Abstract
DNA methylation plays important roles in regulating gene expression and has been reported to be related with epilepsy. This study aimed to define differential DNA methylation patterns in drug-refractory epilepsy patients and to investigate the role of DNA methylation in human epilepsy. We performed DNA methylation profiling in brain tissues from epileptic and control patients via methylated-cytosine DNA immunoprecipitation microarray chip. Differentially methylated loci were validated by bisulfite sequencing PCR, and the messenger RNA (mRNA) levels of candidate genes were evaluated by reverse transcriptase PCR. We found 224 genes that showed differential DNA methylation between epileptic patients and controls. Among the seven candidate genes, three genes (TUBB2B, ATPGD1, and HTR6) showed relative transcriptional regulation by DNA methylation. TUBB2B and ATPGD1 exhibited hypermethylation and decreased mRNA levels, whereas HTR6 displayed hypomethylation and increased mRNA levels in the epileptic samples. Our findings suggest that certain genes become differentially regulated by DNA methylation in human epilepsy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epilepsy is a common and refractory neurological disorder, which affects approximately 0.5–1 % of the world population and often requires lifelong medication (Ngugi et al., 2011). In a previous microarray-based study, we identified 243 genes that were differentially expressed in the anterior temporal neocortex of drug-refractory epilepsy patients relative to control patients (Xi et al., 2009). It is not possible to conclude that the differential expression of these genes either caused or resulted from epileptic seizures. Furthermore, the factors regulating the differentially expressed genes were unclear. Epigenetic mechanisms are one means by which gene expression is regulated, and in the central nervous system (CNS), epigenetic mechanisms are critical for promoting brain development, neural stem cell maintenance, proliferation, neurogenesis, gliogenesis, cellular migration, and synaptic and neural network connectivity (Delcuve et al., 2009). DNA methylation is a major epigenetic modification and plays important roles in regulating gene expression via reversible and dynamic chromatin remodeling processes. Kobow and Blümcke (2011) and Kobow et al. (2013b) first presented a methylation hypothesis for epileptogenesis and pharmacoresistance in epilepsy, and recent experimental studies provide some evidence for a relationship between abnormal DNA methylation and epileptogenesis. DNA methylation is catalyzed by DNA methyltransferases (DNMTs). In our previous study, we showed that DNMT1 and DNMT3a were upregulated in the brain tissue of human epilepsy cases (Zhu et al., 2012). Kobow et al. (2009) examined reelin (RELN) promoter methylation in hippocampal subregions of temporal lobe specimens obtained from epilepsy patients, and deep sequencing revealed increased DNA methylation in chronic rat epilepsy (Kobow et al., 2013a). Miller-Delaney et al. (2012) found genome-wide DNA methylation changes after status epilepticus and in epileptic tolerance, which may contribute to regulating gene expression in the seizure-damaged hippocampus. Machnes et al. (2013) reported that DNA methylation mediates persistent epileptiform activity in vitro and in vivo and that pharmacological agents that block DNA methylation inhibit epileptiform activity raising the prospect of DNA methylation inhibitors in epilepsy therapeutics. Belhedi et al. (2014) described increased CPA6 promoter methylation in focal epilepsy and in febrile seizures.
Differential DNA methylation patterns may contribute to the diversity of phenotypes, pathogenesis, and progression of complex diseases. Strong association has been identified between differential DNA methylation and cancer (Esteller, 2008), and there are now ongoing efforts to investigate links between DNA methylation and other complex diseases. In the current study, we investigated the DNA methylation-based regulation of gene expression in human epilepsy by comparing global DNA methylation profiles in human brain tissue between epileptic and control patients using a methylated-cytosine DNA immunoprecipitation microarray chip.
Materials and Methods
Patients
Human brain tissue was obtained from patients with drug-refractory epilepsy who had undergone surgical removal of the epileptogenic zone in their temporal neocortex. The epileptogenic zones were determined using a combination of ictal semiology, brain magnetic resonance imaging, video-EEG, sphenoidal electrode monitoring, and intracranial EEG before surgery and by intraoperative electrocorticography (EcoG). According to these analyses, the anterior temporal neocortical tissues capable of epileptiform discharges were resected. After resection, the EcoG electrodes were placed on the edges of the remaining tissues to ensure that the dischargeable tissues had been resected completely. All patients were refractory to maximal doses of at least three anti-epilepsy drugs (AEDs). Brain magnetic resonance imaging with routine scan sequences found no progressive lesions in the CNS. For comparison, histologically normal anterior temporal neocortex samples were obtained from patients who were treated for post-trauma intracranial hypertension. These subjects had no history of epilepsy or of exposure to AEDs. Individual patient details are listed in Supplemental Table 1.
Human Brain Tissue Processing
The excised brain tissue was immediately placed in a cryovial containing buffered diethylpyrocarbonate and stored in liquid nitrogen until use.
Methylated-Cytosine DNA Immunoprecipitation Chip
According to previously described protocols (Rakyan et al., 2008, Schumacher et al., 2008), genomic DNA was extracted from brain tissues using the DNeasy Blood & Tissue Kit according to the manufacturer’s instructions (Qiagen, Crawley, UK). Next, 11 μg global DNA (gDNA) of each sample was diluted in 300 μl TE buffer and sonicated into random ~500-bp fragments using a Bioruptor (Diagenode, Belgium). Immunoprecipitation of methylated DNA was performed using a mouse monoclonal anti-5-methyl cytosine antibody (Diagenode) and Biomag magnetic beads (Bangs Laboratories, USA). Five micrograms of anti-5-methyl cytosine antibody was added to the MeDIP sample, 5 μg mouse IgG (Jackson) to the mock IP sample, and nothing to the input sample. The MeDIP quality was assessed by qPCR of H19 (positive control) and GAPDH (negative control). Samples were rotated overnight at 4 °C, and then 50 μl of magnetic beads coupled to anti-mouse IgG was added and mixed. Samples were rotated for an additional 2 h and then washed three times with wash buffers before eluting with 200 μl elution buffer (1 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 10 % SDS). The immunoprecipitated DNA was purified by phenol chloroform extraction and ethanol precipitation.
For each sample, 1 μg of input and IP DNA was labeled with Cy3- and Cy5-labeled random 9-mers, respectively, and co-hybridized to the Nimblegen HG 18 CpG promoter microarray (Nimblegen, WI, USA). This Nimblegen microarray is a single array design with ~385,000 probes that span all known human CpG islands annotated on the UCSC genome browser and all well-characterized Human Refseq gene promoters (24,659) ranging from −2800 to +200 bp relative to the transcription start site (TSS). Scanning was performed with an Axon GenePix 4000B microarray scanner. Raw data were extracted as pair files by NimbleScan software (Nimblegene, USA). Several procedures were then sequentially performed to obtain peak data with specified parameters (sliding window width 750 bp; mini probes per peak 2; p value minimum cutoff 2; maximum spacing between nearby probes per peak 500 bp).
Bisulfite Sequencing PCR
The methylation status of the CpG islands in candidate gene promoters was determined by bisulfite sequencing as described previously (Carr et al., 2007, Movassagh et al., 2010). The primers were designed using Methprimer software (http://www.urogene.org/methprimer). Primer information is shown in Supplemental Table 2. Bisulfite conversion of gDNA was performed using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol. After purification, 5 μl of bisulfite-treated DNA was amplified by PCR. PCR conditions were as follows: 95 °C for 5 min; 42 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 40 s; and a final extension at 72 °C for 10 min. The PCR products were purified and cloned using a Pmd 19-T vector kit (Takara). To determine the methylation status of candidate gene promoters, an average of five positive clones were sequenced using the M13 reverse primer and an automated ABI prism 3730xl Genetic Analyzer (Applied Biosystems, USA). The sequencing results were analyzed using QUMA software (http://quma.cdb.riken.jp).
RT-PCR
Total RNA was isolated from brain tissue using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Paisley, UK). Next, 20 μl complementary DNA (cDNA) was synthesized from 1 μg total RNA using the PrimeScript RT reagent Kit (Takara). Primers were designed with Primer 5.0 and synthesized by Sangon Biotech (Shanghai, China). Primer information is shown in Supplemental Table 3. For PCR reactions, 2 ng cDNA were amplified in a 25-μl reaction volume containing 0.5 μM of each forward and reverse primer and 12.5 μl 2 × PCR TaqMix (TianGen Biotech, Beijing, China) under the following conditions: 5 min of denaturation at 95 °C; followed by 35 cycles of 30 s at 95 °C, 30 s at 60 °C, and 60 s at 72 °C; and a final extension at 72 °C for 10 min. The amplified products were separated on 2 % agarose gels. Densitometry analysis was performed with Quantity One software (Bio-Rad Laboratories, USA).
Data Analysis
Analysis of the Nimblegen methylated-cytosine DNA immunoprecipitation chip (MeDIP-chip) data was performed using the ACME algorithm as previously described (Scacheri et al., 2006). The ACME algorithm can be employed in cases where a fixed-length window is slid along the length of each chromosome, using a one-sided Kolmogorov–Smirnov (KS) test at each probe location to determine whether the surrounding window is enriched for high-intensity probes relative to the rest of the array. Each probe has a corresponding p value score (−log 10), and a threshold is set to select regions enriched in the test sample. The data generated by the software were exported in a spreadsheet format and processed using Microsoft Excel. The differentially methylated genes were analyzed using gene ontology (GO) functional categories based on the DAVID database (Huang da et al., 2009).
Data are reported as means ± standard deviation (SD). Statistical analysis was performed using SPSS 11.5. The Mann–Whitney U test was used to analyze the small sample cohorts, and Student’s t test was used to analyze real-time PCR (RT-PCR) results. A two-tailed p value of less than 0.05 was considered statistically significant.
Results
Demographic and Clinical Characteristics of Subjects
The epileptic patients consisted of 12 males and 13 females and had a mean age of 26.04 ± 8.69 years. Of these patients, 8 % had a history of seizure recurrence of less than 5 years, 56 % had a history of between 5 and 15 years, and 36 % had a clinical history lasting more than 15 years. In addition, 16 % of the patients had less than four seizures per month, 64 % had four to ten seizures per month, and 20 % of patients had more than ten seizures per month. The control group had an average age of 34.50 ± 14.05 years and consisted of six males and four females. Supplemental Table 1 summarizes the demographic and clinical characteristics of all subjects who participated in this study.
DNA Methylation Profiling of Brain Tissues from Epileptic Patients
To obtain a comprehensive profile of aberrant DNA methylation patterns in the epileptic patients, we used the Nimblegen HG 18 CpG promoter microarray chip to analyze samples from three epileptic patients and three controls. Based on the ACEM algorithm, peaks were defined as regions containing DNA methylation. Each probe had a p value, and –log (p value) represented the peak score; when the peak score was more than 2, the probe was selected. If several adjacent probes had p values <0.01, then the group was classified as one peak. We used this cutoff to identity the peaks. By these criteria, we found 1676, 2138, 2371, 2148, 2299, and 2391 peaks for patient 1 (P1), P2, P3, control 1 (C1), C2, and C3, with 578, 645, 740, 656, 613, and 736 peaks in promoters, respectively. In terms of mean number of peaks in promoters, there was no significant difference between epileptic patients and controls (p > 0.05). Figure 1 shows the DNA methylation profile of brain tissues from epileptic patients. Figure 2 displays the methylation profile clustering of the six samples.
DNA methylation profiles of epileptic patients and controls. All probes were mapped onto the human genome (NCBIv35) by BLAST and are depicted as vertical lines. The methylation profile of epileptic patients is shown above the line and that of controls is displayed below the line. Unmethylated regions are marked blue, yellow represents intermediately methylated (IM) regions, red represents densely methylated regions, and green represents centromeres
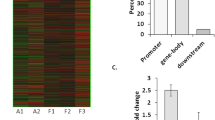
Candidate genes were identified by peaks overlapping the promoter region of the relevant transcript (approximately −800 to +200 bp). Differentially methylated genes were defined as genes with two or three peaks in the promoter region that were differential between epileptic patients and the control group. Under these criteria, 224 genes were selected (see Supplemental Table 5). Differentially methylated genes were classified by gene ontology (Fig. 3 and Supplemental Table 4). Rho guanyl-nucleotide exchange factor activity was the most significant GO term.
DNA Methylation Analysis of Candidate Genes
We used bisulfite sequencing to verify the microarray results for seven selected genes (ADAM15, ARID1A, TUBB2B, ATPGD1, CRMP1, and STK24), which are implicated in epilepsy or seizure according to the literature (Ortiz et al., 2005, Freitas et al., 2009, Jaglin and Chelly, 2009, Luo et al., 2012, Miyaji et al., 2012). The CpG islands were located in the established promoters of these genes. For bisulfite sequencing, regions within CpG islands of approximately 400 bp were selected. ARID1A, TUBB2B, ATPGD1, and CRMP1 were hypermethylated in epileptic patients compared to control samples, whereas HTR6 and STK24 were hypomethylated in epileptic patients (Fig. 4). Figure 5a–g shows the sequencing results of five positive clones from among the six samples for the seven candidate genes. The proportion of methylated CpGs in TUBB2B, ATPGD1, and HTR6 was 6.7, 8.7, and 29.2 % in epileptic patients, respectively, and 5.1, 5, and 50.8 % in controls, respectively (Fig. 5h). For TUBB2B, ATPGD1, and HTR6, the bisulfite sequencing results were consistent with the MeDIP-chip data. Methylation of ADAM15, ARID1A, CRMP1, and STK24 promoters was not significantly different between epileptic and control samples.
The probes of differentially methylated regions for candidate genes in chips. Red represents epileptic patients and black represents controls. ADAM15, ARID1A, TUBB2B, ATPGD1, and CRMP1 appeared hypermethylated in epileptic patients compared to control samples, whereas HTR6 and STK24 were hypomethylated in epileptic patients
Detailed DNA methylation patterns of the candidate genes were determined by bisulfite sequencing PCR. a–g The sequencing results of five positive clones of seven candidate genes among the six samples. Each circle represents a CG dinucleotide, with white indicating unmethylated residues and black indicating methylated residues. h The proportion of methylated CpG islands of candidate genes in epileptic patients and controls
Transcriptional Levels of Candidate Genes
Hypomethylation of DNA in the promoter region of a gene is considered to be associated with increased transcription, whereas hypermethylation in the body of a gene is associated with active transcription. We therefore set out to determine the expression levels of the candidate genes in 25 epileptic samples and 10 control samples by RT-PCR. The messenger RNA (mRNA) levels of candidate genes are shown in Fig. 6. mRNA levels of CRMP1 and HTR6 were upregulated, while those of ADAM15, TUBB2B, and ATPGD1 were downregulated in epileptic samples relative to controls. Expression of ARID1A and STK24 was not significantly different between epileptic and control samples. Combining the microarray and bisulfite sequencing results, we observed that in the epileptic samples, TUBB2B and ATPGD1 exhibited relatively hypermethylated promoters and decreased mRNA levels, while HTR6 displayed a relatively hypomethylated promoter and higher mRNA levels.
Differential mRNA expression levels of candidate genes in epileptic patients (n = 25) and controls (n = 10). Lane 1 is marker, lanes 2–6 represent controls, and lanes 7–11 represent epileptic patients. mRNA levels of CRMP1 and HTR6 were upregulated and those of ADAM15, TUBB2B, and ATPGD1 were downregulated in epileptic samples relative to controls (*p < 0.05; **p < 0.01). Expression of ARID1A and STK24 was not significantly different between epileptic patients and controls
Discussion
In this study, we performed a DNA methylation profile analysis using MeDIP-chip of brain tissues from epileptic patients and controls. This screen detected several genes that were differentially methylated between epileptic patients and controls.
The methylated DNA immunoprecipitation microarray (MeDIP-chip) is a genome-wide, high-resolution approach to detect DNA methylation in the whole genome or specifically in CpG islands. We used this method to profile DNA methylation patterns in the brain tissue of epileptic patients. Surprisingly, the total number of methylated regions was not significantly different between epileptic patients and controls. This result suggests that recurrent epileptic seizures in patients do not induce genome-wide changes in DNA methylation in brain tissues, unlike the general hypomethylation found in cancer (Esteller, 2007). However, hundreds of differentially methylated regions were detected between epileptic patients and controls, with some hypermethylated and some hypomethylated. Some studies reported that the methylation status of the BDNF and RELN promoters in brain tissue correlated with changes in transcriptional activity (Chen et al., 2003, Grayson et al., 2005). Notably, increased RELN promoter methylation was associated with granule cell dispersion in human temporal lobe epilepsy (Kobow et al., 2009). However, we did not detect differential methylation at these promoters in our samples.
GO analysis allows the investigation of functionally linked biological pathways in microarray datasets. In our microarray data, several interesting GO categories were identified among the differentially methylated genes, including regulation of Rho, microtubule-based processes, and mitogen-activated protein kinase (MAPK) scaffold activity. These biological processes play important roles in axonal growth, neurotransmitter release, and synapse reorganization. It was previously reported that RhoA was activated in the cortex and hippocampus after traumatic brain injury and kainic acid-induced seizures (Dubreuil et al., 2006). Likewise, we found increased expression of RhoA in human temporal lobe epilepsy (Yuan et al., 2010). The neuronal cytoskeleton consists of microtubules, actin filaments, intermediate filaments, and associated proteins. A number of mutations and altered levels of microtubule-associated proteins are likely to contribute to the pathogenesis of epilepsy through mechanisms such as increased neurotrophic support to neurons and increased sprouting of mossy fibers (Gardiner and Marc, 2010). Finally, activation of MAPK signaling caused spontaneous epileptic seizures in an animal model (Nateri et al., 2007), and we determined that the extracellular signal-regulated kinases (ERK) 1, ERK2, and phosphorylated ERK were upregulated in human intractable epilepsy (Xi et al., 2007).
Bisulfite sequencing is a method for positioning and quantifying DNA methylation in promoter regions. We performed this technique to verify the results of the MeDIP-chip. The TUBB2B, ATPGD1, and HTR6 promoters exhibited differential methylation between epileptic patients and controls, consistent with the microarray data. However, the methylation status of the other candidate genes was not different between epileptic patients and controls. The microarray is a high-throughput screening technology that is expected to yield a certain amount false-positive results; however, it can still reveal global DNA methylation patterns. Abnormal hypermethylation of promoter CpG islands results in transcriptional silencing, while hypomethylation correlates with increased transcription (El-Osta and Wolffe, 2000). Interestingly, we found that in epileptic patients, hypermethylation of the TUBB2B and ATPGD1 promoters correlated with reduced gene expression and hypomethylation of the HTR6 promoter correlated with increased expression. This result suggests that the expression of TUBB2B, ATPGD1, and HTR6 is regulated by DNA methylation. With regard to the other candidate genes, transcription of CRMP1 was increased, ADAM15 was decreased, and ARID1A and STK24 were not altered in epileptic patients. DNA methylation is not the only epigenetic regulator of gene transcription, as histone acetylation and microRNAs are also important epigenetic mechanisms for modulating gene transcription (Gibney and Nolan, 2010). For these genes, the degree of DNA methylation may, therefore, not necessarily correlate with transcriptional levels because they could be regulated by other mechanisms.
Functionally, the TUBB2B protein belongs to the β-tubulin family, which is the major constituent of microtubules. TUBB2B has been implicated in neuronal migration (Jaglin et al., 2009). TUBB2B mutations and aberrant tubulin heterodimer assembly lead to a large spectrum of neuronal migration disorders, including refractory epilepsy (Uribe, 2010). ATPGD1 is another name for carnosine synthase 1, which is mainly found in skeletal muscle and the CNS and catalyzes the synthesis of carnosine (with the conversion of ATP to AMP and inorganic pyrophosphate). Carnosine inhibits pentylenetetrazol-induced seizures by histaminergic mechanisms in histidine decarboxylase knock-out mice (Zhu et al., 2007). HTR6 is one of several receptors for 5-hydroxytryptamine (serotonin). Functions of HTR6 include modulation of cholinergic and dopaminergic neurotransmission. There is increasing evidence that serotonergic neurotransmission modulates a wide variety of experimentally induced seizures (Bagdy et al., 2007). The roles of HTR1A, HTR2C, HTR3, and HTR7 subtypes in epileptogenesis and/or propagation have been described (Bagdy et al., 2007). Our current findings reveal that DNA methylation regulates TUBB2B, ATPGD1, and HTR6 gene expression and that these genes might play important roles in epileptogenesis by affecting neuronal migration, carnosine metabolism, and serotonergic neurotransmission, respectively.
In the present study, we profiled DNA methylation patterns in brain tissues of epileptic patients. Because of the limitations associated with studying human brain tissue, some experimental details require explanation. As control subjects, we used histologically normal anterior temporal neocortex samples of patients with traumatic brain injury. However, we should consider that epilepsy can be a potential complication of such patients and that injury causes broad molecular changes, including epigenetic modifications (Diaz-Arrastia et al., 2009, Lundberg et al., 2009). Furthermore, DNA methylation is dynamically regulated and influenced by a variety of factors, including gender, age, and social environment (Christensen et al., 2009). Methylation patterns also differ between brain regions and neuronal cell types (Ladd-Acosta et al., 2007, Siegmund et al., 2007). We selected the right anterior temporal neocortex in six males of similar age for DNA methylation analysis to reduce interference factors. However, we used DNA extracted from whole brain tissue homogenates. Brain tissue comprises multiple types of neurons, glia, and other cells, and specific cell populations are thought to be differentially affected in the pathoetiology of epilepsy. We, therefore, only obtained the composite DNA methylation pattern for the epileptic brain tissues. In addition, all patients were refractory to anti-epileptic drugs. Effects of the drugs, especially valproic acid, on DNA methylation cannot be excluded.
Conclusions
We found differential DNA methylation profiles in epileptic patients. The transcription of certain genes was regulated by DNA methylation. Further study of the involvement of DNA methylation and other epigenetic mechanisms in epilepsy represents an important and promising means for identifying novel disease biomarkers for this disorder and for designing rational treatment strategies that overcome the serious limitations of current therapeutic approaches.
References
Bagdy G, Kecskemeti V, Riba P, Jakus R (2007) Serotonin and epilepsy. J Neurochem 100:857–873
Belhedi N, Perroud N, Karege F, Vessaz M, Malafosse A, Salzmann A (2014) Increased CPA6 promoter methylation in focal epilepsy and in febrile seizures. Epilepsy Res 108:144–148
Carr IM, Valleley EM, Cordery SF, Markham AF, Bonthron DT (2007) Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res 35:e79
Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME (2003) Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885–889
Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, Sugarbaker DJ, Yeh RF, Wiencke JK, Kelsey KT (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 5:e1000602
Delcuve GP, Rastegar M, Davie JR (2009) Epigenetic control. J Cell Physiol 219:243–250
Diaz-Arrastia R, Agostini MA, Madden CJ, Van Ness PC (2009) Posttraumatic epilepsy: the endophenotypes of a human model of epileptogenesis. Epilepsia 50(Suppl 2):14–20
Dubreuil CI, Marklund N, Deschamps K, McIntosh TK, McKerracher L (2006) Activation of Rho after traumatic brain injury and seizure in rats. Exp Neurol 198:361–369
El-Osta A, Wolffe AP (2000) DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr 9:63–75
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 8:286–298
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358:1148–1159
Freitas RL, Ferreira CM, Urbina MA, Marino AU, Carvalho AD, Butera G, de Oliveira AM, Coimbra NC (2009) 5-HT1A/1B, 5-HT6, and 5-HT7 serotonergic receptors recruitment in tonic-clonic seizure-induced antinociception: role of dorsal raphe nucleus. Exp Neurol 217:16–24
Gardiner J, Marc J (2010) Disruption of normal cytoskeletal dynamics may play a key role in the pathogenesis of epilepsy. Neuroscientist 16:28–39
Gibney ER, Nolan CM (2010) Epigenetics and gene expression. Heredity 105:4–13
Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E (2005) Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A 102:9341–9346
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Jaglin XH, Chelly J (2009) Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet 25:555–566
Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, Castelnau-Ptakhine L, Odent S, Loget P, Kossorotoff M, Snoeck I, Plessis G, Parent P, Beldjord C, Cardoso C, Represa A, Flint J, Keays DA, Cowan NJ, Chelly J (2009) Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet 41:746–752
Kobow K, Jeske I, Hildebrandt M, Hauke J, Hahnen E, Buslei R, Buchfelder M, Weigel D, Stefan H, Kasper B, Pauli E, Blumcke I (2009) Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol 68:356–364
Kobow K, Blümcke I (2011) The methylation hypothesis: do epigenetic chromatin modifications play a role in epileptogenesis? Epilepsia 52(Suppl 4):15–19
Kobow K, El-Osta A, Blümcke I (2013b) The methylation hypothesis of pharmacoresistance in epilepsy. Epilepsia 54(Suppl 2):41–47
Kobow K, Kaspi A, Harikrishnan KN, Kiese K, Ziemann M, Khurana I, Fritzsche I, Hauke J, Hahnen E, Coras R, Muhlebner A, El-Osta A, Blumcke I (2013a) Deep sequencing reveals increased DNA methylation in chronic rat epilepsy. Acta Neuropathol 126:741–756
Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP (2007) DNA methylation signatures within the human brain. Am J Hum Genet 81:1304–1315
Lundberg J, Karimi M, von Gertten C, Holmin S, Ekstrom TJ, Sandberg-Nordqvist AC (2009) Traumatic brain injury induces relocalization of DNA-methyltransferase 1. Neurosci Lett 457:8–11
Luo J, Zeng K, Zhang C, Fang M, Zhang X, Zhu Q, Wang L, Wang W, Wang X, Chen G (2012) Down-regulation of CRMP-1 in patients with epilepsy and a rat model. Neurochem Res 37:1381–1391
Machnes ZM, Huang TC, Chang PK, Gill R, Reist N, Dezsi G, Ozturk E, Charron F, O'Brien TJ, Jones NC, McKinney RA, Szyf M (2013) DNA methylation mediates persistent epileptiform activity in vitro and in vivo. PLoS ONE 8:e76299
Miller-Delaney SF, Das S, Sano T, Jimenez-Mateos EM, Bryan K, Buckley PG, Stallings RL, Henshall DC (2012) Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 32:1577–1588
Miyaji T, Sato M, Maemura H, Takahata Y, Morimatsu F (2012) Expression profiles of carnosine synthesis-related genes in mice after ingestion of carnosine or ss-alanine. J Int Soc Sports Nutr 9:15
Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS (2010) Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 5:e8564
Nateri AS, Raivich G, Gebhardt C, Da Costa C, Naumann H, Vreugdenhil M, Makwana M, Brandner S, Adams RH, Jefferys JG, Kann O, Behrens A (2007) ERK activation causes epilepsy by stimulating NMDA receptor activity. EMBO J 26:4891–4901
Ngugi AK, Kariuki SM, Bottomley C, Kleinschmidt I, Sander JW, Newton CR (2011) Incidence of epilepsy: a systematic review and meta-analysis. Neurology 77:1005–1012
Ortiz RM, Karkkainen I, Huovila AP, Honkaniemi J (2005) ADAM9, ADAM10, and ADAM15 mRNA levels in the rat brain after kainic acid-induced status epilepticus. Brain Res Mol Brain Res 137:272–275
Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, Howe KL, Jackson DK, Miretti MM, Fiegler H, Marioni JC, Birney E, Hubbard TJ, Carter NP, Tavare S, Beck S (2008) An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 18:1518–1529
Scacheri PC, Crawford GE, Davis S (2006) Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol 411:270–282
Schumacher A, Weinhausl A, Petronis A (2008) Application of microarrays for DNA methylation profiling. Methods Mol Biol 439:109–129
Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S (2007) DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2:e895
Uribe V (2010) The beta-tubulin gene TUBB2B is involved in a large spectrum of neuronal migration disorders. Clin Genet 77:34–35
Xi ZQ, Wang XF, He RQ, Li MW, Liu XZ, Wang LY, Zhu X, Xiao F, Sun JJ, Li JM, Gong Y, Guan LF (2007) Extracellular signal-regulated protein kinase in human intractable epilepsy. Eur J Neurol 14:865–872
Xi ZQ, Xiao F, Yuan J, Wang XF, Wang L, Quan FY, Liu GW (2009) Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy. Synapse 63:1017–1028
Yuan J, Wang LY, Li JM, Cao NJ, Wang L, Feng GB, Xue T, Lu Y, Wang XF (2010) Altered expression of the small guanosine triphosphatase RhoA in human temporal lobe epilepsy. J Mol Neurosci 42:53–58
Zhu Q, Wang L, Zhang Y, Zhao FH, Luo J, Xiao Z, Chen GJ, Wang XF (2012) Increased expression of DNA methyltransferase 1 and 3a in human temporal lobe epilepsy. J Mol Neurosci 46:420–426
Zhu YY, Zhu-Ge ZB, Wu DC, Wang S, Liu LY, Ohtsu H, Chen Z (2007) Carnosine inhibits pentylenetetrazol-induced seizures by histaminergic mechanisms in histidine decarboxylase knock-out mice. Neurosci Lett 416:211–216
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 81201003, 81271445). We thank the patients and their families for their participation in this study. We also thank the KangChen Bio-tech for the technical assistance with the MeDIP-chip.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent was obtained from the participants for the use of their brain tissues for research, and approval was obtained from the ethics committee of the First Affiliated Hospital of Chongqing Medical University.
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, L., Fu, X., Peng, X. et al. DNA Methylation Profiling Reveals Correlation of Differential Methylation Patterns with Gene Expression in Human Epilepsy. J Mol Neurosci 59, 68–77 (2016). https://doi.org/10.1007/s12031-016-0735-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-016-0735-6