Abstract
MAGE-A are normally expressed in testis and placenta. Among MAGEs, the MAGE-A subtype has been the most characterized in cancers. Our study was conducted to assess the expression of (MAGE-A1–MAGE-A6) m-RNA using MMRPs and MAGE-A12 m-RNA in blood for evaluating their clinical implications in breast cancer patients. RT-PCR was carried out to detect the expression of (MAGE-A1–MAGE-A6) m-RNA using MMRPs and MAGE-A12 m-RNA in blood. The study included 100 breast cancer cases aged 41–62 years and 100 controls aged 36–53 years. MAGE m-RNA expression was not detected in healthy donors. In breast cancer patients, the positivity of (MAGE-A1–MAGE-A6) m-RNA was 44 % (44 cases), while MAGE-A12 m-RNA was expressed in 13 % (13 cases). The gene expressions of MAGE-A1–A6 and MAGE-A12 were significantly associated with advanced TNM stages (P = 0.001 and 0.034, respectively). Simultaneous estimation of the gene expressions of MAGE-A1–A6 and MAGE-A12 can detect occult hematogenous dissemination of tumor cells and may help to monitor the effectiveness of the therapy and the development of effective immunotherapeutic strategies in breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is one of the most common causes of cancer-related death in women worldwide [10]. The axillary lymph node metastasis is the most important predictor of distant metastasis [2]. Circulating tumor cells (CTC) detection is considered as a promising noninvasive marker for early prediction of disease prognosis [16].
Melanoma-associated antigen gene (MAGE) was first isolated from an MZ-2 human melanoma cell line by Van der Bruggen et al. [15]. In the following years, dozens of new MAGEs were characterized and classified into 3 subgroups of acidic MAGEs, termed A, B and C, and one basic subgroup, MAGE-D, that involves Necdin, Restin and others [17]. According to expression patterns, the MAGEs were further classified into subgroups I and II. Type I MAGEs include more than 45 chromosome X-clustered genes including MAGE-A, -B and -C subfamilies. They are normally expressed in testis, trophoblast and placenta [4]. During development, type I MAGEs expression is silenced by promoter DNA methylation. However, demethylation of MAGE I promoters during the epigenetic reprogramming that occurs in many tumors triggers their expression [14]. Concerning subgroup II MAGEs, they are normally expressed in various adult human tissues. They have been suggested to play important roles in cell cycle withdrawal, neuronal differentiation and apoptosis [1]. MAGE-A1, 2, 3, 4, 6, 8, 10, 11 and 12 were found to be highly expressed in various tumors [13].
Low frequency of expression of individual MAGE has limited the use of MAGEs in CTC detection. Many researchers reported that at least one MAGE-A is expressed in most carcinomas [9], so for the diagnosis of MAGE-expressing cancer, it may be more valuable to detect expressions of multiple MAGEs together than a single-gene expression. Simultaneous detection of the gene expressions of MAGE-A1–A6 together was allowed by multiple MAGE-recognizing primers (MMRPs) designed by Park et al. [11].
This study was conducted to assess the expression of (MAGE-A1–MAGE-A6) m-RNA using MMRPs and MAGE-A12 m-RNA in blood for evaluating their clinical implications in breast cancer patients.
Subjects and methods
This research was carried out at the Medical Biochemistry and General Surgery Departments, Faculty of Medicine, Zagazig University.
Two hundred individuals were included in this study and classified into two groups. Group I (healthy control): This group included 100 apparently healthy individuals with ages ranging from 36 to 53 years (with mean value ± SD of 43.9 ± 4.4) with no history (either familial or personal). Group II: This group involved 100 breast cancer patients with ages ranging from 41 to 62 years (with mean value ± SD of 51.8 ± 5.5). According to the TNM criteria, patients were classified into four subgroups: 17 cases in stage I, 28 patients in stage II, 34 patients in stage III and 21 patients in stage IV. A written informed consent was obtained from each participant.
The diagnosis of breast cancer was based on history, clinical examination of breast and axilla, radiological examination using ultrasonography and mammogram of both breast and axilla, chest X-ray, abdominal ultrasonography and pathological diagnosis, fine needle aspiration cytology (FNAC) and true cut needle biopsy of breast mass and estrogen and progesterone receptors evaluation.
Research investigations
The expression of (MAGE-A1–MAGE-A6) m-RNA using MMRPs and MAGE-A12 m-RNA were determined in blood by reverse transcription-polymerase chain reaction (RT-PCR).
Collection and handling of samples
Three milliliters of venous blood was collected in sterile heparinized tubes. Peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoflot (Biotest, Dreieich, Germany) which is sterile filtered density gradient for the isolation of lymphocytes. Each blood sample was diluted with 3 ml RPMI1640 medium (Sigma, USA) supplemented by 10 % fetal bovine serum and PBMCs layer was collected after density gradient centrifugation.
RNA extraction
Total RNA was extracted from PBMCs using EZNA total RNA kit according to the protocol provided by the manufacturer (Omega Biotek, USA).
To determine purity and concentration of RNA, the absorbance was measured at 260 and 280 nm in a spectrophotometer. A ratio of 1.8–2.0 corresponds to 90–100 % pure nucleic acid. One OD unit measured at 260 nm corresponds to 40 μg of RNA per ml.
Reverse transcription and PCR amplification
Patient’s RNA is reverse transcribed with reverse transcriptase to synthesize the cDNA using a random primer. Then, the reaction mixture is incubated at 95 °C to inactivate the reverse transcriptase and denature the template. PCR amplification requires Taq DNA polymerase and gene-specific primers.
RT-PCR was performed using
-
Maxime RT-PCR PreMixKit supplied by iNtRON Biotechnology.
Each reaction contains RT System, RT-PCR buffer, dNTPs and Taq DNA polymerase to generate PCR product from an RNA template. It is optimized to allow the first-strand cDNA synthesis and PCR reactions to proceed as a single tube.
-
AmpGene DNA thermal cycler.
The amplified products were visualized using
-
1.
EC 360 Submarine Gel electrophoresis system (Maxicell, EC 360 MEC apparatus Cooperation St. Petersburg. Florida, USA).
-
2.
The PCR products were visualized using 2 % agarose gel and ethidium bromide under UV transillumination. Expression was considered positive when the expected band for each gene was observed.
The reaction pellet was dissolved using diethylpyrocarbonate (DEPC)-ddH2O, and then, 200 ng of total RNA template and PCR primers were added. The volume of the reaction was completed with DEPC-ddH2O to result in a final volume of 20 µl. Reverse transcription reaction was carried out at 42 °C for 60 min, and then, the temperature was adjusted to 95 °C for 5 min to inactivate the reverse transcriptase and completely denature the template.
MAGE-A1–A6 assay
According to Park et al. [11], 0.5 μM of each PCR primer was added. The MMRPs sequences and sizes of PCR products were as follows: sense, 5′-CTGAAGGAGAAGATCTGCCAGTG-3′; antisense, 5′-CCAGCATTTCTGCCTTTGTGA-3′; size, 469–493 base pair (bp). The amplification conditions were thirty cycles of 95 °C for 30 s, 60 °C for 45 s and 72 °C for 45 s followed by final extension incubation at 72 °C for 10 min.
Detection of MAGE-A12 expression
0.4 μM of each PCR primer was added. The primer sequences and size of PCR products were as follows: sense, 5′-TCCGTGAGGAGGCAAGGTTC-3′; antisense, 5′-GAGCCTGCGCACCCACCAA-3′; size, 181 bp. The amplification conditions were 93 °C for 40 s, 58 °C for 30 s and 72 °C for 30 s for thirty cycles followed by a final extension at 72 °C for 2 min [6].
We determined B-actin gene to assess the RNA integrity. The primer sequences and size of PCR products were as follows: sense, 5′-GGCATCGTGATGGACTCCG-3′; antisense, 5′-GCTGGAAGGTGGACAGCGA-3′; size, 613 bp. The amplifications were 28 cycles of 94 °C for 45 s, 65 °C for 45 s and 72 °C for 45 s [8].
Statistical analysis
The results were statistically analyzed using SPSS program (version 16).
The frequencies of positivity of each gene between groups were compared by chi-square test (χ 2 test). P value <0.05 was considered significant. To assess the validity of the screening test, sensitivity and specificity were calculated.
Results
Healthy control
No expression of any of MAGE m-RNA was detected in healthy volunteers.
Breast cancer group
(MAGE-A1–MAGE-A6) m-RNA using MMRPs
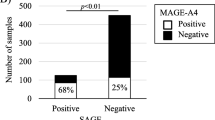
The positive expression of MAGE m-RNA was 44 % (44 out of 100 cases; Table 1; with significant association with advanced TNM stages; P = 0.001; Table 2; Fig. 1a): two out of 17 cases in stage I, 5 out of 28 cases in stage II, 20 out of 34 cases in stage III and 17 out of 21 cases in stage IV. In the early stage (stage I and II), the expressions of MAGE-A1–A6 were detected in 15.6 % (7 out of 45 patients), while in the late stage (stage III and IV), the expression was 67.3 % (37 out of 55 patients; Fig. 1b, with significant increase in the late stage than the early stage; P = 0.001).
MAGE-A12 m-RNA was expressed in 13 % (13 out of 100 cases; Table 1; with significant association with advanced TNM stages; P = 0.034; Table 2; Fig. 2a): no expression was detected in stage I, 1 out of 28 cases in stage II, 7 out of 34 cases in stage III and 5 out of 21 cases in stage IV. In the early stage (stage I and II), the expression of MAGE-A12 was detected in 2.2 % (one out of 45 patients), while in the late stage (stage III and IV) the expression was detected in 21.8 % (12 out of 55 patients; Fig. 2b; with significant increase in late stage than early stage; P = 0.004).
The expression of MAGE-A12 was associated with other MAGE-A1–A6 in nine cases, so the final result was 48 % (48 cases) expressing at least one MAGE (Table 1; Fig. 3). The negative expressions of MAGE-A1–MAGE-A6 and MAGE-A12 among healthy volunteers indicate that they are cancer specific (specificity = 100 %).
Discussion
Several tumor-associated antigens, such as carcinoembryonic antigen (CEA) are expressed in breast cancer cells, but their utility to predict the prognosis is limited by their expression in various normal cells [5].
The MAGE-A subtype has been the most characterized in cancers. Also, detection rate of MAGE-A m-RNA in metastatic tumors is higher than that of the corresponding primary tumors [3]. The dependence on a single-marker assay is limited by high CTC heterogeneity even in the same patient and low levels of individual MAGE-A expression [7]. We estimated the expressions of MAGE-A1, 2, 3, 4a, 4b, 5a, 5b and 6 (MAGE-A1–MAGE-A6) using MMRPs designed by Park et al. [11] and MAGE-A12 to assess their clinical significance in prognosis of breast cancer.
Estimation of MAGE-A1–A6 and MAGE-A12 m-RNA in PBMCs elucidated that 44 % (44 out of 100 breast cancer cases) were positive for MAGE-A1–A6 m-RNA and 13 % (13 out of 100 breast cancer cases) were positive for MAGE-A12. All samples from healthy volunteers were negative indicating that MAGE m-RNA is cancer specific (specificity = 100 %).
Furthermore, we noticed that, MAGE expression in PBMCs is correlated to the pathological stages of breast cancer; the detection rate of micrometastasis of cancer cells in peripheral blood is increased in the late stages; MAGE-A1–A6 transcripts were detected in 67.3 % (37 out of 55 patients) among the late stages, while in the early stages, the expressions of MAGE-A1–A6 were detected in 15.6 % (7 out of 45 patients; P = 0.001). Also, MAGE-A12 transcript was detected in 21.8 % (12 out of 55 patients) among the late stages, while in the early stages, the positivity was 2.2 % (1 out of 45; P = 0.004).
The expression of MAGE-A12 was associated with other MAGE-A1–A6 in nine cases, so the final result was 48 % (48 cases) expressing at least one MAGE, increasing the detection rate of CTC. Also, Sang et al. [12] reported that MAGE-A group is considered as promising targets for cancer immunotherapy because they are tumor specific.
Furthermore, no micrometastasis should have been detected in the breast cancer in the early stages. Detection of MAGE transcripts (tumor-specific markers) revealed the expressions of MAGE-A1–A6 in 15.6 % patients and MAGE-A12 in 2.2 % patients in the early stage indicating micrometastasis to the peripheral blood, so the studied MAGE transcripts may help to detect CTC earlier than other investigations and prefigure breast cancer metastasis.
Conclusion
Simultaneous estimation of the gene expressions of MAGE-A1–A6 and MAGE-A12 can detect occult hematogenous dissemination of tumor cells and may help to monitor the effectiveness of the therapy and the development of effective immunotherapeutic strategies in breast cancer. Furthermore, during follow-up, CTC detection by evaluating the expression of (MAGE-A1–MAGE-A6) m-RNA using MMRPs offers a noninvasive and cost-effective assay to predict the prognosis of breast cancer patients.
Abbreviations
- CTC:
-
Circulating tumor cells
- MAGE:
-
Melanoma-associated antigen gene
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- MMRPs:
-
Multiple MAGE-recognizing primers
- FNAC:
-
Fine needle aspiration cytology
- PBMCs:
-
Peripheral blood mononuclear cells
- DEPC:
-
Diethylpyrocarbonate
- CEA:
-
Carcinoembryonic antigen
References
Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67(6):705–12.
Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–7.
Chen CH, Huang GT, Lee HS, Yang PM, Yan MD, Chen DS. High frequency of expression of MAGE genes in human hepatocellular carcinoma. Liver. 1999;19:110–4.
Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–51.
Häffner AC, Tassis A, Zepter K, Storz M, Tureci O, Burg G, Nestle FO. Expression of cancer/testis antigens in cutaneous T cell lymphomas. Int J Cancer. 2002;97:668–70.
Kufer P, Zippelius A, Lutterbüse R, Mecklenburg I, Enzmann T, Montag A, et al. Heterogeneous expression of MAGE-A genes in occult disseminated tumor cells: a novel multimarker reverse transcription-polymerase chain reaction for diagnosis of micrometastatic disease. Cancer Res. 2002;62:251–61.
Kwon S, Kang SH, Ro J, Jeon CH, Park JW, Lee ES. The melanoma antigen gene as a surveillance marker for the detection of circulating tumor cells in patients with breast carcinoma. Cancer. 2005;104(2):251–6.
Mou DC, Cai SL, Peng JR, Wang Y, Chen HS, Pang XW, Leng XS, Chen WF. Evaluation of MAGE-1 and MAGE-3 as tumor-specific markers to detect blood dissemination of hepatocellular carcinoma cells. Br J Cancer. 2002;86(1):110–6.
Otte M, Zafrakas M, Riethdorf L, Pichlmeier U, Loning T, Janicke F, Pantel K. MAGE-A gene expression pattern in primary breast cancer. Cancer Res. 2001;61:6682–7.
Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358(13):1409–11.
Park JW, Kwon TK, Kim IH, Sohn SS, Kim YS, Kim CI, et al. A new strategy for the diagnosis of MAGE-expressing cancers. J Immunol Methods. 2002;266:79–86.
Sang M, Lian Y, Zhou X, Shan B. MAGE-A family: attractive targets for cancer immunotherapy. Vaccine. 2011;29:8496–500.
Serrano A, Lethé B, Delroisse J, Lurquin C, De Plaen E, Brasseur F, Rimoldi D, Boon T. Quantitative evaluation of the expression of MAGE genes in tumors by limiting dilution of cDNA libraries. Int J Cancer. 1999;83(5):664–9.
Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25.
Van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7.
Zhang L, Riethdorf S, Wu G, Wang T, Yang K, Peng G, Liu J, Pantel K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18:5701–10.
Zhao ZL, Lu F, Zhu F, Yang H, Chai YB, Chen SM. Cloning and biological comparison of Restin, a novel member of Mage superfamily. Sci. China C Life Sci. 2002;45:412–20.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abd-Elsalam, E.AE., Ismaeil, N.A. Melanoma-associated antigen genes: a new trend to predict the prognosis of breast cancer patients. Med Oncol 31, 285 (2014). https://doi.org/10.1007/s12032-014-0285-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0285-0