Abstract
Among noncoding RNAs, microRNAs (miRNAs) have been most extensively studied, and their biology has repeatedly been proven critical for central nervous system pathological conditions. The diagnostic value of several miRNAs was appraised in pediatric dysembryoplastic neuroepithelial tumors (DNETs) using miRNA microarrays and receiving operating characteristic curves analyses. Overall, five pediatric DNETs were studied. As controls, 17 samples were used: the FirstChoice Human Brain Reference RNA and 16 samples from deceased children who underwent autopsy and were not present with any brain malignancy. The miRNA extraction was carried out using the mirVANA miRNA Isolation Kit, while the experimental approach included miRNA microarrays covering 1211 miRNAs. Quantitative real-time polymerase chain reaction was performed to validate the expression profiles of miR-1909* and miR-3138 in all samples initially screened with miRNA microarrays. Our findings indicated that miR-3138 might act as a tumor suppressor gene when down-regulated and miR-1909* as a putative oncogenic molecule when up-regulated in pediatric DNETs compared to the control cohort. Subsequently, both miRNA signatures might serve as putative diagnostic biomarkers for pediatric DNETs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dysembryoplastic neuroepithelial tumors (DNETs) are rare, benign glioneuronal tumors, which become manifest during childhood, and are most commonly located in the temporal lobe [1, 2]. According to the World Health Organization classification system, DNETs have been classified as grade I, and they have been included as mixed neuroglial tumors [3]. Malignant transformation of DNETs has rarely been described [4], while previous reports also suggested infrequent aggressive and recurrent behaviors [5]. As slow-growing tumors though, they have been increasingly documented as a cause of epilepsy in young children [4]. As stated by Qaddoumi et al. [6], clinically, DNETs present as seizures before the age of 20 years in patients with normal intelligence quotient. Seizures add a significant burden, since their presence affords an important risk factor for long-term disability [7]. Moreover, seizures can be at times life-threatening themselves with increasing pediatric brain malignancy survivor’s risk of suicide into adulthood [2, 8].
MicroRNAs (miRNAs) are small noncoding RNAs, which regulate gene expression and silence a wide range of target genes [9]. MicroRNAs impact vital cellular and physiological processes, while their aberrant expression has been linked to a plethora of serious diseases, including cancer [10]. Notably, several previous reports suggested that miRNAs are essential regulators of many of the key pathways implicated in the pathogenesis and progression of pediatric central nervous system (CNS) malignancies [11–13]. Recent evidence also indicated that miRNAs can be used to detain changes in neurons, which makes them ideal putative biomarkers for central nervous system pathology [10]. Previous reports have demonstrated their use as potential diagnostic and prognostic biomarkers as well as therapy-related targets of pediatric CNS neoplasms [11, 14, 15].
Based on these sightings, the current study was undertaken to ascertain putative miRNA signatures in pediatric DNETs to provide additional evidence regarding the early and reliable diagnosis of the disease. To our knowledge, the present report is the first to identify miRNA signatures in DNETs. Our findings seemed to provide novel insights into the potential diagnostic properties of certain miRNAs in pediatric DNETs.
Materials and methods
Patients and tumor samples
Overall, resected brain tumors were studied from children diagnosed with DNETs (n = 5) diagnosed according to the 2007 WHO criteria [16]. As controls, 17 samples were used; the FirstChoice Human Brain Reference RNA was used (Ambion, Austin, TX, USA), and 16 samples were obtained from deceased children who underwent autopsy and were not present with any brain distortion, including the following anatomic locations: cerebellum (n = 4), medulla oblongata (n = 4), parietal lobe (n = 4) and temporal lobe (n = 4). The patient cohort included two males and three females, aged from 4.02 to 12.05 years. The median age of DNET patients was 7.07 years, while the median age of the non-malignant cohort was 9 years. The patients’ clinicopathologic characteristics are presented in Table 1. All samples were snap-frozen during resection and stored at −80 °C until use. The present study was conducted with the approval of “Aghia Sophia” Children’s Hospital Ethics Committee (Protocol No. 11685/11-8-2004). Informed consent was obtained from the parents of all children included in the study.”
MicroRNA profiling
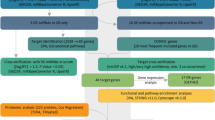
The miRNA profiling was performed as previously described by Braoudaki et al. [11]. Total RNA and miRNAs were extracted using the TRIzol standard protocol (Invitrogen, Carlsbad, CA) and the mirVANA miRNA Isolation Kit (Ambion, Austin, TX). Labeling and hybridization were carried out using the Label IT miRNA Labeling Kit (Mirus Bio LLC, USA) following the manufacturer’s instructions. Samples were hybridized to Applied MicroArrays (miRlink Bioarray 300054-3PK) platform, while images were scanned using Agilent Microarray Scanner (G2565CA) controlled by Agilent Scan Control 7.0 software. The total gene signals were extracted using the ImaGene 6.0 software (BioDiscovery Inc., USA). MicroRNAs were considered significantly differentially expressed (DE) if they obtained a p value <0.05 and an FDR ≤0.05.
MiRNA signature validation by quantitative real-time polymerase chain reaction (qRT-PCR)
Expression measurements of selected miRNAs: hsa-miR-3138 and hsa-miR-1909* targets were studied in all samples screened (n = 22) with miRNA microarrays using qRT-PCR, as described in Braoudaki et al. [11]. Briefly, qRT-PCR was performed using a standard TaqMan PCR kit procedure on a LC480 LightCycler system (Roche GmbH, Switzerland). RNU44 was used as a reference gene. Relative expression was calculated using the comparative ΔΔCt method.
Statistical analysis
The multiparameter analyses were performed with MATLAB® simulation environment (The MathWorks, Inc., Natick, MA). The two-tailed Student’s t test was used to test the mean differences between two groups. Continuous variables are expressed as median ± standard deviation unless indicated differently. Receiver operating characteristic (ROC) curves were established to evaluate the diagnostic value of deregulated miRNAs for differentiating between tumors (PAs and EPs) and controls. MiRNA expression analyses of association with clinical variables were conducted with the Kruskal–Wallis test. Time to relapse and overall survival were also analyzed from different groups of clinical variables with the Kruskal–Wallis test.
Results
MicroRNA expression and patient diagnosis
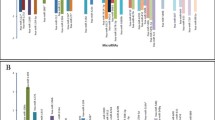
In the current study, we identified a total of 120 DE miRNAs (p < 0.05 and FDR < 0.05) in the DNET tumor group when compared to the non-malignant group of patients. Overall, the majority of the DE miRNAs observed in DNETs were down-regulated with a total of 70 miRNAs (58.3 %) exhibiting decreased expression and 50 miRNAs (41.6 %) showing increased expression (Fig. 1).
ROC analysis
ROC analysis of all miRNAs in each tumor type, using the microarray expression data, evaluated the extent to which they could separate the tumor entity from the control group. MicroRNAs with a p < 0.05 and an AUC >0.7 were selected as successful distinguishing markers between DNETs and the control group (Fig. 2). More specifically, the ROC curves yielded the following AUCs: miR-101* (AUC = 0.81, down-regulated), miR-186* (AUC = 0.92, down-regulated), miR-1909* (AUC = 0.99, up-regulated), miR-224* (AUC = 0.86, down-regulated), miR-3138 (AUC = 0.89, down-regulated), miR-363* (AUC = 0.81, down-regulated) and miR-572 (AUC = 0.94, down-regulated) were found to discriminate DNETs from controls. In particular, miR-1909* and miR-3138 once again appeared to discriminate DNETs from controls.
ROC analysis using miRNA data. ROC curves of the most imperative DE miRNAs in DNETs identified in this study using the microarray expression data. Selected miRNAs could significantly separate control from DNET samples. In particular, miRNAs presented included miR-101* with AUC = 0.81 (a), miR-186* with AUC = 0.92 (b), miR-1909* with AUC = 0.99 (c), miR-224* with AUC = 0.86 (d), miR-3138 with AUC = 0.89 (e), miR-363* with AUC = 0.81 (f) and miR-572 with AUC = 0.94 (g)
MiRNA signature validation by qRT-PCR
We examined by qRT-PCR the expression levels of miR-3138 and miR-1909* in a total of 22 samples including the patients and control cohorts (Fig. 3). According to our results, miR-3138 was found down-regulated in all DNET samples screened, verifying the initial miRNA microarray findings. Regarding miR-1909*, qRT-PCR again verified the increased expression levels initially found in all DNET samples examined by miRNA microarrays.
Discussion
To date, a surge of interest is shown on the role of miRNAs in cancer, since several previous reports have suggested that the deregulation of miRNA molecules can have a dramatic biological and clinical impact. In addition, as has previously been reported, miRNA expression profiles can be used to distinguish between closely related tumor tissue subtypes and might provide a consistent diagnosis of the disease. In the current study, we examined miRNA signatures in pediatric patients with DNETs in an attempt to identify putative biomarkers predictive for the diagnosis of these infrequent epileptogenic tumors, which might inflict significant neurological and cognitive damage, despite the favorable survival rates.
Specifically, herein, we compared the differential miRNA expression profiles between brain tissues resected from children diagnosed with DNETs and those collected during autopsy from diseased children that were not present with any brain distortion and were not diagnosed with any type of malignancy. Such comparisons are highly prominent, since the heterogeneity of the compared tissues might lead to the identification of putative reliable and robust miRNA biomarkers.
By performing miRNA microarrays, overall 120 differentially expressed miRNAs between DNETs and the control cohort were identified. Quantitative RT-PCR measurement was used to validate the expression levels of two miRNAs, miR-3138 and miR-1909*, whose differential expression was more pronounced. More specifically, miR-3138 was found down-regulated in the DNET cohort when compared to the control group. MiR-3138 was selected as a successful distinguishing marker between DNETs and the control cohort, according to ROC analyses findings (AUC = 0.898). To our knowledge, limited reports have been found regarding the role of miR-3138 in cancer. Zhang et al. [17] indicated that miR-3138 could enhance radioresistance in human cervical cancer cells.
Regarding miR-1909*, markedly elevated levels of expression were detected in the DNET cohort, suggesting that it might possess oncogenic activities when overexpressed. It is also noteworthy that mir-1909* was also detected following a fairly strict filtering approach in the ROC tests (AUC = 0.998), verifying the possibility that it affords a distinguishing marker between the tumor types and the normal tissues. Yet again, to the best of our knowledge, thus far, no studies are available describing the value of miR-1909* in pediatric brain malignancies, whereas inadequate reports were found regarding their role in other types of cancer. In a similar context, Della Vittoria Scarpati et al. [18] who studied miRNA signatures in locally advanced rectal cancer found that miR-1909* was significantly up-regulated in patients with less aggressive rectal cancer of tumor regression grade 1.
It is notable that by conducting ROC curves analyses, the diagnostic value of several additional miRNAs: miR-101*, miR-186*, miR-363*, miR-224*, and miR-572 was appraised with AUCs ranging from 0.807 to 0.941. Of note, all aforementioned miRNAs were found to possess tumor suppressor properties in DNETs, and for the majority of them, this is in line with other investigations which proposed similar properties in other types of cancers. For instance, previous reports have shown that miR-101* participates in the carcinogenesis and tumor progression in various cancers. More specifically, decreased levels of expression of miR-101* have been reported in natural killer/T cell lymphoma (NKTL) [19], in rhabdomyosarcoma [20] and in laryngeal squamous cell carcinoma [21]. Further on, the diminished expression of miR-186* has been detected in non-small cell lung cancer [22, 23] and in bladder cancer [24]. The tumor suppressor properties of miR-363* have also been reported in hepatocellular carcinoma [25], in human head and neck squamous cell carcinoma [26], in NKTL [19] and in neuroblastoma tumorigenesis [27].
Our findings though are not in line with similar reports investigating the role of miR-224* and miR-572 in malignant diseases. According to selected studies, miR-574 possesses oncogenic properties in human ovarian cancer [28] and in early-stage renal cell carcinoma [29]. As far as it concerns miR-224*, it has been found overexpressed in diverse brain malignancies and notably in gliomas and medulloblastomas. More specifically, Lu et al. [30] suggested that miR-224* was associated with advanced pathological grade and was linked to inferior prognosis, while Kunder et al. [31] reported that in medulloblastomas in the WNT subgroup, significant overexpression of miR-224* was noted.
Collectively, as previously reported by Braoudaki et al. [11], it is probable if not assured that miRNAs play tissue-specific roles and do not possess global tumor properties. Therefore, it is essential to investigate their potential role in every tissue before entitling them as either oncogenes or tumor suppressor genes. Herein, our findings provided novel evidence regarding the potential role of several miRNAs and markedly for miR-3138 and miR-1909* in the pathogenesis of DNETs alone. Confirmation of larger-scale prospective studies of children with DNETs and additional confirmatory experimentation are necessitated to unravel the described role of the aforementioned miRNAs.
References
Chassoux F, Daumas-Duport C. Dysembryoplastic neuroepithelial tumors: where are we now? Epilepsia. 2013;54(Suppl 9):129–34. doi:10.1111/epi.12457.
Ranger A, Diosy D. Seizures in children with dysembryoplastic neuroepithelial tumors of the brain–A review of surgical outcomes across several studies. Childs Nerv Syst. 2015;31(6):847–55. doi:10.1007/s00381-015-2675-9.
Spalice A, Ruggieri M, Grosso S, Verrotti A, Polizzi A, Magro G, et al. Dysembryoplastic neuroepithelial tumors: a prospective clinicopathologic and outcome study of 13 children. Pediatr Neurol. 2010;43(6):395–402. doi:10.1016/j.pediatrneurol.2010.06.017.
Ozlen F, Gunduz A, Asan Z, Tanriverdi T, Ozkara C, Yeni N, et al. Dysembryoplastic neuroepithelial tumors and gangliogliomas: clinical results of 52 patients. Acta Neurochir (Wien). 2010;152(10):1661–71. doi:10.1007/s00701-010-0696-4.
Thom M, Toma A, An S, Martinian L, Hadjivassiliou G, Ratilal B, et al. One hundred and one dysembryoplastic neuroepithelial tumors: an adult epilepsy series with immunohistochemical, molecular genetic, and clinical correlations and a review of the literature. J Neuropathol Exp Neurol. 2011;70(10):859–78. doi:10.1097/NEN.0b013e3182302475.
Qaddoumi I, Ellison DW, Morris EB, Broniscer A, Boop F, Merchant T, et al. Dysembryoplastic neuroepithelial tumors and cognitive outcome: cure at a price? Cancer. 2010;116(23):5461–9. doi:10.1002/cncr.25528.
Maschio M, Sperati F, Dinapoli L, Vidiri A, Fabi A, Pace A, et al. Weight of epilepsy in brain tumor patients. J Neurooncol. 2014;118(2):385–93. doi:10.1007/s11060-014-1449-7.
Brinkman TM, Liptak CC, Delaney BL, Chordas CA, Muriel AC, Manley PE. Suicide ideation in pediatric and adult survivors of childhood brain tumors. J Neurooncol. 2013;113(3):425–32. doi:10.1007/s11060-013-1130-6.
Tessitore A, Cicciarelli G, Mastroiaco V, Vecchio FD, Capece D, Verzella D, et al. Therapeutic use of MicroRNAs in cancer. Anti-Cancer Agents Med Chem. 2015;16(1):7–19.
Braoudaki M, Lambrou GI. MicroRNAs in pediatric central nervous system embryonal neoplasms: the known unknown. J Hematol Oncol. 2015;8(1):6. doi:10.1186/s13045-014-0101-5.
Braoudaki M, Lambrou GI, Giannikou K, Milionis V, Stefanaki K, Birks DK, et al. Microrna expression signatures predict patient progression and disease outcome in pediatric embryonal central nervous system neoplasms. J Hematol Oncol. 2014;7:96. doi:10.1186/s13045-014-0096-y.
Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–27. doi:10.1038/emboj.2008.172.
Birks DK, Barton VN, Donson AM, Handler MH, Vibhakar R, Foreman NK. Survey of MicroRNA expression in pediatric brain tumors. Pediatr Blood Cancer. 2011;56(2):211–6. doi:10.1002/pbc.22723.
Wang F, Ren X, Zhang X. Role of microRNA-150 in solid tumors. Oncol Lett. 2015;10(1):11–6. doi:10.3892/ol.2015.3170.
Wang M, Deng X, Ying Q, Jin T, Li M, Liang C. MicroRNA-224 targets ERG2 and contributes to malignant progressions of meningioma. Biochem Biophys Res Commun. 2015;460(2):354–61. doi:10.1016/j.bbrc.2015.03.038.
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi:10.1007/s00401-007-0243-4.
Zhang B, Chen J, Ren Z, Chen Y, Li J, Miao X, et al. A specific miRNA signature promotes radioresistance of human cervical cancer cells. Cancer cell Int. 2013;13(1):118. doi:10.1186/1475-2867-13-118.
Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83(4):1113–9. doi:10.1016/j.ijrobp.2011.09.030.
Ng SB, Yan J, Huang G, Selvarajan V, Tay JL, Lin B, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood. 2011;118(18):4919–29. doi:10.1182/blood-2011-07-364224.
Vella S, Pomella S, Leoncini PP, Colletti M, Conti B, Marquez VE, et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin Epigenet. 2015;7(1):82. doi:10.1186/s13148-015-0107-z.
Li M, Tian L, Ren H, Chen X, Wang Y, Ge J, et al. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J Transl Med. 2015;13:271. doi:10.1186/s12967-015-0626-6.
Cai J, Wu J, Zhang H, Fang L, Huang Y, Yang Y, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73(2):756–66. doi:10.1158/0008-5472.can-12-2651.
Cui G, Cui M, Li Y, Liang Y, Li W, Guo H, et al. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol J Int Soc Oncodev Biol Med. 2014;35(9):8933–7. doi:10.1007/s13277-014-2168-6.
Yao K, He L, Gan Y, Zeng Q, Dai Y, Tan J. MiR-186 suppresses the growth and metastasis of bladder cancer by targeting NSBP1. Diagn Pathol. 2015;10:146. doi:10.1186/s13000-015-0372-3.
Zhou P, Huang G, Zhao Y, Zhong D, Xu Z, Zeng Y, et al. MicroRNA-363-mediated downregulation of S1PR1 suppresses the proliferation of hepatocellular carcinoma cells. Cell Signal. 2014;26(6):1347–54. doi:10.1016/j.cellsig.2014.02.020.
Jamali Z, Aminabadi NA, Attaran R, Pournagiazar F, Oskouei SG, Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321–31.
Qiao J, Lee S, Paul P, Theiss L, Tiao J, Qiao L, et al. miR-335 and miR-363 regulation of neuroblastoma tumorigenesis and metastasis. Surgery. 2013;154(2):226–33. doi:10.1016/j.surg.2013.04.005.
Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu J, et al. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6(17):15180–93.
Wang C, Hu J, Lu M, Gu H, Zhou X, Chen X, et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci Rep. 2015;5:7610. doi:10.1038/srep07610.
Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Upregulation of microRNA-224 confers a poor prognosis in glioma patients. Clin Transl Oncol. 2013;15(7):569–74. doi:10.1007/s12094-012-0972-2.
Kunder R, Jalali R, Sridhar E, Moiyadi A, Goel N, Goel A, et al. Real-time PCR assay based on the differential expression of microRNAs and protein-coding genes for molecular classification of formalin-fixed paraffin embedded medulloblastomas. Neuro-Oncology. 2013;15(12):1644–51. doi:10.1093/neuonc/not123.
Author Contributions
MB conceived and designed the study, performed all experiments, evaluated and interpreted data analyses and drafted the manuscript. GIL performed all data analyses and participated in interpretation of data analyses. SAP performed resections of the postmortem specimens, KS performed tumor diagnosis, NP performed tumor resections and EK participated in the coordination and supervision of the study. All authors approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial or non-financial interests.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Rights and permissions
About this article
Cite this article
Braoudaki, M., Lambrou, G.I., Papadodima, S.A. et al. MicroRNA expression profiles in pediatric dysembryoplastic neuroepithelial tumors. Med Oncol 33, 5 (2016). https://doi.org/10.1007/s12032-015-0719-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-015-0719-3