Abstract
Introduction of selectivity/specificity into viral-based gene delivery systems, such as lentiviral vectors (LVs), is crucial in their systemic administration for cancer gene therapy. The pivotal role of tumor-associated endothelial cells (TAECs) in tumor angiogenesis and overexpression of vascular endothelial growth factor receptor-2 (VEGFR2 or KDR) in TAECs makes them a potent target in cancer treatment. Herein, we report the development of VEGFR2-targeted LVs pseudotyped with chimeric sindbis virus E2 glycoprotein (cSVE2s). For this purpose, either sequence of a VEGFR2-specific nanobody or its natural ligand (VEGF121) was inserted into the binding site of sindbis virus E2 glycoprotein. In silico modeling data suggested that the inserted targeting motifs were exposed in the context of cSVE2s. Western blot analysis of LVs indicated the incorporation of cSVE2s into viral particles. Capture ELISA demonstrated the specificity/functionality of the incorporated cSVE2s. Transduction of 293/KDR (expressing VEGFR2) or 293T cells (negative control) by constructed LVs followed by fluorescent microscopy and flow cytometric analyses indicated selective transduction of 293/KDR cells (30 %) by both targeting motifs compared to 293T control cells (1–2 %). These results implied similar targeting properties of VEGFR2-specific nanobody compared to the VEGF121 and indicated the potential for transductional targeting of tumor vasculature by the nanobody displaying LVs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer remains to be one of the main causes of morbidity and mortality worldwide. Advances in viral vector-based gene delivery systems have profoundly accelerated the viral-based gene therapy approaches in cancer [1]. A major drawback of these systems, however, is the lack of selectivity/specificity to tumor cells which impedes their “systemic administration.” This shortcoming restricted most of the clinical trials to ex vivo or intratumoral (in the case of solid tumors) studies [2, 3]. Therefore, a targeted viral vector that could selectively home in onto tumor cells while sparing the normal cells is the crucial tool for cancer gene therapy.
Lentiviral vectors (LVs) are among the most popular gene delivery tools, mainly due to the ease of manipulation and production, low genotoxicity, promising results of recent clinical trials on their use in gene therapy and their ability to stably transduce both dividing and nondividing cells, a property of utmost importance in cancer gene therapy [4]. However for their “systemic administration,” LVs should be engineered to specifically target the tumor cells too.
The natural tropism of LVs, similar to other enveloped viruses, is defined by their surface glycoproteins (GPs) [5]. Alteration, restriction, or refinement of natural tropism toward the desired cells might be an appealing strategy for targeted cancer therapy, a process known as “transductional targeting” [6]. The common practice for alteration of LVs’ natural tropism is through the substitution of original lentiviral GP with other viral GPs, a process known as “pseudotyping” [7]. Viral GPs are also artificially engineered to harbor a specific “targeting moiety” for recognition of the targeted cell(s). To this end, different targeting moieties, including peptides, natural ligands and specific antibodies (Abs), and Ab fragments, have been exploited for targeting of LVs [2]. Although Abs are among the most recognized targeting moieties, their large size (150 kDa) could restrict their application. Even the use of smaller fragments of Abs like single-chain variable fragments (ScFvs) is usually associated with lower affinity and stability [8]. The heavy-chain antibodies (HcAbs) of Camelidae family which are composed of only two heavy chains recognize antigens via the variable domain of each heavy chain. The variable domain of HcAbs, called nanobody or VHH, is the smallest natural antigen-binding fragment. Nanobodies have affinities equal to or even higher than Abs (in the range of nanomolar and picomolar), and considering their small size and stability, might represent an intriguing alternative targeting modality [8–10].
In addition to binding and attachment to specific cell surface receptors (recognition), viral GPs also perform “fusogenic activity” to implement the fusion between virus envelope and the membrane of target cells to complete the process of viral entry (infection) [11]. While LVs need proper interaction and consecutive conformational change of binding (gp120) and fusion (gp41) portions of the same GP protein to start the infection [12], some other viruses like sindbis make use of two functionally separate GPs for binding (E2) and membrane fusion (E1) [13]. Thus, considering the complexity of the interactions between targeting moiety, envelope, and receptor in viral infectivity, manipulations on sindbis virus-binding glycoprotein E2, may not affect the fusogenic activity. Accordingly, this strategy has been successfully used in the production of LVs, engineered to bear sindbis virus E2 GP harboring IgG-binding domain of protein A (ZZ domain) [14–17].
Angiogenesis, the formation of new blood vessels from preexisting vasculature, is a key step in the growth and metastasis of cancer [18, 19] and thus an obvious target for cancer treatment [20]. The major cell type involved in angiogenesis is tumor-associated endothelial cells (TAECs) which show differential expression of cell surface-associated markers and a higher proliferation rate than normal endothelial cells. Furthermore, every endothelial cell supports nearly 50–100 tumor cells [21, 22], and thus, targeting TAECs has the additional advantage of obviating the need to deliver transgenes to every tumor cell while providing a common target to all angiogenic tumors [23]. Of note, vascular endothelial growth factor (VEGF) and its cognate receptors (VEGFRs) on endothelial cells, the key regulators of angiogenesis, are upregulated on TAECs [24]. Among VEGFRs, VEGFR2 (KDR) plays the most important role in pathologic angiogenesis [25] and lymphangiogenesis [26], and thus it is an important candidate for targeted cancer therapy [27, 28]. To this end, few attempts to develop TAECs-targeted retro/lentiviral vectors by means of targeting peptide [29] or adaptor-based strategy [17] and VEGFR2-targeted retro/lentiviral vectors via antibody [30] or its natural ligand, VEGF [31] were reported. However, in these prior studies, targeting via the use of adaptors or externally introduced Abs could limit applications in clinical practice.
To address these concerns, in the present study, we constructed two LVs displaying chimeric sindbis virus E2 GP (cSVE2), engineered to harbor either a specific nanobody against VEGFR2 (3VGR19) [32] or a short isoform of VEGF (VEGF121), which is the natural ligand of VEGFR2 [33] as targeting motifs against VEGFR2-expressing cells. Using this strategy, we analyzed the transductional targeting properties of nanobody harboring cSVE2 compared to that of VEGF harboring for the final aim of targeted cancer therapy by direct delivery (i.e., without application of adaptors and externally introduced Abs) to VEGFR2-expressing cells.
Methods
Primary Assessment of Targeting Ligands’ Exposure in the Context of cSVE2 GPs via Modeling (In Silico Analyses)
To gain primary insights on the structural stability/functionality (exposure) of the cSVE2 GPs for the selected insertion sites (between amino acids 71 and 74) of E2 for the targeting moieties (Nb and VEGF121), the in silico modeling was employed. To this end, FASTA sequence formats of cSVE2s (E2-Nb and E2-VEGF121) were generated using the sequences corresponding to 3VGR19 nanobody [32] and VEGF121 (Uniprot identifier: P15692-9). Subsequently, the sequences of cSVE2s were submitted to I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), and modeling runs were performed based on the secondary structure-enhanced profile–profile threading alignment and the iterative implementation of the threading assembly refinement program [34]. The energy minimization of the selected models was performed using GROMACS package version 5.1 on Linux operating system and OPLS-AA/M force field in a cubic box. The SPC/E water model (extended simple point charge model) was used, and the charge of the systems was neutralized by adding proportional chloride ions. The periodic boundary conditions were applied to all three dimensions. The temperature and pressure of systems were coupled at 300 °K and 1 bar using V-rescale and Parrinello–Rahman algorithms, respectively. Visualization of structures were performed using VMD version 1.8.7. All graphs were plotted using Grace Software version 5.1.23 on Linux operating system. Phi and psi angles' analysis of the energy-minimized structures was performed using RAMPAGE online server (mordred.bioc.cam.ac.uk/~rapper/rampage.php).
Plasmids and Chimeric Vectors Construction
Vectors harboring the coding sequences of 3VGR19 (the 382 bp, small size nanobody with high affinity (Kd = 3.4 nM) for VEGFR2 [32] and VEGF121 [35] were used as templates to amplify the corresponding genes by PCR using primers designed to contain Eco91I restriction sites at their 5′ ends. Plasmid 2.2 (gifted by Irvin Chen; Addgene plasmid # 34885; [16]) was used as a backbone for the construction of all chimeric plasmids employed in this study. Plasmid 2.2 encodes an engineered form of sindbis virus glycoprotein which contains several mutations to diminish the natural tropism of sindbis while preserving its fusogenic activity. One of the mutations built in this plasmid is the insertion of Fc-binding domain (ZZ domain) between amino acids 71 and 74 to eliminate its original receptor-binding capacity and further confer upon it the ability to bind to Fc domains of antibodies [36]. As shown in Fig. 1, cSVE2-encoding plasmids (p2.2-Nb and p2.2-VEGF121) were constructed by replacing the ZZ domain (Fc-binding region of S. aureus protein A) in plasmid 2.2 with 3VGR19 and VEGF121 encoding PCR products, respectively. As a nontargeted negative control vector, p2.2-L was constructed by replacing ZZ domain with a synthetic linker consisted of His-tag and HA-tag (Biomatik, Canada). All cloning procedures were performed according to the standard protocols [37]. Schematic diagrams and detailed explanations for cloning steps and construction of p2.2-Nb, p2.2-VEGF121 and p2.2-L are provided in supplementary materials (Figs. 1, 2, and 3 supplemental, respectively).
Schematic representation of final recombinant constructs encoding chimeric sindbis E2 GPs. The plasmid 2.2 (backbone vector) encodes sindbis virus GPs (E3, E2, 6 K and E1). To construct chimeric sindbis glycoprotein containing vectors (cSVE2s: p2.2-Nb and p2.2-VEGF121) and nontargeted negative control vector (p2.2-L), ZZ domain was excised by digestion with Eco91I enzyme and sequences corresponding to 3VGR19 nanobody, VEGF121, and a synthetic linker (comprising two peptide tags (HA-tag and His-tag) were inserted at the same site to generate p2.2-Nb, p2.2-VEGF121, and p2.2-L, respectively. E1 is fusogenic protein. E2 recognizes and binds to the cell receptor. E3 and 6 K denote leader sequences for E2 and E1, respectively; G4S is a flexible linker which consisted of four Glycine and one Serine; His-tag (HHHHHH); HA-tag (YPYDVPDYA); ZZ denotes the sequences encoding the IgG-binding domain of protein A from S. aureus. Complete schematic diagrams and detailed explanations for cloning steps and construction of p2.2-Nb, p2.2-VEGF121, and p2.2-L are provided in supplementary section (Figs. 1, 2, and 3 supplemental, respectively)
Cells, Transfection, and Production of Viruses
293T cells were obtained from the National Cell Bank of Iran (Pasteur Institute of Iran). 293/KDR cells, which express VEGFR2, were purchased from Sibtech (USA). Cells were cultured in DMEM supplemented with 10 % FBS, Pen/Strep (100 units/mL of penicillin and 100 µg/mL of streptomycin) and 1 % (v/v) Glutamax in humidified CO2 (5 %) incubator under the standard aseptic procedure. Cell transfections were performed by Turbofect transfection reagent (Thermoscientific, Lithuania) according to the manufacturer’s instructions. Briefly, 1 × 106 of 293T cells were seeded in 6-cm cell culture plates one night prior to transfection, and next day, the cell culture plates were transfected with 6 µg of total plasmids used in the production of LVs.
To produce LVs pseudotyped with targeted cSVE2s (2.2-Nb, 2.2-VEGF121) or nontargeted negative control (2.2-L), 293T cells were transfected with 3 µg lentiviral transfer vector pLOX-CWgfp (a LV transfer vector encoding GFP as a reporter gene) [38], 2 µg packaging plasmid, psPAX2 (a gift from Didier Trono, Addgene plasmid #12260), and 1 µg of p2.2-Nb, p2.2-VEGF121 or p2.2-L, respectively, using Turbofect as described above. In addition, LVs pseudotyped with vesicular stomatitis virus glycoprotein G (VSV-G) were produced as the transduction positive control. After 48 and 72 h of transfection, viral supernatants were collected and centrifuged at 3000×g for 15 min at 4 °C to remove cell debris. To concentrate viral vectors, centrifugation was performed at 48000×g for 3 h at 4 °C, and viral pellets were resuspended in cold PBS. Physical titration of viral vectors was performed via p24 (capsid) ELISA (Pasto Lentivirus HIV p24, Pasteur Institute of Iran) according to the manufacturer’s instruction. Briefly, ELISA wells pre-coated with anti-p24 Ab were incubated with produced LVs, and p24 contents of viruses were determined using a biotinylated p24 Ab followed by streptavidin-HRP [39].
Western Blotting
Analyses for the expression of cSVE2s (p2.2-Nb, p2.2-VEGF121) and p2.2-L in virus-producing cells and their proper incorporation in the produced LVs were performed by western blotting (WB) according to standard protocols [37].
To detect the expression of cSVE2s, 1 × 106 of each virus-producing cell (which were transfected with transfer, packaging and the corresponding envelope plasmid for each virus) was lysed by boiling in the appropriate volume of loading buffer, and equal amounts of each cell lysate was loaded onto a 12 % SDS-PAGE gel. Subsequently, protein bands were transferred to polyvinylidene fluoride (PVDF) membrane, and envelope proteins were detected using Rabbit antiserum raised against sindbis E2 glycoprotein (1:1000 dilution) and Mouse anti-rabbit HRP (Sigma, USA) (1:10000 dilution).
In the second step, to evaluate the incorporation of cSVE2s on LVs, equal amount of each concentrated virus was subjected to SDS-PAGE followed by immunoblotting as described above.
Capture ELISA
Extracellular domain of human VEGFR2 (G&P Biosciences, USA) or bovine serum albumin (BSA) (Sigma, USA) was coated in a 96-well ELISA plate (2.5 µg/ml in carbonate/bicarbonate buffer pH 9.5). Wells were blocked overnight at 4 °C with PBS with 3 % BSA. Equal amounts of concentrated lentiviral particles (determined by p24 amount) were added to each well and incubated for 1 h at 37 °C. Wells were subsequently washed four times with PBS with 2 % FBS. Captured viruses were lysed and transferred to a p24 pre-coated ELISA plate (Pasto Lentivirus HIV p24, Pasteur Institute of Iran), and p24 contents were determined according to the Pasto Lentivirus HIV p24 ELISA kit’s protocol as described above.
Microscopy and Flow Cytometric Analyses
Cells (293T and 293/KDR) were seeded in 24-well cell culture plates to reach 70 % confluency and incubated with equal amount of viruses in 250 µl medium (20 ng p24) for 8 h. Subsequently, viruses were replaced with fresh medium, and finally 96 h later, the cells were observed using fluorescent microscope (INVERSO TC100 Epi Fluor, Medline Scientific, UK) and images were taken at ×200 magnification. Following the microscopic evaluations, cells were detached and subjected to flow cytometry using Cyflow analyzer (Partec, Germany) for the analysis of GFP expression.
Statistical Analyses
All reported numbers are means of three replicates. Analysis of variance with Tukey’s multiple comparison test was used to compare multiple groups. A t-student test was utilized to show the difference between the test samples and BSA control in ELISA. A p < 0.05 was considered a statistically significant difference. Free 30-day trial version of GraphPad Prism™ for windows (version 6.01, Graphpad Software Inc.) was used for all statistical analysis.
Results
Targeting Ligands Were Exposed in the Context of cSVE2 GPs (In silico Data)
Based on prior reports, residues between amino acids 71 and 74 of E2 were considered as a candidate region for insertion of targeting moieties [40–42], and the structure of the targeting ligands for proper exposure (to interact with the corresponding receptors) was assessed using bioinformatics tools. The generated models for each fused protein were subjected to energy minimization and structure refinement using GROMACS package (Fig. 4 supplemental) at 300 °K and 1 bar (Fig. 5 supplemental). Visualization of refined structures suggested that the adapted ligands (3VGR19 nanobody and VEGF121 which were inserted between the amino acids 71 and 74 of E2 GP) were exposed in the context of cSVE2s and might be able to recognize their corresponding receptor (Fig. 2). Besides, phi and psi analyses also confirmed the quality of refined structures (Fig. 6 supplemental).
Tube representation of modeled cSVE2s. The conformation of the inserted targeting moieties, a nanobody and b VEGF121, in the final structure of cSVE2s (a 2.2-Nb and b 2.2-VEGF121) appears to be properly exposed for efficient interaction with their corresponding receptor (VEGFR2). The targeting moieties are shown in gray, and the sindbis E2 GP is depicted in black
Constructed cSVE2s and Their Incorporation on LVs
As shown in Fig. 1, to generate cSVE2 encoding plasmids (p2.2-Nb and p2.2-VEGF121), the ZZ domain in 2.2 plasmid was substituted with 3VGR19 nanobody or VEGF121 fragments. p2.2-L was constructed in the same manner by substituting the ZZ domain with the synthetic linker. Schematic diagrams for the construction of p2.2-Nb, p2.2-VEGF121, and p2.2-L are provided in supplementary materials (Figs. 1, 2, and 3 supplemental, respectively). All the three constructed vectors were verified by restriction enzyme digestion and sequencing analyses (data not shown). Comparison of viral titers by p24 ELISA following the production of LVs pseudotyped with cSVE2 constructs showed that the production titer of LVs pseudotyped with 2.2-Nb and 2.2-VEGF121 were 66.5 and 70 % of LVs bearing VSV-G, respectively (Fig. 7 supplemental).
Western blotting (WB) of each group of virus-producing cells demonstrated proper expression of cSVE2s and 2.2-L GPs with expected sizes. As shown in Fig. 3a, three bands with approximate sizes of 67, 75, and 75 kDa, corresponding to the expression of 2.2-L, 2.2-Nb, and 2.2-VEGF121 GPs, respectively, were detected. As expected, WB of concentrated LVs also demonstrated the same expected bands (Fig. 3b) verifying the incorporation of cSVE2s or 2.2-L GPs on the corresponding LVs, respectively.
Western blotting of chimeric sindbis E2 GPs. To evaluate the expression of chimeric sindbis E2 GPs in virus-producing cells (293T) transfected with transfer, packaging, and the corresponding envelope plasmid for each virus (p2.2-Nb, p2.2-VEGF121, or p2.2-L). 48 h post transfection, equal number of cells were subjected to western blotting. Three bands with approximate lengths of 67, 75, and 75 kDa (indicated by arrows), corresponding to 2.2-L, 2.2-Nb, and 2.2-VEGF121, respectively (lanes 1, 2, and 3), were detected (Fig. 3a). To evaluate the incorporation of chimeric sindbis GPs on produced LVs, equal amounts of each concentrated virus mixed with loading buffer was subjected to SDS-PAGE followed by immunoblotting as described above (lanes 1, 2, and 3 correspond to 2.2-L, 2.2-Nb, and 2.2-VEGF121, respectively) (Fig. 3b). Western blotting of virus-producing cells a and LVs b demonstrated successful expression and incorporation of chimeric sindbis GPs in cells and viruses, respectively
Functionality of Targeting Moieties (3VGR19 and VEGF121) in cSVE2-Pseudotyped LVs by Capture ELISA
To investigate the functionality of the incorporated cSVE2s on LVs, a virus capture experiment was performed. Briefly, equal amounts of each virus (17 ng based on P24 content) were incubated with the extracellular domain of VEGFR2 or BSA (as negative control) coated in a 96-well ELISA plate. Following extensive washing, the amount of captured LVs was determined by p24 ELISA. As shown in Fig. 4, while LVs pseudotyped with cSVE2s could selectively bind to VEGFR2 coated wells (compared to BSA), no significant binding to VEGFR2 was observed for 2.2-L- (negative control) pseudotyped virus (P ≤ 0.0001). These results demonstrated that inserted targeting moieties (3VGR19 and VEGF121), could retain their functionality in cSVE2-pseudotyped LVs.
Virus capture ELISA assay. Viral supernatants were incubated in ELISA wells coated with either the extracellular domain of human VEGFR2 or BSA. The bound viruses were then measured by commercial p24 ELISA kit. The results shown here are the representatives of two independent experiments. In each panel, the white histograms (bars) correspond to the virus incubated in VEGFR2- (KDR)-coated wells, and the gray histograms (bars) represent the BSA (negative control) groups. The mean and standard deviation from two independent experiments (where each was performed in duplicate) are shown. These results demonstrated a significant increase in attachment of chimeric LVs to VEGFR2 in comparison to 2.2-L-pseudotyped LVs (P ≤ 0.0001) which emphasizes the functionality of LV surface displaying targeting moieties in the context of sindbis virus GPs. 2.2-L denotes that LVs containing nontargeted negative controls 2.2-L GP, 2.2-Nb, and 2.2-VEGF121 are the representatives of LVs harboring 2.2-Nb and 2.2-VEGF121 GPs, respectively
Targeted Transduction Efficiency of cSVE2-Pseudotyped LVs for VEGFR2-Expressing Cells
To determine the targeted transduction efficiency of cSVE2-pseudotyped LVs, 293/KDR cells (which express VEGFR2 on their surface) were used as target cells, and 293T cells used as a negative control were infected by the produced viruses. The virus-encoded GFP emission by cells was evaluated by fluorescent microscopy and flow cytometry.
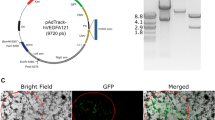
As shown in Fig. 5, fluorescent microscopy demonstrated selective and efficient transduction of 293/KDR cells by cSVE2-pseudotyped LVs harboring the targeting moieties in contrast to control 293T cells which only showed a small background of GFP expression. Accordingly, flow cytometric data indicated that approximately 30 % of 293/KDR cells were transduced with cSVE2-pseudotyped LVs harboring 3VGR19 or VEGF121 as targeting moieties, while 293T cells showed around 1 % transduction efficiencies with both viruses (P = 0.0021). As expected, 293/KDR and 293T cells transduced with control 2.2-L-pseudotyped LVs harboring no targeting moiety also showed only background GFP expression around 3–5 % (Fig. 6). This final result demonstrated that LVs pseudotyped with both cSVE2 GPs could target and transduce VEGFR2-expressing cells with comparable efficiencies.
Fluorescence microscopy images of transduction assays. VEGFR2-expressing 293/KDR cells (upper images) and 293T cells (lower images) as nontarget cells were transduced with LVs pseudotyped with 2.2-L, VSV-G, 2.2-Nb, and 2.2-VEGF121 from left to right, respectively. 96 h after transduction, cells were analyzed by fluorescence microscopy (×200 magnification). For each cell, the bright field images are represented in the upper row, and the fluorescent images are depicted in the lower row. These results demonstrated selective transduction of 293/KDR cells by chimeric viruses (cSVE2-pseudotyped viruses: 2.2-Nb and 2.2-VEGF121 containing LVs), and small off-target (background) transduction by nontargeted negative control (2.2-L)-pseudotyped LVs
Analysis of transduction efficiency by flow cytometry. 293/KDR cells (which express VEGFR2) (upper images) and 293T cells (lower images) as nontarget cells were transduced with LVs (20 ng of p24) pseudotyped with 2.2-L, VSV-G, 2.2-Nb, and 2.2-VEGF121 from left to right, respectively. 96 h after transduction, the cells were analyzed by flow cytometry. Results indicated that both cSVE2-pseudotyped LVs harboring 3VGR19 or VEGF121 as targeting moieties could target and efficiently transduce VEGFR2-expressing 293/KDR cells
Discussion
In the present study, for the final aim of transductional targeting of tumor vasculature by LVs, two chimeric sindbis virus glycoproteins (cSVE2s) harboring either a specific nanobody against VEGFR2 (designated 2.2-Nb) or natural ligand of VEGFR2 (designated 2.2-VEGF121) were constructed and compared for their transductional targeting efficiencies against VEGFR2-expressing cells. Our results demonstrated that cSVE2-pseudotyped LVs harboring the nanobody could target and transduce VEGFR2-expressing cells with comparable efficiencies to that of cSVE2 LVs containing the natural ligand and indicated the proper specificity and functionality of the employed VEGFR2-specific nanobody in the context of cSVE2-pseudotyped LVs for potential targeting of tumor vasculature in cancer therapy.
Contrary to longer isoforms of VEGF (like VEGF165), VEGF121 used in the present study lacks a heparin-binding domain which reduces nonspecific transduction. Similarly, Snitkovsky et al. adapted a truncated form of VEGF165 to avoid nonspecific heparin binding to target avian leukosis virus to VEGFR2-expressing cells [31]. However, contrary to their study, which was based on adaptor strategy, a genetically incorporated VEGF121 in our study might be better suited for clinical applications [6].
In one pioneering study, retroviral vectors mosaically pseudotyped with amphotropic murine leukemia virus (MLV) envelope (which recognizes both human and mouse cells), and chimeric envelopes containing targeting peptides could enhance the transduction efficiency of human endothelial cells compared to retroviral vectors pseudotyped with sole amphotropic envelope [29]. In another study, mosaically pseudotyped (co-pseudotyped) retroviral vectors with both wild-type MLV envelope (performing the fusion) and a modified MLV envelope containing IgG-binding motif of protein A (ZZ domain) were constructed to enhance transduction of VEGFR2-expressing cells via the application of an externally provided anti-VEGFR2 Ab [30]. However, in these early studies, co-expression of wild-type amphotropic envelope to render the viruses fully infectious would compromise targeting specificity [43]. Moreover, the application of adaptor-based targeting strategy via ZZ domain requires the external application of Abs for indirect targeting of VEGFR2-expressing cells. This approach might not be applicable in an in vivo setting, where serum Abs would compete with the externally introduced Abs for binding with ZZ domain [42].
Our strategy in insertion of targeting moieties between amino acids 71 and 74 of sindbis virus E2 GP (Fig. 1) was consistent with prior reports for retaining the functional properties (recognition and binding to the target receptors) of the motifs incorporated in this region [14–17, 42, 44]. Preserving the functional properties of the targeting motifs in the context of chimeric GPs is a critical step in transductional targeting studies. For instance, a prior attempt to insert human angiogenin sequence into MLV envelope GP for transduction of endothelial cells was disappointing, apparently due to the improper conformational change of chimeric envelope upon binding to cognate cell surface receptors [43]. To address this concern, we first undertook in silico modeling studies, and the results provided a primary proof of suitability of 3VGR19 nanobody and VEGF121 as targeting moieties in the context of E2 GP (Fig. 2 and Figs. 4–6 supplemental) which were in agreement with our subsequent experimental results. The production titers of cSVE2 LVs were relatively high and comparable to VSV-G pseudotype LVs which is considered standard among other GPs.
Western blotting analyses for expression of constructed vectors in both virus-producing cells and cSVE2s-pseudotyped LVs (Fig. 3) demonstrated successful expression and incorporation of chimeric sindbis GPs in cells and viruses, respectively. The sizes of the observed bands were as expected for each construct and consistent with prior reports on the approximate size of chimeric sindbis GPs [14, 40].
The results of virus capture ELISA (Fig. 4) indicated a significant increase in the attachment of cSVE2-pseudotyped LVs harboring the targeting moieties to VEGFR2 in comparison to 2.2-L- (negative control) pseudotyped viruses, demonstrating that both incorporated targeting moieties are functional and could specifically recognize and bind to VEGFR2. On the contrary, cSVE2-pseudotyped LVs could not bind to BSA, further emphasizing their specific and selective mode of recognition. Accordingly, results of microscopic evaluation of transduction experiments (Fig. 5) also demonstrated selective and efficient transduction of 293/KDR cells by cSVE2-pseudotyped LVs harboring the targeting moieties in contrast to control 293T cells which only showed a small background of GFP expression. To quantify efficiency of this targeting, flow cytometry was employed in similar transduction experiments. As shown in Fig. 6, flow cytometric data indicated efficient (30 %) transduction of VEGFR2-expressing 293/KDR cells by cSVE2-pseudotyped LVs harboring 3VGR19 or VEGF121 as targeting moieties compared to controls, which showed only <5 % background GFP expression. These results were consistent with a study on dual-targeted lentiviral vectors where 36 % transduction efficiency of primary endothelial cells was achieved [17]. However, Utilization of adaptor-based strategy (ZZ domain) would limit the in vivo applications of this prior study. Our results were also in agreement with the report of Morizono et al. [42], where insertion of integrin targeting peptides between amino acid 71 and 74 of E2 GP resulted in efficient transduction of desired cells. However, in the aforementioned study, high transduction efficiency (around 50 %) was attained only when a high amount of virus (40 ng p24 of virus for 5 × 104 cells) was used, while application of lower amount of virus (4 ng p24 of virus for 5 × 104 cells) resulted in transduction efficiencies as low as 5 % [42]. Although, in our study, lower amount of LVs (20 ng p24 of virus for 1 × 105 cells) were used to obtain comparable transduction efficiencies with the above-mentioned studies, but it should be noted that they have used primary endothelial cells compared to easy-to-transduce 293 derivative cells in our study, and therefore the obtained transduction efficiencies should be further confirmed in primary endothelial cells or cell lines before any exact conclusion for enhanced efficiencies obtained in our study could be drawn. However, it is worth mentioning that transduction efficiency of even <10 % could result in eradication of whole cancer cell populations [45] especially if transgenes with bystander effects (like herpes virus thymidine kinase) are used.
As it could be concluded from the results of capture ELISA (Fig. 4) and transductional experiments (Figs. 5 and 6 respectively), both cSVE2-pseudotyped LVs harboring the targeting moieties (3VGR19 and VEGF121) showed similar efficacies for transducing the VEGFR2-expressing cells. But VEGF121 has a critical limitation as a targeting moiety against tumor vasculature, since it also has high affinity for VEGFR1, also present on some normal cells like endocrine cells of pituitaries [46]. Therefore, it might be concluded that the smaller size (382 bp) and high affinity (Kd = 3.4 nM) of 3VGR19 nanobody used in this study led to proper incorporation of this targeting moiety in sindbis virus E2 GP and efficient recognition of VEGFR2 on target cells, respectively. Our results are consistent with reports on successful application of specific nanobodies to redirect LVs toward different subsets of immune cells [47] and further confirmed the prior report on potential efficacy of 3VGR19 nanobody as a specific targeting moiety for VEGFR2-expressing cells [48]. However, it should be noted that (although not the primary concern of this study) the potential of even nonantigen-presenting cells (APCs), such as endothelial cells, to internalize antigens [49] and the possibility of LV uptake by endothelial cells might affect the precise dose of gene delivery vectors. Therefore, for final in vivo applications, the possible uptake of untargeted LVs by endothelial cells might be carefully evaluated.
Taken together, to the best of our knowledge, this is the first report on direct retargeting of LVs toward VEGFR2-expressing cells using nanobody as targeting moiety. The constructed cSVE2-pseudotyped LVs carrying VEGFR2-specific nanobody in this study might be further armed by the capability to express cytotoxic genes with bystander effect [45] for the final aim of cancer therapy via abolishing tumor vasculature.
Conclusion
In silico analyses and the employed strategy in insertion of cSVE2 GPs resulted in proper exposure of the targeting moieties (3VGR19 Nb and VEGF121) in LVs for efficient interaction with the corresponding receptor (VEGFR2). Application of VEGFR2-specific nanobody as the targeting moiety provided the possibility of direct delivery of the engineered virus to the VEGFR2-expressing cells. Our results indicated that 3VGR19 Nb-harboring cSVE2 GPs conferred upon LVs have similar targeting properties as the natural ligand, VEGF121, while avoiding the shortcomings of the natural ligand in interaction with VEGFR1. The acquired targeting efficiency in 293/KDR cells (~30 %) is well suited for transductional strategies employing cytotoxic genes with bystander effect (like herpes virus thymidine kinase) for potential systemic applications in cancer therapy (if comparable efficiencies could be earned with endothelial cells too).
References
Collins, M., & Thrasher, A. (2015). Gene therapy: Progress and predictions. Proceedings of the Royal Society B, 282, 20143003.
Buchholz, C. J., Friedel, T., & Buning, H. (2015). Surface-engineered viral vectors for selective and cell type-specific gene delivery. Trends in Biotechnology, 33, 777–790.
Zhao, L., Wu, J., Zhou, H., Yuan, A., Zhang, X., Xu, F., et al. (2011). Local gene delivery for cancer therapy. Current Gene Therapy, 11, 423–432.
Ou, W., Marino, M. P., Suzuki, A., Joshi, B., Husain, S. R., Maisner, A., et al. (2012). Specific targeting of human interleukin (IL)-13 receptor alpha2-positive cells with lentiviral vectors displaying IL-13. Human Gene Therapy Methods, 23, 137–147.
Sakuma, T., Barry, M. A., & Ikeda, Y. (2012). Lentiviral vectors: Basic to translational. Biochemical Journal, 443, 603–618.
Goyvaerts, C., Liechtenstein, T., Bricogne, C., Escors, D., & Breckpot, K. (2013). Targeted lentiviral vectors: current applications and future potential, in gene therapy. In F. Martin (Ed.), Tools and potential applications (pp. 343–386). Rijeka: InTech Open Access Publisher.
Levy, C., Verhoeyen, E., & Cosset, F. L. (2015). Surface engineering of lentiviral vectors for gene transfer into gene therapy target cells. Current Opinion in Pharmacology, 24, 79–85.
Kijanka, M., Dorresteijn, B., Oliveira, S., & en Henegouwen, P. M. V. B. (2015). Nanobody-based cancer therapy of solid tumors. Nanomedicine (London), 10, 161–174.
Muyldermans, S., & Smider, V. V. (2016). Distinct antibody species: structural differences creating therapeutic opportunities. Current Opinion in Immunology, 40, 7–13.
Turner, K. B., Alves, N. J., Medintz, I. L., & Walper, S. A. (2016). Improving the targeting of therapeutics with single-domain antibodies. Expert Opinion on Drug Delivery, 13, 561–570.
Cosset, F. L., & Lavillette, D. (2011). Cell entry of enveloped viruses. Advances in Genetics, 73, 121–183.
Checkley, M. A., Luttge, B. G., & Freed, E. O. (2011). HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. Journal of Molecular Biology, 410, 582–608.
Leung, J. Y., Ng, M. M., & Chu, J. J. (2011). Replication of alphaviruses: a review on the entry process of alphaviruses into cells. Advance Virology, 2011, 249640.
Morizono, K., Bristol, G., Xie, Y. M., Kung, S. K., & Chen, I. S. (2001). Antibody-directed targeting of retroviral vectors via cell surface antigens. Journal of Virology, 75, 8016–8020.
Morizono, K., Xie, Y., Ringpis, G. E., Johnson, M., Nassanian, H., Lee, B., et al. (2005). Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nature Medicine, 11, 346–352.
Pariente, N., Morizono, K., Virk, M. S., Petrigliano, F. A., Reiter, R. E., Lieberman, J. R., et al. (2007). A novel dual-targeted lentiviral vector leads to specific transduction of prostate cancer bone metastases in vivo after systemic administration. Molecular Therapy, 15, 1973–1981.
Pariente, N., Mao, S. H., Morizono, K., & Chen, I. S. (2008). Efficient targeted transduction of primary human endothelial cells with dual-targeted lentiviral vectors. The Journal of Gene Medicine, 10, 242–248.
Huang, Y., & Carbone, D. P. (2015). Mechanisms of and strategies for overcoming resistance to anti-vascular endothelial growth factor therapy in non-small cell lung cancer. Biochimica et Biophysica Acta, 1855, 193–201.
Fan, F., Schimming, A., Jaeger, D., & Podar, K. (2012). Targeting the tumor microenvironment: Focus on angiogenesis. Journal of Oncology, 2012, 281261.
Jayson, G. C., Kerbel, R., Ellis, L. M. & Harris, A. L. (2016) Antiangiogenic therapy in oncology: current status and future directions.The Lancet.
Persano, L., Crescenzi, M., & Indraccolo, S. (2007). Anti-angiogenic gene therapy of cancer: Current status and future prospects. Molecular Aspects of Medicine, 28, 87–114.
Aird, W. C. (2012). Endothelial cell heterogeneity. Cold Spring Harbor Perspectives in Medicine, 2, a006429.
Trepel, M., Stoneham, C. A., Eleftherohorinou, H., Mazarakis, N. D., Pasqualini, R., Arap, W., et al. (2009). A heterotypic bystander effect for tumor cell killing after adeno-associated virus/phage-mediated, vascular-targeted suicide gene transfer. Molecular Cancer Therapeutics, 8, 2383–2391.
Sullivan, L. A., & Brekken, R. A. (2010). The VEGF family in cancer and antibody-based strategies for their inhibition. MAbs, 2, 165–175.
Ferrara, N., & Kerbel, R. S. (2005). Angiogenesis as a therapeutic target. Nature, 438, 967–974.
Secker, G. A., & Harvey, N. L. (2015). VEGFR signaling during lymphatic vascular development: From progenitor cells to functional vessels. Developmental Dynamics, 244, 323–331.
Liu, D., Liu, F., Liu, Z., Wang, L., & Zhang, N. (2011). Tumor specific delivery and therapy by double-targeted nanostructured lipid carriers with anti-VEGFR-2 antibody. Molecular Pharmaceutics, 8, 2291–2301.
Miettinen, M., Rikala, M. S., Rys, J., Lasota, J., & Wang, Z. F. (2012). Vascular endothelial growth factor receptor 2 as a marker for malignant vascular tumors and mesothelioma: an immunohistochemical study of 262 vascular endothelial and 1640 nonvascular tumors. American Journal of Surgical Pathology, 36, 629–639.
Liu, L., Anderson, W. F., Beart, R. W., Gordon, E. M., & Hall, F. L. (2000). Incorporation of tumor vasculature targeting motifs into moloney murine leukemia virus env escort proteins enhances retrovirus binding and transduction of human endothelial cells. Journal of Virology, 74, 5320–5328.
Masood, R., Gordon, E. M., Whitley, M. D., Wu, B. W., Cannon, P., Evans, L., et al. (2001). Retroviral vectors bearing IgG-binding motifs for antibody-mediated targeting of vascular endothelial growth factor receptors. International Journal of Molecular Medicine, 8, 335–343.
Snitkovsky, S., Niederman, T. M. J., Mulligan, R. C., & Young, J. A. T. (2001). Targeting avian leukosis virus subgroup a vectors by using a TVA-VEGF bridge protein. Journal of Virology, 75, 1571–1575.
Behdani, M., Zeinali, S., Khanahmad, H., Karimipour, M., Asadzadeh, N., Azadmanesh, K., et al. (2012). Generation and characterization of a functional nanobody against the vascular endothelial growth factor receptor-2; angiogenesis cell receptor. Molecular Immunology, 50, 35–41.
Kazemi, M., Carrer, A., Moimas, S., Zandona, L., Bussani, R., Casagranda, B., et al. (2016). VEGF121 and VEGF165 differentially promote vessel maturation and tumor growth in mice and humans. Cancer Gene Therapy, 23, 125–132.
Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9, 40.
Kazemi-Lomedasht, F., Behdani, M., Pooshang Bagheri, K., Habibi Anbouhi, M., Abolhassani, M., Khanahmad, H., et al. (2014). Expression and purification of functional human vascular endothelial growth factor-a121; the most important angiogenesis factor. Advanced Pharmaceutical Bulletin, 4, 323–328.
Morizono, K., Ku, A., Xie, Y., Harui, A., Kung, S. K., Roth, M. D., et al. (2010). Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins. Journal of Virology, 84, 6923–6934.
Sambrook, J., & Russell, D. W. (2006). The condensed protocols from molecular cloning : A laboratory manual. New York: Cold Spring Harbor Laboratory Press.
Salmon, P., Oberholzer, J., Occhiodoro, T., Morel, P., Lou, J., & Trono, D. (2000). Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Molecular Therapy, 2, 404–414.
Barde, I., Salmon, P., & Trono, D. (2010). Production and titration of lentiviral vectors. Current Protocols in Neuroscience, 4, 4–21.
Aires da Silva, F., Costa, M. J., Corte-Real, S., & Goncalves, J. (2005). Cell type-specific targeting with sindbis pseudotyped lentiviral vectors displaying anti-CCR5 single-chain antibodies. Human Gene Therapy, 16, 223–234.
Morizono, K., Xie, Y., Helguera, G., Daniels, T. R., Lane, T. F., Penichet, M. L., et al. (2009). A versatile targeting system with lentiviral vectors bearing the biotin-adaptor peptide. The Journal of Gene Medicine, 11, 655–663.
Morizono, K., Pariente, N., Xie, Y., & Chen, I. S. (2009). Redirecting lentiviral vectors by insertion of integrin-targeting peptides into envelope proteins. The Journal of Gene Medicine, 11, 549–558.
Gornikiewicz, A., Zommer, A., Jakesz, R., Gnant, M., & Brostjan, C. (2005). Retroviral targeting of proliferating endothelial cells. Acta Biochimica Polonica, 52, 731–735.
Leoh, L. S., Morizono, K., Kershaw, K. M., Chen, I. S., Penichet, M. L., & Daniels-Wells, T. R. (2014). Gene delivery in malignant B cells using the combination of lentiviruses conjugated to anti-transferrin receptor antibodies and an immunoglobulin promoter. The Journal of Gene Medicine, 16, 11–27.
Karjoo, Z., Chen, X., & Hatefi, A. (2016). Progress and problems with the use of suicide genes for targeted cancer therapy. Advanced Drug Delivery Reviews, 99, 113–128.
Onofri, C., Theodoropoulou, M., Losa, M., Uhl, E., Lange, M., Arzt, E., et al. (2006). Localization of vascular endothelial growth factor (VEGF) receptors in normal and adenomatous pituitaries: Detection of a non-endothelial function of VEGF in pituitary tumours. Journal of Endocrinology, 191, 249–261.
Goyvaerts, C., Dingemans, J., De Groeve, K., Heirman, C., Van Gulck, E., Vanham, G., et al. (2013). Targeting of human antigen-presenting cell subsets. Journal of Virology, 87, 11304–11308.
Behdani, M., Zeinali, S., Karimipour, M., Khanahmad, H., Schoonooghe, S., Aslemarz, A., et al. (2013). Development of VEGFR2-specific nanobody pseudomonas exotoxin a conjugated to provide efficient inhibition of tumor cell growth. New Biotechnology, 30, 205–209.
Goyvaerts, C., & Breckpot, K. (2015). Pros and cons of antigen-presenting cell targeted tumor vaccines. Journal of Immunology Research, 2015(785634), 1–18.
Acknowledgments
This work was supported by Pasteur Institute of Iran in fulfillment of the Ph.D. thesis of R. Ahani. We are grateful to Ms Dorsa Torabi for her technical assistance with western blotting.
Funding
Funding was provided by Pasteur Institute of Iran (Grant No. BP-8918).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahani, R., Roohvand, F., Cohan, R.A. et al. Sindbis Virus-Pseudotyped Lentiviral Vectors Carrying VEGFR2-Specific Nanobody for Potential Transductional Targeting of Tumor Vasculature. Mol Biotechnol 58, 738–747 (2016). https://doi.org/10.1007/s12033-016-9973-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-016-9973-7