 193–197.
Tayal, M. K., & Chauhan, T. P. S. (2017). Silkworm diseases and pests. Industrial entomology (pp. 265–289). Springer.
Xia, Q., Li, S., & Feng, Q. (2014). Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annual Review of Entomology, 59, 513–536.
Jiang, L., & Xia, Q. (2014). The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochemistry and Molecular Biology, 48, 1–7.
Jiang, L., Goldsmith, M. R., & Xia, Q. (2021). Advances in the arms race between silkworm and baculovirus. Frontiers in Immunology, 12, 628151.
Kanginakudru, S., Royer, C., Edupalli, S. V., Jalabert, A., Mauchamp, B., Chandrashekaraiah, Prasad, S. V., Chavancy, G., Couble, P., & Nagaraju, J. (2007). Targeting ie-1 gene by RNAi induces baculoviral resistance in Lepidopteran cell lines and in transgenic silkworms. Insect Molecular Biology, 16, 635–644.
Jiang, L., Wang, G., Cheng, T., Yang, Q., Jin, S., Lu, G., Wu, F., Xiao, Y., Xu, H., & Xia, Q. (2012). Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Archives of Virology, 157, 1323–1328.
Jiang, L., Cheng, T., Zhao, P., Yang, Q., Wang, G., Jin, S., Lin, P., Xiao, Y., & Xia, Q. (2012). Resistance to BmNPV via overexpression of an exogenous gene controlled by an inducible promoter and enhancer in transgenic silkworm Bombyx mori. PLoS ONE, 7, e41838.
Yang, J. G., Liu, T. H., Dong, X. L., Wu, Y. F., Zhang, Q., Zhou, L., Chen, P., Lu, C., & Pan, M. H. (2017). In vivo RNA interference of BmNHR96 enhances the resistance of transgenic silkworm to BmNPV. Biochemical and Biophysical Research Communications, 493, 332–339.
Zhao, P., Xia, F., Jiang, L., Guo, H., Xu, G., Sun, Q., Wang, B., Wang, Y., Lu, Z., & Xia, Q. (2018). Enhanced antiviral immunity against Bombyx mori cytoplasmic polyhedrosis virus via overexpression of peptidoglycan recognition protein S2 in transgenic silkworms. Developmental & Comparative Immunology, 87, 84–89.
Dong, Z., Long, J., Huang, L., Hu, Z., Chen, P., Hu, N., Zheng, N., Huang, X., Lu, C., & Pan, M. (2019). Construction and application of an HSP70 promoter–inducible genome editing system in transgenic silkworm to induce resistance to Nosema bombycis. Applied Microbiology and Biotechnology, 103, 9583–9592.
Jiang, L., Liu, W., Guo, H., Dang, Y., Cheng, T., Yang, W., Sun, Q., Wang, B., Wang, Y., Xie, E., & Xia, Q. (2019). Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Frontiers in Immunology, 10, 776.
Xu, J., Luo, X., Fang, G., Zhan, S., Wu, J., Wang, D., & Huang, Y. (2020). Transgenic expression of antimicrobial peptides from black soldier fly enhance resistance against entomopathogenic bacteria in the silkworm, Bombyx mori. Insect Biochemistry and Molecular Biology, 127, 103487.
Guo, H., Sun, Q., Wang, B., Wang, Y., Xie, E., Xia, Q., & Jiang, L. (2019). Spry is downregulated by multiple viruses to elevate ERK signaling and ensure viral reproduction in silkworm. Development and Comparative Immunology, 98, 1–5.
Jiang, L. (2021). Insights into the antiviral pathways of the silkworm Bombyx mori. Frontiers in Immunology, 12, 639092.
Hedengren, M., Dushay, M. S., Ando, I., Ekengren, S., Wihlborg, M., & Hultmark, D. (1999). Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Molecular Cell, 4, 827–837.
Schluns, H., & Crozier, R. H. (2007). Relish regulates expression of antimicrobial peptide genes in the honeybee, Apis mellifera, shown by RNA interference. Insect Molecular Biology, 16, 753–759.
Antonova, Y., Alvarez, K. S., Kim, Y. J., Kokoza, V., & Raikhel, A. S. (2009). The role of NF-κB factor REL2 in the Aedes aegypti immune response. Insect Biochemistry and Molecular Biology, 39, 303–314.
Lemaitre, B., Reichhart, J. M., & Hoffmann, J. A. (1997). Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proceedings of the National Academy of Sciences, 94, 14614–14619.
Tanaka, H., Matsuki, H., Furukawa, S., Sagisaka, A., Kotani, E., Mori, H., & Yamakawa, M. (2007). Identification and functional analysis of Relish homologs in the silkworm, Bombyx mori. Biochimica et Biophysica Acta, 1769, 559–568.
Tanaka, H., Ishibashi, J., Fujita, K., Nakajima, Y., Sagisaka, A., Tomimoto, K., Suzuki, N., Yoshiyama, M., Kaneko, Y., Iwasaki, T., & Sunagawa, T., Yamaji, K., Asaoka, A., Mita, K., & Yamakawa, M. (2008). A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochemistry and Molecular Biology, 38, 1087–1110.
Fehlbaum, P., Bulet, P., Michaut, L., Lagueux, M., Broekaert, W. F., Hetru, C., & Hoffmann, J. A. (1994). Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. Journal of Biological Chemistry, 269, 33159–33163.
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., & Hoffmann, J. A. (1996). The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell, 86, 973–983.
Landon, C., Sodano, P., Hetru, C., Hoffmann, J., & Ptak, M. (1997). Solution structure of Drosomycin, the first inducible antifungal protein from insects. Protein Science, 6, 1878–1884.
Ullal, S., R., & Narasimhanna, M. N. (1994). Hand book of sericulture. In Sampath, J. (Ed). Central Silk Board.
Tamura, T., Thibert, C., Royer, C., Kanda, T., Abraham, E., Kamba, M., Komoto, N., Thomas, J. L., Mauchamp, B., Chavancy, G., Shirk Transgenic Res 123 P, Fraser, M., Prudhomme, J. C., & Couble, P. (2000). Germline transformation of the silkworm Bombyx mori. using a piggyBac transposon-derived vector. Nature Biotechnology, 8, 81–84.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols., 3, 1101–1108.
Handler, A. M., McCombs, S. D., Fraser, M. J., & Saul, S. H. (1998). The Lepidopteran transposon vector, piggyBac, mediates germline transformation in the Mediterranean fruit fly. Proceedings of the National Academy of Sciences, 95, 7520–7525.
Guo, X. Y., Dong, L., Wang, S. P., Guo, T. Q., Wang, J. Y., & Lu, C. D. (2004). Introduction of foreign genes into silkworm eggs by electroporation and its application in transgenic vector test. Acta Biochimia et Biophysia Sinica, 36, 323–330.
Huang, L., Cheng, T., Xu, P., Fang, T., & Xia, Q. (2012). Bombyx mori transcription factors: Genome-wide identification, expression profiles and response to pathogens by microarray analysis. Journal of Insect Science, 12, 1–24.
Li, Z. H., Pan, G. Q., Ma, Z. G., Han, B., Sun, B., Ni, Q., Chen, J., Li, T., Liu, T. B., Long, M. X., Li, C. F., & Zhou, Z. (2017). Comparative proteomic analysis of differentially expressed proteins in the Bombyx mori fat body during the microsporidia, Nosema bombycis infection. Invertebrate Pathology, 149, 36–43.
Hua, X., Li, B., Song, L., Hu, C., Li, X., Wang, D., Xiong, Y., Zhao, P., He, H., Xia, Q., & Wang, F. (2018). Stimulator of interferon genes (STING) provides insect antiviral immunity by promoting Dredd caspase-mediated NF-κB activation. Journal of Biologial Chemistry, 293, 11878–11890.
Hungund, S. P., Pradeep, A. N. R., Makwana, P., Sagar, C., & Mishra, R. K. (2020). Cellular defence and innate immunity in the larval ovarian disc and differentiated ovariole of the silkworm Bombyx mori induced by microsporidian infection. Invertebrate Reprodution and Development, 64, 10–21.
Hua, X., Xu, W., Ma, S., & Xia, Q. (2021). STING-dependent autophagy suppresses Nosema bombycis infection in silkworm, Bombyx mori. Developmental and Comparative Immunology, 115, 103862.
Dong, Y., Das, S., Cirimotich, C., Souza-Neto, J. A., McLean, K. J., & Dimopoulos, G. (2011). Engineered Anopheles immunity to Plasmodium infection. PLoS Pathogen, 7, e1002458.
De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M., & Lemaitre, B. (2002). The Toll and Imd pathways are the major regulators of the immune response in Drosophila. The EMBO Journal, 21, 2568–2579.
Tanaka, H., Sagisaka, A., Nakajima, Y., Fujita, K., Imanishi, S., & Yamakawa, M. (2009). Correlation of differential expression of silkworm antimicrobial peptide genes with different amounts of rel family proteins and their gene transcriptional activity. Bioscience, Biotechnology, and Biochemistry, 73, 599–606.
Keshavarz, M., Jo, Y. H., Park, K. B., Ko, H. J., Edosa, T. T., Lee, Y. S., & Han, Y. S. (2019). Tm DorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Scientific Reports, 9, 1–19.
Subbaiah, E. V., Royer, C., Kanginakudru, S., Satyavathi, V. V., Babu, A. S., Sivaprasad, V., Chavancy, G., Darocha, M., Jalabert, A., Mauchamp, B., Basha, I., Couble, P., & Nagaraju, J. (2013). Engineering silkworms for resistance to baculovirus through multigene RNA interference. Genetics, 193, 63–75.
Jiang, L., Peng, Z., Guo, H., Sun, J., Sun, Q., Xia, F., Huang, C., Xu, G., & Xia, Q. (2017). Enhancement of antiviral capacity of transgenic silkworms against cytoplasmic polyhedrosis virus via knockdown of multiple viral genes. Developmental & Comparative Immunology, 77, 138–140.
Sun, Q., Jiang, L., Guo, H., Xia, F., Wang, B., Wang, Y., Xia, Q., & Zhao, P. (2018). Increased antiviral capacity of transgenic silkworm via knockdown of multiple genes on Bombyx mori bidensovirus. Developmental & Comparative Immunology, 87, 188–192.
Valanne, S., Wang, J. H., & Ramet, M. (2011). The Drosophila toll signaling pathway. The Journal of Immunology, 186, 649–656.
Tian, C., Gao, B., del Carmen Rodriguez, M., Lanz-Mendoza, H., Ma, B., & Zhu, S. (2008). Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Molecular Immunology, 45, 3909–3916.
Keeling, P. (2009). Five questions about microsporidia. PLoS Pathogen, 5, e1000489.
Roxstrom-Lindquist, K., Terenius, O., & Faye, I. (2004). Parasite-specific immune response in adult Drosophila melanogaster: A genomic study. EMBO Reports, 5, 207–212.
Tzou, P., Reichhart, J. M., & Lemaitre, B. (2002). Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proceedings of the National Academy of Sciences, 99, 2152–2157.
Michel, T., Reichhart, J. M., Hoffmann, J. A., & Royet, J. (2001). Drosophila Toll is activated by gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature, 414, 756–759.
Ma, Z., Li, C., Pan, G., Li, Z., Han, B., Xu, J., Lan, X., Chen, J., Yang, D., Chen, Q., & Sang, Q. (2013). Genome-wide transcriptional response of silkworm (Bombyx mori) to infection by the microsporidian Nosema bombycis. PLoS ONE, 8, e84137.
Zambon, R. A., Nandakumar, M., Vakharia, V. N., & Wu, L. P. (2005). The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Sciences, 102, 7257–7262.
Acknowledgements
This work was supported by a Grant (No. AIT 3540) to RG and UN from Central Silk Board (CSB), Bengaluru, India. RSY, DST, CM, and GR are thankful to CSB for providing research fellowships.
Author information
Authors and Affiliations
Contributions
RSY performed experiments, data organization and analysis, and writing-first draft. DST, CM, and GR assisted in experiments, data organization and analysis, and writing-review. VK analyzed the data and review. RKM performed funding acquisition, resources and coordination. UN guided in experiments, supervision, writing-review and editing. RG did conceptualization, funding acquisition, guidance in experiments, supervision, and writing-review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rasalkar Sandhya Yashwant—Research Scholar of Jain University, Bengaluru, India under the guidance of RG.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yashwant, R.S., Thomas, D.S., Manoharan, C. et al. Transgenic Silkworms Overexpressing Relish and Expressing Drosomycin Confer Enhanced Immunity to Multiple Pathogens. Mol Biotechnol 64, 711–724 (2022). https://doi.org/10.1007/s12033-021-00438-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00438-0