Abstract
l-Arabinose isomerase (L-AI, EC 5.3.1.4) catalyzes the isomerization between l-arabinose and l-ribulose, and most of the reported ones can also catalyze d-galactose to d-tagatose, except Bacillus subtilis L-AI. In this article, the L-AI from the psychrotolerant bacterium Pseudoalteromonas haloplanktis ATCC 14393 was characterized. The enzyme showed no substrate specificity toward d-galactose, which was similar to B. subtilis L-AI but distinguished from other reported L-AIs. The araA gene encoding the P. haloplanktis L-AI was cloned and overexpressed in E. coli BL21 (DE3). The recombinant enzyme was purified by one-step nickel affinity chromatography . The enzyme displayed the maximal activity at 40 °C and pH 8.0, and showed more than 75 % of maximal activity from pH 7.5–9.0. Metal ion Mn2+ was required as optimum metal cofactor for activity simulation, but it did not play a significant role in thermostability improvement as reported previously. The Michaelis–Menten constant (K m), turnover number (k cat), and catalytic efficiency (k cat/K m) for substrate l-arabinose were measured to be 111.68 mM, 773.30/min, and 6.92/mM/min, respectively. The molecular docking results showed that the active site residues of P. haloplanktis L-AI could only immobilize l-arabinose and recognized it as substrate for isomerization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-Arabinose isomerase (L-AI, EC 5.3.1.4), catalyzing the isomerization between l-arabinose and l-ribulose, is an important enzyme included in the microbial l-arabinose degradation pathway [1]. In many kinds of microorganisms, L-AI converts l-arabinose to l-ribulose, and then l-ribulose is further converted by L-ribulokinase and l-ribulose-5-phosphate 4-epimerase to d-xylulose-5-phosphate, an intermediate in microbial pentose phosphate pathway [2]. Therefore, l-arabinose can be used as a sole carbon source for these microorganisms. Due to the importance in microbial l-arabinose metabolic pathway, L-AI is widely distributed in various microorganisms [3, 4].
In addition to converting l-arabinose to l-ribulose, L-AI can also catalyze d-galactose to its ketose isomer, d-tagatose, based on the structural similarity of the similar substrate configuration. d-tagatose is a functional low-calorie bulk sweetener, which has 92 % of relative sweetness but only 1/3 of calories of sucrose [3]. It has many health benefits, such as prebiotic activity [5], low glycemic index [6], anti-hyperglycemia effect [3], treatment for Type 2 diabetes [7], and noncariogenicity [8]. d-tagatose has been approved as GRAS (Generally Recognized as Safe) by the Food and Drug Administration of the United States since 2001 [9]. It has also been approved to be used as a food ingredient in Japan, Korea, New Zealand, Australia, Brazil, the European Union, and China. Due to the great potential for broad industrial and pharmaceutical applications, d-tagatose is widely believed to have a very good market prospect, and has been industrially manufactured by many international companies, such as Spherix (Tysons Corner, VA, US), Arla Foods (Aarhus Denmark), and Nutrilab (Bekkevoort, Belgium).
As mentioned above, L-AI exists in a wide range of microorganisms, likely for starting l-arabinose degradation. Through the literature search from Web of Sciences database, more than 30 of microbial L-AIs have been identified and characterized from various sources up to date, and the microorganisms for L-AI sources include mesophilic, thermophilic, hyperthermophilic, and thermoacidophilic bacteria. For the industrial production of d-tagatose by L-AI, there are several issues needed to be considered. Isomerization of d-galactose to d-tagatose by L-AI is an equilibrium reaction, and elevated reaction temperatures (>60 °C) favor the equilibrium to shift toward d-tagatose during the enzymatic reaction process [3, 4]. However, higher temperatures (>70 °C) and alkaline pHs easily introduce undesired nonenzymatic reactions such as browning and lead to unwanted by-products. In addition, d-galactose is industrially obtained from the cheap raw material lactose by lactase with pH 5–6 [4]. Therefore, slightly acidic pHs are suitable for d-tagatose bioconversion to both reduce the side reactions and enable coupling with the lactose hydrolysis in one step [3, 4]. As a result, the thermostability and slightly acidic pH stability are the two main issues related to biological production of d-tagatose. Thermostable L-AIs have been widely identified from the thermophilic and hyperthermophilic bacteria, such as Anoxybacillus flavithermus [10], Bacillus stearothermophilus US100 [11], Thermotoga neapolitana 5608 [12], andThermotoga maritima DSM3109 [13]. The low pH active and stable L-AIs have been characterized from thermoacidophilic bacteria, such as Alicyclobacillus acidocaldarius ATCC 43030 [14], Acidothermus cellulolyticus ATCC 43068 [15], Alicyclobacillus hesperidum URH17-3-68 [16], and Bacillus coagulans 2-6 [17], and some food-grade lactic acid bacteria (LAB), such as Lactobacillus plantarum NC8 [18], Lactobacillus sakei 23 K [19], Lactobacillus fermentum CGMCC2921 [20], Lactobacillus plantarum SK-2 [21], and Lactobacillus reuteri 100-23 [22].
Recently, the L-AIs from food-grade LAB have been proved attractive for d-tagatose production using lactose as a cheaper material in dairy products. The acidic pH optimum of LAB L-AIs is an advantage for d-tagatose production under the pH condition of milk fermentation and storage. However, the process also requires an efficient L-AI with high activity at low temperature. An acid-tolerant L-AI from Shewanella sp. ANA-3 [23] has been characterized showing high activity at low temperatures. It displays maximal activity within a large range of temperatures from 15 to 35 °C, and interestingly preserves nearly 90 % of maximal activity at 4 °C, showing great potential for application in dairy industry.
Substrate specificity is another important factor for biotechnological production of d-tagatose. So far, almost all of the characterized L-AIs are reported having substrate specificity toward both l-arabinose and d-galactose; however, the L-AI from Bacillus subtilis str. 168 has unique specificity toward only l-arabinose [24]. In addition, among the L-AIs with specificity toward both substrates, except A. flavithermus L-AI [10], all the L-AIs show much higher substrate specificity toward l-arabinose than d-galactose.
Psychrotolerant organisms are widely considered as an important source for cold-adapted enzymes, which have good potential in some low-temperature processing fields. Although there are so many reports of L-AIs identified from various microorganism sources, the L-AI has not yet been characterized from psychrotolerant bacteria. In the present article, an L-AI was characterized from a psychrotolerant bacterium, Pseudoalteromonas haloplanktis ATCC 14393. However, the P. haloplanktis L-AI was not cold adapted and was characterized having no substrate specificity toward d-galactose. Although this specific L-AI could not be used as a biocatalyst candidate for d-tagatose production, this report may be helpful to provide valuable information for the study on the molecular mechanism of substrate specificity of L-AI.
Materials and Methods
Chemicals and Reagents
Chelating Sepharose Fast Flow resin was from GE Healthcare (Uppsala, Sweden). Electrophoresis reagents were purchased from Bio-Rad. Yeast extract and tryptone were obtained from Sangon Biological Engineering Technology & Services Co. Ltd. (Shanghai, China). Isopropyl β-D-1-thiogalactopyranoside (IPTG) is obtained from Sigma (St Louis, MO, USA). All chemicals used for enzyme assays and characterization in this study were of at the least analytical grade were obtained from Sinopharm Chemical Reagent (Shanghai, China).
Plasmid and Culture Condition
The expression vector pET-22b(+) was from Novagen (Darmstadt, Germany). E. coli strains, DH5α and BL21 (DE3), were obtained from Sangon Biological Engineering Technology and Services (Shanghai, China), and used as host cells for plasmid transformation and foreign gene expression, respectively. Both the E. coli strains were cultured in Luria–Bertani (LB) medium (5 g/l yeast extract, 10 g/l tryptone, and 10 g/l NaCl) supplemented with 100 μg/ml ampicillin at 37 °C at 200 rpm.
Gene Cloning and Expression of the araA Gene from P. haloplanktis
The complete genome of P. haloplanktis ATCC 14393 has been recently sequenced and released in GenBank (NCBI accession number: NZ_AHCA01000001.1) [25]. From the information of genomic sequence, a putative araA gene encoding an L-AI is included in the genome (gene locus tag: PHAL_02715, protein ID: WP_016706880.1). The full-length nucleotide sequence of the araA gene from P. haloplanktis was synthesized and incorporated with NdeI and XhoI sites in the 5′- and 3′-terminal of the gene, respectively, and then was cloned into pET-22b(+) expression vector by Shanghai Generay Biotech Co., Ltd (Shanghai, China), to give a reconstructed plasmid, pET-Phsa-araA, which may express P. haloplanktis L-AI as a protein fused to the C terminus of a His6 tag under the control of the T7 promoter.
The recombinant plasmid harboring the putative P. haloplanktis araA gene was then transformed into E. coli BL21(DE3) for expression. Recombinant E. coli cells were cultivated at 37 °C by shaking (200 rpm). IPTG (0.5 mM) was supplemented to the culture when the OD600 reached 0.6 to induce the expression of the foreign araA gene at 28 °C. After cultivating for another 8 h, the cells were harvested by centrifugation at 10,000g for 10 min at 4 °C.
Protein Purification
To purify the recombinant P. haloplanktis L-AI, the cell pellets were resuspended in a lysis buffer (50 mM Tris–HCl, 100 mM NaCl, pH 7.5) and then were disrupted by sonication for 15 min at 4 °C (pulse on 1 s and pulse off 3 s) using a Vibra-Cell™ 72405 Sonicator. The cell fragments were precipitated by centrifugation at 10, 000×g for 15 min at 4 °C. The centrifuged supernatant was collected and then filtered through a 0.45-μm filter. The filtered extract was loaded onto Chelating Sepharose Fast Flow resin column (1 × 10 cm), previously chelating NiSO4 and equilibrated with a binding buffer (50 mM sodium phosphate buffer, 500 mM NaCl, pH 7.0). A washing buffer (50 mM sodium phosphate buffer, 500 mM NaCl, 20 mM imidazole, pH 7.0) was used to elute the unbound proteins from the column. At last, the His-tagged L-AI protein was eluted from the column with an elution buffer (50 mM sodium phosphate buffer, 500 mM imidazole, 500 mM NaCl, pH 7.0). The purified enzyme solution was dialyzed against sodium phosphate buffer (50 mM, pH 7.0) containing 10 mM of ethylenediaminetetraacetic acid (EDTA) for 12 h at 4 °C, and then dialyzed against EDTA-free sodium phosphate buffer (50 mM, pH 7.0) for 12 h at 4 °C.
Protein concentration was measured by the Folin-phenol reagent method using bovine serum albumin (BSA) as a standard. The molecular mass of the purified enzyme was analyzed, under the condition of denaturing treatment, by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme Assay
The P. haloplanktis L-AI activity was measured by the determination of the amount of L-ribulose produced from l-arabinose. The standard reaction mixture included 50 mM l-arabinose, 100 mM sodium phosphate buffer (pH 7.0), 1 mM MnCl2, and 0.5 U/mL purified enzyme. The reaction mixture was incubated for 15 min at 40 °C, and was terminated by cooling samples on ice for 30 min. The generated L-ribulose was determined by cysteine–carbazole–sulfuric acid method with absorbance measured at 540 nm [26]. One unit of P. haloplanktis L-AI activity was defined as the amount of enzyme catalyzing the production of 1 μmol L-ribulose per minute.
Effect of Temperature, pH, and Metal Ions on Enzyme Activity
The optimum temperature of the enzyme was determined by assaying the enzyme samples over the range of 20–60 °C at pH 7.0 (sodium phosphate buffer, 100 mM), following the procedures described above. And, in order to study the effect of pH on the enzyme activity, two buffer systems, sodium phosphate buffer (50 mM, pH 6.0–8.0), and Glycine-NaOH buffer solution (50 mM, pH 8.5–10.0), were used.
To examine the influence of various divalent metal ions on enzymatic activity, the purified enzyme solvent was dialyzed against sodium phosphate buffer (50 mM, pH 7.0) containing 10 mM of ethylenediaminetetraacetic acid (EDTA) for 24 h at 4 °C, and then was dialyzed against EDTA-free sodium phosphate buffer (50 mM, pH 7.0) four times at 4 °C intervals. The enzyme activity was measured in the presence of different divalent metal ions at 1 mM: ZnSO4, NiSO4, CoCl2, MnCl2, MgCl2, CaCl2, BaCl2, or CuSO4, at 40 °C. The P. haloplanktis L-AI activity without adding any divalent metal ion was taken as 100 %, and all experiments were performed in triplicate.
Effect of Temperature and pH on the Enzyme Stability
To study the effect of temperature on the enzyme stability, the enzyme solution was preserved in sodium phosphate (100 mM, pH 7.0) with or without added Mn2+ (1 mM) at various temperatures for the given period of time. Then, the enzyme solution was taken out, and the residual activity was measured under standard assay conditions, according to the procedure described above. The activity of untreated P. haloplanktis L-AI was set as 100 %.
The effect of pH on the enzyme stability was studied with the enzyme solution preserved in two different buffer systems: sodium phosphate buffer (50 mM, pH 6.0–8.0) and Glycine-NaOH buffer solution (50 mM, pH 8.5–10.0) for 10 h at 4 °C. Then, the enzyme solution was withdrawn to be measured the residual activity under standard assay conditions as described above. The activity of untreated P. haloplanktis L-AI was set as 100 %.
Kinetic Parameters
The kinetic parameters of P. haloplanktis L-AI were determined in sodium phosphate buffer (50 mM, pH 8.0) containing 1 mM Mn2+, with l-arabinose as the substrate from 10 to 400 mM. According to the Lineweaver–Burk equation, Michaelis–Menten constant (K m) and turnover number (k cat) values were measured.
Docking Study
The homology model of the P. haloplanktis L-AI was created with the template of E. coli L-AI complexed with D-ribitol (PDB: 4F2D) using SWISS-MODEL protein-modeling server (http://www.expasy.ch/swissmod/ SWISS-MODEL.html).
Molecular docking of l-arabinose or d-galactose to the P. haloplanktis L-AI model was performed by the AutoDock3.0.5 software package. All the torsion angles in the small molecules were set free to perform flexible docking. The protein was prepared for docking by adding polar hydrogen atoms with the Hydrogen module in AutoDock Tools. The docking process was executed with Lamarckian genetic algorithm (LGA) and empirical free energy function . The relating parameters were set as follows: energy evaluations of 25, 000,000 (maximal number), randomly placed individuals of 150, mutation rate of 0.02, a crossover rate of 0.80, and an elitism value of 1 [27]. Forty-three independent docking runs were carried out for l-arabinose ligand, and fifty-five were for d-galactose ligand. The root-mean-square deviation (RMSD) criterion was used to classify the docked conformations. The best docked conformations were selected as the initial conformations to evaluate the molecular docking between the enzyme and the ligands. The output from AutoDock was then rendered with Discovery Studio Visuallizer 4.0 (Accelrys, USA).
Results and Discussion
Purification of the Recombinant P. haloplanktis L-AI
The araA gene (1500 bp) was obtained by the full-length gene synthesis, sub-cloned into pET-22b(+) expression vector, and then heterologously expressed in E. coli BL21(DE3). The recombinant P. haloplanktis L-AI was purified by nickel affinity chromatography to homogeneity (Fig. 1). The molecular weight of the purified enzyme was measured to be approximately 55 kDa by SDS-PAGE.
The standard reaction mixture included 50 mM l-arabinose, 100 mM sodium phosphate buffer (pH 7.0), 1 mM MnCl2, and 0.5 U/mL purified enzyme. The reaction mixture was incubated for 15 min at 40 °C and was terminated by cooling samples on ice for 30 min. The P. haloplanktis L-AI exhibited specific activity of 5.2 U/mg toward l-arabinose; however, it could not convert d-galactose to d-tagatose, even after the reaction for 10 h, suggesting that the enzyme had no substrate specificity toward d-galactose. So far, only the L-AI from B. subtilis str. 168 was identified having specificity toward only l-arabinose, although almost all of the L-AIs had isomerization activity toward both l-arabinose and d-galactose. In this study, a novel L-AI without d-galactose isomerase activity was identified from P. haloplanktis ATCC 14393.
Sequence Analysis of P. haloplanktis L-AI
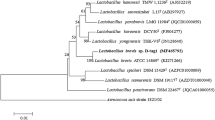
Sequence analysis of P. haloplanktis L-AI revealed an open reading frame consisting of 1500 bp, which encoded a 499 amino acid protein with a calculated molecular weight of 55,014 Da. The phylogenetic tree of the L-AIs from various microorganisms was constructed based on the amino acid sequence of each protein. The P. haloplanktis L-AI showed much closer phylogenetic relationship with the L-AI from Shewanella sp. ANA-3 [23] (Fig. 2). Homologous comparison of amino acid sequences of L-AIs from various microorganisms was also measured by Basic Local Alignment Search Tool (BLAST). The P. haloplanktis L-AI showed the highest identity (74 %) with the L-AI from Shewanella sp. ANA-3 [23], which was the unique cold-adapted L-AI reported so far, but exhibited only 22 and 23 % of identity with the L-AIs from hyperthermophilic bacteria Thermoanaerobacterium saccharolyticum NTOU1 [28] and Thermoanaerobacter mathranii DSM 11426 [29], respectively. Besides, the P. haloplanktis L-AI showed 41–56 % of identity with other reported L-AIs, including the d-galactose isomerase-free L-AI from B. coagulans 2-6 [17], B. licheniformis ATCC 14580 [38], and B. subtilis str. 168 [24], with exact 46, 47, and 50 % of identity, respectively (Table 1).
Effect of Temperature and pH on the Recombinant L-AI Activity
The optimum temperature for P. haloplanktis L-AI to catalyze the isomerization reaction was at 40 °C (Fig. 3a). And, it still retained 94 % of maximum relative activity at 45 °C, but the relative activity decreased to less than 15 % at 60 °C. In addition, although the enzyme source was from the psychrotolerant bacterium P. haloplanktis, the enzyme was obviously not cold adapted and retained only approximately 30 % of maximum relative activity at 20 °C. The results showed that the optimum temperature of L-AI from the psychrotolerant bacteria P. haloplanktis was even higher than some L-AIs from mesophilic microorganisms, such as B. subtilis str. 168 [24], L. sakei 23K [19], and Shewanella sp. ANA-3 [23] (Table 1). By comparison, the L-AI from mesophilic microorganism Shewanella sp. ANA-3 displayed the maximal activity at low temperatures comprised between 15 and 35 °C, and could show 90–95 % of its maximum relative activity at 4–10 °C. The original intention of this study was actually to obtain a cold-adapted L-AI from a psychrotolerant bacterium, for the potential application in d-tagatose production during cold food processing. But unfortunately, the above-mentioned findings obviously indicated that the P. haloplanktis L-AI could impossibly be used as a biocatalysis for d-tagatose conversion during cold food processing.
The optimum pH for isomerization of l-arabinose by P. haloplanktis L-AI was determined to be pH 8.0 (Fig. 3b). P. haloplanktis L-AI displayed a relatively wide pH spectrum, showing about 80 % of maximum activity from pH 7.5–8.5. It was similar to many other reported L-AIs, showing relatively high activity in alkaline zone, such as the L-AIs from A. flavithermus [10], Thermus sp. IM6501 [30], B. halodurans DSM 497 [31], T. mathranii DSM 11426 [29], and B. stearothermophilus US100 [11].
Effect of Temperature and pH on the Recombinant L-AI Stability
P. haloplanktis L-AI displayed significant stability with or without metal ion addition below 30 °C and retained more than 80 % of its initial activity in 2 h of preservation at 30 °C (Fig. 4a). In previous works, metal ions, especially Mn2+ and Co 2+, could significantly increase the thermostability of many L-AIs. But, in this study, the Mn2+ just slightly improved the stability of P. haloplanktis L-AI at 50 °C and almost did not change its thermostability at 30 and 40 °C. When incubated at 50 °C, the metal-free enzyme lost 40 and 70 % of its initial activity after 0.5 and 4 h of incubation, and the enzyme lost half of its initial activity after 4 h of incubation at the same temperature in the presence of 1 mM of Mn2+ (Fig. 4b). On the whole, the thermostability of P. haloplanktis L-AI was not good compared to majority of reported L-AIs.
Effect of temperature on stability of P. haloplanktis L-AI. The thermal stability was analyzed by exposing the enzyme without metal ion (a) at 30 (closed square), 40 (closed circle), and 50 °C (closed triangle), or with 1 mM of Mn2+ (b) at 30 (open square), 40 (open triangle), and 50 °C (open circle), for different time intervals. Values are means of three replications ± standard deviation
The study on the effect of pH on the enzyme stability showed that the enzyme was relatively stable at the pH range of 7.0–8.5 for 10 h (Fig. 5). After preservation for 10 h in different pH buffer systems at range of 7.0–8.5, the residual enzyme activity was 70–90 % of the untreated activity.
Effect of Metal Ions on the Recombinant L-AI Activity
The P. haloplanktis L-AI activity was determined in the presence of different metal ions at the concentration of 1 mM. The catalytic activity was significantly activated by Co2+ and Mn2+, giving 340 and 420 % of the relative activity compared to its initial activity (Fig. 6). And, when both Co2+ and Mn2+ were added at the concentration of 0.5 and 1 mM, respectively, the activity was 380 % of the initial activity. Among all the reported L-AIs, most of the L-AIs could be significantly activated by Co2+ or Mn2+, but a few L-AIs were different, such as A. flavithermus L-AI [10], which was significantly activated by Ni2+. In addition to Co2+ and Mn2+, other mental ions, Mg2+, Ca2+, and Ni2+, were also found to slightly activate the catalytic activity of P. haloplanktis L-AI in our study; Zn2+ and Ba2+ did not change the activity much (less than 10 %); however, Cu2+ obviously inhibited the enzyme activity up to 70 % (Fig. 6). The results of the study showed some metal ions could greatly increase the enzyme activity, which was consistent with most of the reported L-AIs. But, there were also some metal-independent L-AIs, such as the L-AIs from B. stearothermophilus US100 [11], Arthrobacter sp. 22c [32], and B. halodurans DSM 497 [31].
Kinetics of the Recombinant L-AI
The kinetic parameters of P. haloplanktis L-AI toward l-arabinose was determined, according to the Lineweaver–Burk plots, under the optimum reaction conditions (pH 8.0 and 40 °C). The K m and V max were determined to be 111.68 mM and 13.91 U/mg toward l-arabinose, respectively (Table 2). The value of K m was close to that of many reported L-AIs, such as the ones from A. hesperidum URH17-3-68 [16], B. coagulans 2-6 [17], B. longum [33], and T. neapolitana 5608 [34]. As a result, the catalytic efficiency (k cat/K m) was calculated to be 6.92/mM/min toward l-arabinose. The catalytic efficiency toward l-arabinose was much lower than that of most of the reported L-AIs (Table 2), except A. flavithermus L-AI [10], which showed significantly higher catalytic efficiency toward d-galactose than l-arabinose.
Molecular Docking
The homology model of P. haloplanktis L-AI (protein ID: WP_016706880.1), using crystal structure of E. coli L-AI complexed with D-ribitol (PDB entry 4F2D) as template, was created with SWISS-MODEL protein-modeling server. The amino acid sequences of P. haloplanktis L-AI showed 55 % of identity to that of E. coli L-AI, and the location of proposed active sites (E305, E332, H348, and H446) from the P. haloplanktis L-AI model was also similar to the reported E. coli L-AI (E306, E333, H350, and H450) [35]. Although the crystal structure of E. coli L-AI was identified, unfortunately, there was still no information available about the crystal structure of L-AI in complex with the substrate l-arabinose or d-galactose. The comparison with known structures revealed that E. coli L-AI structure was the most similar to the structure of E. coli L-fucose isomerase. It was reported that the residues D361 and E337 of E. coli L-fucose isomerase active site were involved in proton transfer in an enediol mechanism [36]. In the P. haloplanktis L-AI model, E305 and E332 were at the similar positions as that of E337 and D361 of E. coli L-fucose isomerase, suggesting that E305 and E332 of P. haloplanktis L-AI probably transferred proton via an enediol intermediate.
Then, the model was docked with two different ligands (d-galactose and l-arabinose), and forty-three and fifty-five conformations were obtained, respectively. The best ligand docking conformation was selected based on the docking energy and the root-mean-square deviation (RMSD) criterion. When l-arabinose was used as substrate to be docked into the binding site of P. haloplanktis L-AI (Fig. 7a), potential hydrogen bonds (1.9 and 2.5Å) were formed by the hydrogens in the hydroxyl groups at C2 and C5 sites of l-arabinose and the oxygens in carboxyl groups of E332 and E305, respectively. When the binding site of P. haloplanktis L-AI was docked with d-galactose (Fig. 7b), the hydrogens in the hydroxyl groups at C5 and C6 sites of d-galactose both formed hydrogen bonds with oxygen in carboxyl groups of E305, but the distance between the oxygen in carboxyl groups of E332 and the hydrogen in the hydroxyl groups of C2 of d-galactose was 5.4 Å, not forming any hydrogen bonds at this site. These docking results were similar to the molecular modeling of B. subtilis L-AI to some extent [24]. Hydrogen bonds formed between B. subtilis L-AI active residues (E330, E305) and hydroxyl groups at C2 and C5 sites of substrate l-arabinose. When the active site of B. subtilis L-AI was docked with d-galactose, the results did not show any potential hydrogen bond interactions between B. subtilis L-AI active residues and hydroxyl groups at C2 and C5 sites of d-galactose. The authors speculated that the hydrogen bonds, especially at C2 of the substrate, played an important role in stabilizing the transition state intermediate and keeping proper orientation to perform the isomerization reaction.
Molecular docking of L-AIs with l-arabinose or d-galactose in the active site pocket. a, b showed the active sites of P. haloplanktis L-AI docked with l-arabinose and d-galactose, respectively. L-arabinose was bound into the active site through hydrogen bonds (green dotted lines) with E305 (2.5 Å) and E332 (1.9 Å). The hydroxyls of C5 and C6 of d-galactose formed hydrogen bonds (green dotted lines) with E305; however, the distance between hydrogen in the hydroxyl groups of C2 of d-galactose and oxygen in carboxyl groups of E332 was 5.4 Å (orange straight line). C and D showed the the active sites of A. cellulolytics L-AI docked with l-arabinose and d-galactose, respectively. l-Arabinose was bound into the active site through hydrogen bonds (green dotted lines) with E307 (1.4 Å) and E334 (2.2 Å), and d-galactose was bound into the active site through hydrogen bonds (green dotted lines) with E334 (2.0 Å) (Color figure online)
To support the speculation, the homology model of the L-AI from Acidothermus cellulolytics ATCC 43068, which could catalyze the isomerization of d-galactose to d-tagatose, was also created using the same method. For A. cellulolytics L-AI, hydrogens in the hydroxyl groups at C2 and C5 sites of substrate l-arabinose formed potential hydrogen bonds (2.2Å and 1.4Å) with the oxygens in carboxyl groups of E334 and E307, respectively (Fig. 7c). The hydrogens in the hydroxyl groups of C2 of d-galactose also formed hydrogen bonds (2.0 Å) with oxygen in the carboxyl groups of E334 (Fig. 7d). The results could also exhibit that C2 of the substrate played an important role for isomerization reaction.
As mentioned above, E305 and E332 of P. haloplanktis L-AI (also E307 and E334 of A. cellulolytics L-AI) probably transferred proton via an enediol intermediate for aldose–ketose isomerization. Although there were hydrogen bonds between d-galactose and the active site residues in our docking models, hydrogen in the hydroxyl groups of C2 d-galactose did not form hydrogen bonds with E332 which played an important role in the forming of enediol intermediate. But for A. cellulolytics L-AI, hydrogen in the hydroxyl groups of C2 d-galactose could form hydrogen bonds with E334, and the experimental results also showed that this isomerase could catalyze the isomerization of d-galactose to d-tagatose [15]. Therefore, it was suggested that the hydrogen bonds between hydrogen in the hydroxyl groups of C2 of the ligand with the active site residues E332 could effectively immobilize the ligand, and the enzyme would recognize the immobilized ligand as a substrate for isomerization.
In conclusion, the L-AI was characterized for the first time from a psychrotolerant bacterium, P. haloplanktis ATCC 14393. The enzyme was the second reported d-galactose isomerase-free L-AI, after the L-AI from B. subtilis str. 168, with only substrate specificity toward l-arabinose. The enzyme was not cold adapted showing optimum temperature at 40 °C, although it was from the psychrotolerant bacterium source. Based on the molecular docking simulation studies of the enzyme with both l-arabinose and d-galactose, it was suggested that the active site residues of P. haloplanktis L-AI could recognize only l-arabinose but not d-galactose as substrate for isomerization. This specific L-AI might not be a good candidate for d-tagatose production, while these interesting findings provided some valuable information for the study on the molecular mechanism of substrate specificity of L-AI.
References
Lin, H. C., Lei, S. P., & Wilcox, G. (1985). The araBAD operon of Salmonella typhimurium LT2. II. Nucleotide sequence of araA and primary structure of its product, l-arabinose isomerase. Gene, 34, 123–128.
Kawaguchi, H., Sasaki, M., Vertes, A. A., Inui, M., & Yukawa, H. (2008). Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Applied Microbiology and Biotechnology, 77, 1053–1062.
Oh, D. K. (2007). Tagatose: properties, applications, and biotechnological processes. Applied Microbiology and Biotechnology, 76, 1–8.
Kim, P. (2004). Current studies on biological tagatose production using l-arabinose isomerase: a review and future perspective. Applied Microbiology and Biotechnology, 65, 243–249.
Buemann, B., Toubro, S., Raben, A., Blundell, J., & Astrup, A. (2000). The acute effect of d-tagatose on food intake in human subjects. British Journal of Nutrition, 84, 227–231.
Bär, A., Lina, B., De Groot, D., De Bie, B., & Appel, M. (1999). Effect of d-tagatose on liver weight and glycogen content of rats. Regulatory Toxicology and Pharmacology, 29, S11–S28.
Donner, T., Wilber, J., & Ostrowski, D. (1999). d-Tagatose, a novel hexose: acute effects on carbohydrate tolerance in subjects with and without type 2 diabetes. Diabetes, Obesity and Metabolism, 1, 285–291.
Bertelsen, H., Jensen, B. B., & Buemann, B. (1999). d-Tagatose—a novel low-calorie bulk sweetener with prebiotic properties. World Review of Nutrition and Dietetics, 85, 98–109.
Levin, G. V. (2002). Tagatose, the new GRAS sweetener and health product. Journal of Medicinal Food, 5, 23–36.
Li, Y., Zhu, Y., Liu, A., & Sun, Y. (2011). Identification and characterization of a novel l-arabinose isomerase from Anoxybacillus flavithermus useful in d-tagatose production. Extremophiles, 15, 441–450.
Rhimi, M., & Bejar, S. (2006). Cloning, purification and biochemical characterization of metallic-ions independent and thermoactive l-arabinose isomerase from the Bacillus stearothermophilus US100 strain. Biochimica et Biophysica Acta, 1760, 191–199.
Hong, Y. H., Lee, D. W., Lee, S. J., Choe, E. A., Kim, S. B., Lee, Y. H., et al. (2007). Production of d-tagatose at high temperatures using immobilized Escherichia coli cells expressing l-arabinose isomerase from Thermotoga neapolitana. Biotechnology Letters, 29, 569–574.
Lee, D. W., Jang, H. J., Choe, E. A., Kim, B. C., Lee, S. J., Kim, S. B., et al. (2004). Characterization of a thermostable l-arabinose (d-galactose) isomerase from the hyperthermophilic eubacterium Thermotoga maritima. Applied and Environmental Microbiology, 70, 1397–1404.
Lee, S. J., Lee, D. W., Choe, E. A., Hong, Y. H., Kim, S. B., Kim, B. C., et al. (2005). Characterization of a thermoacidophilic l-arabinose isomerase from Alicyclobacillus acidocaldarius: role of Lys-269 in pH optimum. Applied and Environmental Microbiology, 71, 7888–7896.
Cheng, L., Mu, W., Zhang, T., & Jiang, B. (2010). An l-arabinose isomerase from Acidothermus cellulolytics ATCC 43068: cloning, expression, purification, and characterization. Applied Microbiology and Biotechnology, 86, 1089–1097.
Fan, C., Liu, K., Zhang, T., Zhou, L., Xue, D., Jiang, B., et al. (2014). Biochemical characterization of a thermostable l-arabinose isomerasefrom a thermoacidophilic bacterium, Alicyclobacillus hesperidum URH17-3-68. Journal of Molecular Catalysis B-Enzymatic, 102, 120–126.
Zhou, X., & Wu, J. C. (2012). Heterologous expression and characterization of Bacillus coagulans l-arabinose isomerase. World Journal of Microbiology and Biotechnology, 8, 2205–2212.
Chouayekh, H., Bejar, W., Rhimi, M., Jelleli, K., Mseddi, M., & Bejar, S. (2007). Characterization of an l-arabinose isomerase from the Lactobacillus plantarum NC8 strain showing pronounced stability at acidic pH. FEMS Microbiology Letters, 277, 260–267.
Rhimi, M., Ilhammami, R., Bajic, G., Boudebbouze, S., Maguin, E., Haser, R., et al. (2010). The acid tolerant l-arabinose isomerase from the food grade Lactobacillus sakei 23 K is an attractive d-tagatose producer. Bioresource technology, 101, 9171–9177.
Xu, Z., Qing, Y., Li, S., Feng, X., Xu, H., & Ouyang, P. (2011). A novel l-arabinose isomerase from Lactobacillus fermentum CGMCC2921 for d-tagatose production: Gene cloning, purification and characterization. Journal of Molecular Catalysis B-Enzymatic, 70, 1–7.
Zhang, H., Jiang, B., & Pan, B. (2007). Purification and characterization of l-arabinose isomerase from Lactobacillus plantarum producing d-tagatose. World Journal of Microbiology and Biotechnology, 23, 641–646.
Staudigl, P., Haltrich, D., & Peterbauer, C. K. (2014). l-Arabinose isomerase and d-Xylose isomerase from Lactobacillus reuteri: characterization, coexpression in the food grade Host Lactobacillus plantarum, and application in the conversion of d-galactose and d-glucose. Journal of Agricultural and Food Chemistry, 62, 1617–1624.
Rhimi, M., Bajic, G., Ilhammami, R., Boudebbouze, S., Maguin, E., Haser, R., et al. (2011). The acid-tolerant l-arabinose isomerase from the mesophilic Shewanella sp. ANA-3 is highly active at low temperatures. Microbial Cell Factories, 10, 96.
Kim, J. H., Prabhu, P., Jeya, M., Tiwari, M. K., Moon, H. J., Singh, R. K., et al. (2010). Characterization of an l-arabinose isomerase from Bacillus subtilis. Applied Microbiology and Biotechnology, 85, 1839–1847.
Xie, B. B., Shu, Y. L., Qin, Q. L., Rong, J. C., Zhang, X. Y., Chen, X. L., et al. (2012). Genome sequences of type strains of seven species of the marine bacterium Pseudoalteromonas. Journal of Bacteriology, 194, 2746–2747.
Nakamura, M. (1968). Determination of fructose in the presence of a large excess of glucose. Agricultural and Biological Chemistry, 32, 701–706.
Lu, S. Y., Jiang, Y. J., Lv, J., Wu, T. X., Yu, Q. S., & Zhu, W. L. (2010). Molecular docking and molecular dynamics simulation studies of GPR40 receptor–agonist interactions. Journal of Molecular Graphics and Modelling, 28, 766–774.
Hung, X. G., Tseng, W. C., Liu, S. M., Tzou, W. S., & Fang, T. Y. (2014). Characterization of a thermophilic l-arabinose isomerase from Thermoanaerobacterium saccharolyticum NTOU1. Biochemical Engineering Journal, 83, 121–128.
Jorgensen, F., Hansen, O. C., & Stougaard, P. (2004). Enzymatic conversion of d-galactose to d-tagatose: heterologous expression and characterisation of a thermostable l-arabinose isomerase from Thermoanaerobacter mathranii. Applied Microbiology and Biotechnology, 64, 816–822.
Kim, J. W., Kim, Y. W., Roh, H. J., Kim, H. Y., Cha, J. H., Park, K. H., et al. (2003). Production of tagatose by a recombinant thermostable l-arabinose isomerase from Thermus sp. IM6501. Biotechnology Letter, 25, 963–967.
Lee, D. W., Choe, E. A., Kim, S. B., Eom, S. H., Hong, Y. H., Lee, S. J., et al. (2005). Distinct metal dependence for catalytic and structural functions in the l-arabinose isomerases from the mesophilic Bacillus halodurans and the thermophilic Geobacillus stearothermophilus. Archives of Biochemistry and Biophysics, 434, 333–343.
Wanarska, M., & Kur, J. (2012). A method for the production of d-tagatose using a recombinant Pichia pastoris strain secreting beta-d-galactosidase from Arthrobacter chlorophenolicus and a recombinant l-arabinose isomerase from Arthrobacter sp. 22c. Microbial Cell Factories, 11, 113.
Salonen, N., Nyyssola, A., Salonen, K., & Turunen, O. (2012). Bifidobacterium longum l-arabinose isomerase–overexpression in Lactococcus lactis, purification, and characterization. Applied Biochemistry and Biotechnology, 168, 392–405.
Kim, B. C., Lee, Y. H., Lee, H. S., Lee, D. W., Choe, E. A., & Pyun, Y. R. (2002). Cloning, expression and characterization of l-arabinose isomerase from Thermotoga neapolitana: bioconversion of d-galactosed-galactose to d-tagatose using the enzyme. FEMS Microbiology Letter, 212, 121–126.
Manjasetty, B. A., & Chance, M. R. (2006). Crystal structure of Escherichia coli l-arabinose isomerase (ECAI), the putative target of biological tagatose production. Journal of Molecular Biology, 360, 297–309.
Seemann, J. E., & Schulz, G. E. (1997). Structure and mechanism of l-fucose isomerase from Escherichia coli. Journal of Molecular Biology, 273, 256–268.
Seo, M. J. (2013). Characterization of an l-arabinose isomerase from Bacillus thermoglucosidasius for d-tagatose production. Bioscience, Biotechnology, and Biochemistry, 77, 385–388.
Prabhu, P., Tiwari, M. K., Jeya, M., Gunasekaran, P., Kim, I. W., & Lee, J. K. (2008). Cloning and characterization of a novel l-arabinose isomerase from Bacillus licheniformis. Applied Microbiology and Biotechnology, 81, 283–290.
Yoon, S. H., Kim, P., & Oh, D. K. (2003). Properties of l-arabinose isomerase from Escherichia coli as biocatalyst for tagatose production. World Journal of Microbiology and Biotechnology, 19, 47–51.
Torres, P. R., Manzo, R. M., Rubiolo, A. C., Batista-Viera, F. D., & Mammarella, E. J. (2014). Purification of an l-arabinose isomerase from Enterococcus faecium DBFIQ E36 employing a biospecific affinity strategy. Journal of Molecular Catalysis B-Enzymatic, 102, 99–105.
Cheng, L., Mu, W., & Jiang, B. (2010). Thermostable l-arabinose isomerase from Bacillus stearothermophilus IAM 11001 for d-tagatose production: gene cloning, purification and characterisation. Journal of the Science of Food and Agriculture, 90, 1327–1333.
Kim, H. J., Kim, J. H., Oh, H. J., & Oh, D. K. (2006). Characterization of a mutated Geobacillus stearothermophilus l-arabinose isomerase that increases the production rate of d-tagatose. Journal of Applied Microbiology, 101, 213–221.
Kim, H. J., & Oh, D. K. (2005). Purification and characterization of an L-arabinose isomerase from an isolated strain of Geobacillus thermodenitrificans producing d-tagatose. Journal of Biotechnology, 120, 162–173.
Nakamatu, T., & Yamanaka, K. (1969). Crystallization and properties of l-arabinose isomerase from Lactobacillus gayonii. Biochimica et Biophysica Acta, 178, 156–165.
Takata, G., Poonperm, W., Rao, D., Souda, A., Nishizaki, T., Morimoto, K., et al. (2007). Cloning, expression, and transcription analysis of l-arabinose isomerase gene from Mycobacterium smegmatis SMDU. Bioscience, Biotechnology, and Biochemistry, 71, 2876–2885.
Men, Y., Zhu, Y., Zhang, L., Kang, Z., Izumori, K., Sun, Y., et al. (2014). Enzymatic conversion of d-galactose to d-tagatose: cloning, overexpression and characterization of l-arabinose isomerase from Pediococcus pentosaceus PC-5. Microbiological Research, 169, 171–178.
Acknowledgments
This work was supported by the NSFC Project (No. 21276001), the 863 Project (No. 2013AA102102), the Support Project of Jiangsu Province (No. BK20130001 and 2015-SWYY-009), and the project of Outstanding Scientific and Technological Innovation Group of Jiangsu Province (Jing Wu).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, W., Fan, C., Zhang, T. et al. Cloning, Expression, and Characterization of a Novel l-Arabinose Isomerase from the Psychrotolerant Bacterium Pseudoalteromonas haloplanktis . Mol Biotechnol 58, 695–706 (2016). https://doi.org/10.1007/s12033-016-9969-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-016-9969-3