Abstract
Forensic genotyping can be impeded by γ-irradiation of biological evidence in the event of radiological crime; that is, criminal activity involving radioactive material. Oxidative effects within the mitochondria of living cells elicits greater damage to mitochondrial DNA (mtDNA) than nuclear DNA (nuDNA) at low doses. This study presents a novel approach for the assessment of nuDNA versus mtDNA damage from a comparison of genotype and quantity data, while exploring likely mechanisms for differential damage after high doses of γ-irradiation. Liquid (hydrated) and dried (dehydrated) whole blood samples were exposed to high doses of γ-radiation (1–50 kilogray, kGy). The GlobalFiler PCR Amplification Kit was used to evaluate short tandem repeat (STR) genotyping efficacy and nuDNA degradation; a comparison was made to mtDNA degradation measured using real-time PCR assays. Each assay was normalized before comparison by calculation of integrity indices relative to unirradiated controls. Full STR profiles were attainable up to the highest dose, although DNA degradation was noticeable after 10 and 25 kGy for hydrated and dehydrated blood, respectively. This was manifested by heterozygote imbalance more than allele dropout. Degradation was greater for mtDNA than nuDNA, as well as for hydrated than dehydrated cells, after equivalent doses. Oxidative effects due to water radiolysis and mitochondrial function are dominant mechanisms of differential damage to nuDNA versus mtDNA after high-dose γ-irradiation. While differential DNA damage was reduced by cell desiccation, its persistence after drying indicates innate differences between nuDNA and mtDNA radioresistance and/or continued oxidative effects within the mitochondria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High-dose exposure of DNA evidence to γ-radiation may be caused by γ-emitting radionuclides present at a radiological crime. Such crimes involve the abandonment, theft, or trafficking of radioactive material and could lead to the construction of crude radiological weapons, such as a dirty bomb [1]. While such an attack has not yet taken place, extremists have previously demonstrated interest in the use of such unconventional weaponry [2, 3]. The doses received by a forensic sample in such cases may be well beyond several kilogray (kGy); dose rates up to 4.6 kGy/h are expected within a meter of an unshielded Category 1 cobalt-60 γ-emitter with typical activity of 150 terabecquerel (TBq) [4]. Similarly, doses of γ-radiation necessary for biological agent decontamination may be upwards of 10 kGy [5,6,7]. Due to its high probative value, DNA evidence is the most reliable means of identification available today, and hence may be critical for the identification of victims or perpetrators of such crimes.
Genotyping of autosomal ‘length polymorphic’ short tandem repeats (STRs) is the current standard for forensic identity testing [8,9,10]. This relies on the polymerase chain reaction (PCR) to facilitate DNA target selection. Ionizing irradiation of DNA evidence can disrupt the PCR by introducing a variety of DNA lesions, including base modifications, abasic sites, crosslinkages, and strand breaks [11,12,13]. These lesions can prevent strand uncoiling, alter primer binding sites, and/or block DNA polymerase during PCR [14, 15]. This results in allelic dropout, particularly for longer amplicons that incur DNA damage lesions with greater frequency [16], following sufficiently high doses (> 10 kGy) of γ-radiation [17, 18].
γ-irradiation interacts with DNA constituents via direct ionization events, as well as through secondary oxidative reactions mediated by reactive oxygen species (ROS). The latter are produced from the radiolysis of cellular water molecules, as well as by mitochondrial hyperfunction of viable cells [11, 19,20,21,22].
The ‘sequence polymorphic’ hypervariable regions (HVRs) located within the mitochondrial DNA (mtDNA) control region (D-loop) are alternatives to STR genotyping for degraded DNA. They are present in higher copy number than nuclear DNA (nuDNA) and enable identification from maternal lineage, although the discrimination power of multiplexed STRs is unrivalled by HVR sequencing [23, 24]. Further, the role of mitochondria in mediating ROS production subjects mtDNA to greater oxidative damage than nuDNA [25,26,27]. Mitochondrial content/volume and oxidative function may also be upregulated by ionizing radiation exposure [21, 22], where increased mitochondrial volume may lead to more frequent ionization events than the nucleus [28].
The aim of this study was to evaluate the degradation of STR genotypes after high doses (1–50 kGy) of γ-irradiation to both liquid (hydrated) and dried (dehydrated) whole blood samples, as well as the relative impact of γ-irradiation upon nuDNA and mtDNA targets. Integrity indices for mtDNA were determined from the quantity ratios of different sized amplicons targeted by quantitative real-time PCR (qPCR) assays. Similarly, peak height ratios between STRs of equivalent size to the mtDNA targets were used to provide an index of nuDNA integrity.
Methodology
DNA samples
Whole blood was collected by venipuncture from 10 individuals in 4 mL Vacutainers (Becton Dickinson, Franklin Lakes, USA) coated with 7.2 mg of dipotassium ethylenediaminetetraacetic acid (EDTA). Aliquots of 150 μL were transferred into sterile 1.5 mL glass vials with polyethylene push caps (liquid/hydrated samples) or air dried onto sterile glass microscope slides (dried/dehydrated samples). A sterile glass cover slip was secured over dried blood flakes with adhesive tape. Blood collection and sample preparation was performed for all samples (including unirradiated controls) the day prior to sample irradiation and stored at 4 °C until irradiation.
Sample irradiation
γ-irradiation of whole blood samples was conducted at the Australian Nuclear Science and Technology Organisation (ANSTO) using the Gamma Technology Research Irradiator (GATRI). Irradiations with cobalt-60 to approximate absorbed doses of 1, 5, 10, 25 and 50 kGy were performed independently at ambient temperature (~24.0 °C). For each dose, the dose rate was confirmed by two ceric-cerous sulphate dosimeters, except at 1 kGy, which relied on a dose rate previously determined by a dose mapping study (data not shown). Samples received a dose rate of approximately 2 kGy per hour over a continuous period until the target dose was reached. Exposure times ranged from approximately 30 min (1 kGy) to 24 h (50 kGy), correcting times to account for radioactive source decay. Samples were immediately stored at −20 °C post-irradiation.
Sample irradiations took place over three days batched by dose. To evaluate any impact of storage time on DNA integrity, two sets of unirradiated controls were prepared for each individual and sample type (i.e. liquid or dried). These controls were stored under the same conditions as the first and last irradiation batch for subsequent comparison of DNA integrity.
DNA extraction
Whole blood samples were extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) [29]. Sample lysis was carried out directly in the glass vials of liquid samples or by transferring dried blood flakes into 1.5 mL microcentrifuge tubes. Extracted DNA was eluted into 100 μL elution buffer (10 mM Tris-chloride, pH 9.0, 0.5 mM EDTA). Aliquots of the DNA extracts were stored at −20 °C prior to use.
Quantitative real-time PCR (qPCR)
Quantification of nuDNA was performed with the Quantifiler Human DNA Quantification Kit (Applied Biosystems, Foster City, USA) [30]. Three mtDNA rRNA coding region targets of different length (86, 190 and 452 base pairs, bp) were quantified by SYBR Green-based qPCR assays [31]. All assays were performed on a 7500 Real-Time PCR System with HID Real-Time PCR Analysis Software v1.1 (Applied Biosystems). Internal PCR controls (IPCs) were included with both the Quantifiler and mtDNA assays.
STR genotyping
A panel of 23 forensic STR markers and amelogenin were genotyped using the GlobalFiler PCR Amplification Kit (Applied Biosystems). The standard 25 μL reaction chemistry was applied [32], with products amplified from 1 ng template DNA (29 cycle protocol) on a Veriti Thermal Cycler (Applied Biosystems). Sample dilutions were in TE buffer (10 mM Tris-chloride, pH 8.0, 0.1 mM EDTA). Positive controls were Control DNA 007 (Applied Biosystems).
Capillary electrophoresis was performed for GlobalFiler [32] using GeneScan 600 LIZ dye Size Standard v2.0 (Applied Biosystems) and Hi-Di Formamide (Applied Biosystems). Electrophoresis was performed on a 3500xl Genetic Analyser with 3500 Series Data Collection Software 2 (Applied Biosystems), run module ‘HID36_POP4xl’. The capillary was 36 cm filled with POP-4 Polymer (Applied Biosystems). Spectral calibration was performed with DS-36 Matrix Standard (Dye Set J6; Applied Biosystems). Analysis of genotypes was conducted in GeneMapper ID-X v1.4 (Applied Biosystems) with a detection limit of 225 relative fluorescence units (RFU), corresponding to 10 standard deviations above baseline. Stochastic thresholds of 500 and 1000 RFU were empirically determined for heterozygote and homozygote alleles, respectively, with a heterozygote peak imbalance threshold of 70% for each locus.
DNA degradation assays
An index of DNA integrity was determined for both nuDNA and mtDNA from the amplification of long versus short targets. For nuDNA, a subset of autosomal forensic STR markers were selected for relative size consistency with the three mtDNA qPCR targets (86, 190 and 452 bp), including loci of low molecular weight (D2S441, ~75–110 bp), intermediate molecular weight (vWA and D1S1656, ~150–210 bp), and high molecular weight (TPOX and SE33, ~310–450 bp). For STR size groups containing multiple loci, the average peak heights of alleles for each marker were determined. Integrity indices were calculated from peak height or quantity ratios comprising intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C). Integrity indices for irradiated samples were normalized against those for unirradiated samples of equivalent DNA type. This ‘relative integrity index’ was used for comparison of nuDNA versus mtDNA integrity to account for any differences in PCR efficiency and/or template damage prior to irradiation.
Statistical analysis
To account for any variation between the two sets of unirradiated controls (stored under the same conditions as the first and last irradiation batch), the irradiated samples were compared against both sets of controls and the data pooled for statistical analysis using SPSS Statistics 23 (IBM, Armonk, USA). A p value of less than 0.05 was considered statistically significant. Divergence from a normal Gaussian distribution was assessed using Shapiro-Wilk normality tests and quantile-quantile (Q-Q) plots. Equality of variances was checked using Levene’s test. Outliers were removed if they were beyond the first or third quartile of the dataset by more than 1.5× the interquartile range.
Nonparametric tests for related samples (repeated measures) were applied for evaluation of any dose-effect differences within data grouped by integrity index (A, B or C), sample preparation (hydrated or dehydrated), or DNA type (nuDNA or mtDNA). These analyses were conducted using Friedman’s tests, with multiple comparisons made by Wilcoxon Signed-Rank tests. Mann-Whitney U tests were used to compare the effect of sample hydration status at equivalent doses. Sequential Bonferroni (Holm-Bonferroni) corrected p-values were applied to mitigate against chance significance due to multiple comparisons [33].
Results
DNA quantification and PCR inhibitor detection
The presence of inhibitors was tested using a TaqMan-based IPC multiplexed with the Quantifiler chemistry (nuDNA) and a separate SYBR Green-based IPC reaction designed for use with the mtDNA assays. Both inhibitor assays did not detect inhibition in any sample; the IPC amplified after ~25 cycles using Quantifiler and ~ 30 cycles using the SYBR Green IPC assay, consistent with positive and negative controls. The nuDNA concentrations of pooled unirradiated controls ranged from 10 to 34 ng/μL for dehydrated samples and 31 to 94 ng/μL for hydrated samples, while mtDNA ranged from 5000 to 90,000 copies/μL (10 to 210 fg/μL) for dehydrated samples and 32,000 to 150,000 copies/μL (74 to 350 fg/μL) for hydrated samples.
Integrity of unirradiated genotypes
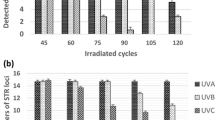
The duplicate unirradiated controls for each individual and matrix were pooled to compare the integrity of genotypes prior to irradiation with that of positive genotyping controls (Fig. 1). Mean peak heights (± 95% confidence interval) of controls were 9200 ± 500 RFU (dehydrated controls), 8900 ± 480 RFU (hydrated controls), and 7300 ± 660 RFU (Control DNA 007). While all heterozygote alleles were well balanced for Control DNA 007, heterozygote imbalance (< 70% peak height ratio) was observed in half of the dehydrated controls (at up to two loci) and in 80% of hydrated controls (at up to three loci). No correlation with amplicon size was discernible.
Peak heights and heterozygote balance of short tandem repeat (STR) genotypes for unirradiated samples and positive controls. Samples included dried (dehydrated) and liquid (hydrated) whole blood (n = 20 for autosomal STRs and 10 for Y-STRs), as well as Control DNA 007 (n = 4). Vertical bars (left vertical axis) represent the average (± 95% confidence interval) relative fluorescence units (RFU) of peak heights, while dots (right vertical axis) represent the median (± minimum/maximum) heterozygote imbalance. A heterozygote imbalance threshold of 70% is indicated by a solid line. STR loci are arranged in approximate size order (Y indel < SE33)
Effect of γ-irradiation on forensic STR genotyping
A panel of STRs was amplified from whole blood (liquid/hydrated and dried/dehydrated) using the GlobalFiler PCR Amplification Kit and evaluated for signs of degradation after high-dose (1–50 kGy) γ-irradiation (Fig. 2). Peak height averages (± 95% confidence interval) across all loci and individuals were consistent for dehydrated samples at 1 kGy (11,000 ± 750 RFU), 5 kGy (12,000 ± 760 RFU), and 10 kGy (12,000 ± 790 RFU). This declined after 25 kGy (9700 ± 680 RFU) and 50 kGy (5800 ± 470 RFU). Similarly, peak heights of hydrated samples were consistent between 1 kGy (13,000 ± 820 RFU) and 5 kGy (11,000 ± 690 RFU), declining after 10 kGy (9100 ± 622 RFU), 25 kGy (5800 ± 530 RFU), and 50 kGy (3000 ± 400 RFU).
Peak heights and heterozygote balance of short tandem repeat (STR) genotypes for γ-irradiated samples. Dried (dehydrated: left) and liquid (hydrated: right) whole blood (n = 9 to 10 for autosomal STRs and 5 for Y-STRs) were irradiated to doses from 1 to 50 kilogray (kGy). Vertical bars (left vertical axis) represent the average (± 95% confidence interval) relative fluorescence units (RFU) of peak heights, while dots (right vertical axis) represent the median (± minimum/maximum) heterozygote imbalance. A heterozygote imbalance threshold of 70% is indicated by a solid line. STR loci are arranged in approximate size order (Y indel < SE33)
Compared to unirradiated controls (Fig. 3), peak heights were significantly increased after 1 kGy for hydrated (by 41 ± 4.0%) and dehydrated samples (by 18 ± 2.9%), 5 kGy for hydrated (by 24 ± 3.9%) and dehydrated samples (by 34 ± 3.7%), and 10 kGy for dehydrated samples only (by 31 ± 3.7%). Peak heights were not statistically different from controls at 10 kGy for hydrated samples or 25 kGy for dehydrated samples. A significant decline in peak height relative to controls occurred for hydrated samples at 25 kGy (by 35 ± 2.8%) and 50 kGy (by 68 ± 2.5%), which did not occur for dehydrated samples until 50 kGy (by 37 ± 2.3%). TH01, D2S1338 and DYS391 produced markedly higher relative changes in comparison to other loci; thus, these loci were excluded from statistical analysis.
Change in the peak height of short tandem repeat (STR) loci after γ-irradiation. Dried (dehydrated) and liquid (hydrated) whole blood (n = 16 to 20 for autosomal STRs and 9 to 10 for Y-STRs) were irradiated to doses from 1 to 50 kilogray (kGy). The average (± 95% confidence interval) change in the peak height of each locus was determined for irradiated samples relative to unirradiated controls (indicated by a solid line at 0% change). STR loci are arranged in approximate size order (Y indel < SE33)
Employing heterozygote and homozygote thresholds of 500 and 1000 RFU, respectively, full profiles were attained for all dehydrated samples, as well as hydrated samples up to 25 kGy. Partial profiles attained for 50 kGy hydrated samples were above-threshold for 86 ± 4.1% (mean ± 95% confidence interval) of alleles. Alleles below peak height thresholds (dropout) were above approximately 225 bp (≥ D16S539), with 5.3% of these alleles (all from SE33) being undetectable (below 225 RFU). Genotype nonconcordance (relative to unirradiated genotypes) was found at 21% of nonreportable (subthreshold) loci, consisting of dropout for a single heterozygous allele (miscalled homozygote).
Heterozygote imbalance (< 70% peak height ratio) contributed to greater levels of nonreportable alleles, particularly as dose increased. Out of 10 profiles, no cases of imbalance were observed in dehydrated samples at 1 kGy, 1–3 profiles were imbalanced at up to two loci each from 5 to 25 kGy, and seven profiles had imbalances at up to four loci each at 50 kGy. Hydrated samples exhibited imbalances for 3–4 profiles at up to two loci each from 1 to 10 kGy, nine profiles with up to three loci at 25 kGy, and eight profiles with 2–6 imbalanced loci at 50 kGy. The frequency of imbalances was less than unirradiated controls at 1–25 kGy for dehydrated samples (by 40–100%) and 1–10 kGy for hydrated samples (by 50–63%). Imbalances were more prevalent than in controls beyond these doses; 40% more for dehydrated samples at 50 kGy and 13% more for hydrated samples at 25 and 50 kGy. Imbalances predominantly affected amplicons above 200–300 bp.
Effect of γ-irradiation on nuDNA integrity
Significant increases in nuDNA integrity index relative to unirradiated controls occurred at 1 kGy (dehydrated and hydrated) and 5 kGy (dehydrated only) (Fig. 4). For dehydrated and hydrated samples, respectively, this transpired with frequencies of 77 and 78% at 1 kGy and 56 and 42% at 5 kGy (data not shown); such cases diminished with increasing dose, with no cases beyond 10 kGy for hydrated samples, which did not substantiate statistically significant effects. Corresponding changes to relative integrity were nonsignificant at 5 kGy (hydrated) and 10 kGy (dehydrated) and declined significantly as dose increased.
Change in nuclear DNA integrity indices relative to those for unirradiated controls at each dose. Dried (dehydrated: left) and liquid (hydrated: right) whole blood (n = 15 to 20) were γ-irradiated to doses from 1 to 50 kilogray (kGy). Integrity indices comprised peak height ratios for intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C), expressed as the average (± 95% confidence interval) percentage change relative to those for unirradiated controls (indicated by a solid line at 0% change). * = significantly different
The relative nuDNA integrity indices were compared for hydrated samples relative to those for dehydrated samples (Fig. 5). Cell hydration significantly lowered relative integrity after 5, 10, or 25 kGy, dependent on the index applied. These differences became more pronounced as dose increased.
Change in nuclear DNA dose-response of hydrated samples relative to dehydrated samples. Dried (dehydrated) and liquid (hydrated) whole blood (n = 15 to 20) were γ-irradiated to doses from 1 to 50 kilogray (kGy). Integrity indices were comprised of peak height ratios for intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C). The relative integrity indices of irradiated to unirradiated samples were expressed for hydrated samples as the average (± 95% confidence interval) percentage change relative to that for dehydrated samples (indicated by the solid line at 0% change). * = significantly different
Effect of γ-irradiation on mtDNA integrity
Integrity indices of mtDNA were more often reduced relative to unirradiated controls without substantive increases (Fig. 6). Significant losses of relative integrity were possible after 5 kGy independent of cell hydration; however, this was dependent on the integrity index applied when cells were dehydrated. Increases in relative integrity index were observed in near 50% of all samples at 1 kGy (data not shown), diminishing as dose increased with no such effects beyond 5 kGy (hydrated) or 25 kGy (dehydrated); this did not result in any statistical significance.
Change in mitochondrial DNA integrity indices relative to those for unirradiated controls at each dose. Dried (dehydrated: left) and liquid (hydrated: right) whole blood (n = 14 to 18) were γ-irradiated to doses from 1 to 50 kilogray (kGy). Integrity indices comprised quantity ratios for intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C), expressed as the average (± 95% confidence interval) percentage change relative to those for unirradiated controls (indicated by a solid line at 0% change). * = significantly different
The relative mtDNA integrity indices of hydrated samples were compared relative to those for dehydrated samples (Fig. 7). Hydrated samples produced significantly lower relative integrity indices after 5 kGy. These differences due to sample hydration increased with dose.
Change in mitochondrial DNA dose-response of hydrated samples relative to dehydrated samples. Dried (dehydrated) and liquid (hydrated) whole blood (n = 14 to 18) were γ-irradiated to doses from 1 to 50 kilogray (kGy). Integrity indices comprised quantity ratios for intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C). The relative integrity indices of irradiated to unirradiated samples were expressed for hydrated samples as the average (± 95% confidence interval) percentage change relative to that for dehydrated samples (indicated by the solid line at 0% change). * = significantly different
Comparison of nuDNA and mtDNA degradation
The mtDNA relative integrity indices were compared with those for nuDNA at each dose (Fig. 8). Integrity of mtDNA was significantly lower than nuDNA after equivalent doses, which occurred after a minimum of 1 kGy, dependent on the integrity index applied. Only Index A of dehydrated cells did not demonstrate any significant effects. Differences generally increased with dose and were more extensive when cells remained hydrated.
Change in dose-response of mitochondrial DNA (mtDNA) relative to nuclear DNA (nuDNA). Dried (dehydrated: left) and liquid (hydrated: right) whole blood (n = 13 to 20) were γ-irradiated to doses from 1 to 50 kilogray (kGy). Integrity indices were comprised of peak height ratios (nuDNA) or quantity ratios (mtDNA) for intermediate/short (Index A), long/intermediate (Index B), and long/short loci (Index C). The relative integrity indices of irradiated to unirradiated samples were expressed for mtDNA as the average (± 95% confidence interval) percentage change relative to that for nuDNA (indicated by the solid line at 0% change). * = significantly different
Discussion
Forensic DNA evidence may be exposed to γ-radiation doses beyond 10 kGy in the event of a radiological crime, or during decontamination of biological agents from forensic evidence in cases concerning biosecurity [4,5,6,7]. This study examined γ-irradiation of whole blood to doses ranging 1 to 50 kGy from a cobalt-60 source. Dried (dehydrated) blood samples were selected to represent typical forensic material, while liquid (hydrated) blood samples were included to preserve cell integrity and water content prior to irradiation. This enabled contributions to DNA damage from indirect mechanisms (e.g. ROS induction) to be evaluated; localized sample heating during irradiation may also contribute [34].
These experiments were designed to imitate a scenario where biological evidence is continuously exposed to γ-radiation for up to 24 h (achieving a dose of 50 kGy), before collection and flash freezing. Continued ROS generation or cell death mechanisms initiated by irradiation can contribute to greater levels of DNA degradation where rapid sample processing or freezing does not occur, which is observed in live cells below 1 kGy [35,36,37]. For γ-irradiation beyond 1 kGy, inherent analytical variation of STR genotypes was greater than the effect of up to four weeks delayed analysis [38], although this likely depends on cell hydration level and sample storage conditions.
In the present study, genotypes at forensic autosomal STR loci did not show signs of degradation pre-irradiation, although heterozygote peak imbalances were prevalent (Fig. 1). Doses above 10 and 25 kGy for hydrated and dehydrated samples, respectively, produced greater frequencies of imbalances than the unirradiated controls, which increased progressively up to 50 kGy (Fig. 2). The level of imbalance was consistent with a significant reduction in overall peak height for hydrated and dehydrated samples after respective doses of 25 and 50 kGy (Figs. 2 and 3). This was associated with a progressive decline in peak height as amplicon size increased, typical of degradation [16]. However, the impact on genotype reporting based on peak height and heterozygote imbalance thresholds was minor, affecting a maximum of six loci due to imbalance, with partial profiles (due to dropout of less than 15% of alleles) prevalent for only 50 kGy hydrated samples. Genotype nonconcordance due to dropout of single heterozygous alleles affected one-fifth of subthreshold loci.
The robustness of STRs to γ-radiation has been demonstrated for several STR kits and cell substrates (e.g. blood, saliva) capable of full profiles up to 50 kGy [18, 38, 39]. This has also been achieved for dried bloodstains up to 90 kGy [40], although reductions in peak height are typical after 10 kGy [17, 18, 38]. Successful STR genotypes and HV1 sequences from dried saliva are also possible after 51.6 kGy electron beam (beta) irradiation, another common biological decontaminant [41]. However, another study found 56.4 kGy γ-irradiation to produce only 40% full profiles from dried saliva, while a 50 kGy electron beam resulted in 70% full profiles [42]. This demonstrates γ-radiation to be more damaging than beta-radiation at similar dose, although highlights potential for significant points of difference, such as STR kit, sample type and/or post-irradiation sample storage conditions, to influence the consistency of findings between such studies. Another consideration to profiling success, not discussed by these studies, is heterozygote allele imbalance.
Heterozygote imbalances are typical of PCR inhibition or degradation, particularly of longer targets [43, 44]. Imbalances in unirradiated controls, without peak height characteristics of degradation [16], indicated that inhibition was likely to have impacted genotypes pre-irradiation (Fig. 1). γ-radiation then improved genotypes relative to unirradiated samples at lower doses; imbalances were reduced by doses of up to 10 and 25 kGy, while peak heights were increased at doses of ≤5 and 10 kGy for hydrated and dehydrated samples, respectively (Fig. 2). This result is unusual and not demonstrated by prior studies exploring similar effects [17, 18, 38,39,40, 42]. It is unlikely that inherent template damage or cellular function is responsible for these observations, since the doses applied are beyond those expected to initiate any adaptive DNA repair response [45,46,47]. It is more likely that degradation of potential PCR inhibitors after γ-irradiation, including heme and EDTA that are degraded by respective doses below 1 and 5 kGy [48, 49], lends to improved genotypes at lower doses that are inconsequential to DNA integrity. While inhibition was unconfirmed by qPCR, differences in assay chemistries, length of targets and/or primer design (GC content / melting temperature) can lead to differential sensitivity of PCR assays, and individual amplicons, to inhibition [50,51,52,53]. Confirmation of this hypothesis is required via inhibitor-spiking experiments.
At higher doses the sensitivity of integrity indices to degradation proceeded, generally, in order of Index A < Index B < Index C, consistent with greater degradation of longer amplicons. Doses of 10 and 25 kGy were sufficient to cause a significant loss of hydrated and dehydrated nuDNA integrity, respectively (Fig. 4). Only 5 kGy was required to significantly reduce mtDNA integrity of hydrated samples, with up to 25 kGy required for dehydrated samples (Fig. 6). A radioprotective effect was therefore conferred by cellular desiccation (Figs. 5 and 7), demonstrating ROS generation from water radiolysis or other cellular interaction to be a prime contributor to DNA damage from γ-radiation, which is more greatly localized to the mitochondria than the nucleus (Fig. 8). This is consistent with mitochondrial hyperfunction after ionizing-irradiation coupled with a reduced DNA repair capacity [21, 22, 27], or upregulation of mitochondrial ROS from cell death [54, 55]; however, such studies include dose regimens well below 1 kGy.
Differential nuDNA versus mtDNA damage was reduced by cell drying, but not completely removed (except for Index A), indicating a capacity for such effects to continue (i.e. due to residual moisture) or for innate differences in radiosensitivity to exist. This may be due to structural arrangements, such as chromatin compaction or DNA interaction with nuclear histones versus mitochondrial transcription factor A (TFAM) [56,57,58], or the frequency of nuclear versus mitochondrial ionization events [22, 28]. Inclusion of naked (cell-free) DNA after both desiccation and dissolution into aqueous medium could test these hypotheses by evaluation of cellular versus non-cellular effects. Such controls have been applied to support a mechanism for DNA damage from continued activity in desiccated cells after UV-B irradiation [59].
Further radiosensitivity is anticipated for HVRs due to D-loop susceptibility to oxidative damage, as demonstrated for X-rays at low doses (up to 8 Gy) [60]; thus, an equal distribution of damage across the entire mitochondrial genome cannot be assumed. While mtDNA target selection within the rRNA coding region offers multiplexing potential [31], this location causes them to be indirect indicators of HVR sequencing success, despite similar lengths to HVR amplicons. However, no loss of sequencing fidelity has been demonstrated for both HV1 and HV2 of single hairs after a γ-radiation dose of 90 kGy; this was consistent with, although not directly comparable, to STR genotyping success of dried blood [40].
Conclusion
At a radiological crime scene, successful GlobalFiler STR genotypes can be expected from biological evidence exposed to substantial doses of γ-radiation, at least in the absence of additional degradative factors. While peak heights are reduced with increasing dose and accompanied by heterozygote peak imbalance, full profiles are possible from whole blood up to a dose of 50 kGy; at this dose, allelic dropout is prone for hydrated samples, where peak imbalance is liable to cause genotype nonconcordance. Thus, such genotyping thresholds must be carefully considered for γ-irradiated samples to ensure reliability, especially beyond 50 kGy. The success of STR genotyping suggests there is little to be gained from HVR sequencing at the doses examined; however, at higher doses that may be sufficient to cause autosomal DNA degradation, our evaluation of mtDNA damage suggests poor prospects for HVR sequencing, although this was not attempted.
γ-irradiation of liquid and dried blood demonstrated significantly greater damage to mtDNA than nuDNA at equivalent doses, which was more substantial without desiccation. This implicates ROS induction from water radiolysis and mitochondrial function as causal of DNA damage when sample integrity and water content are preserved during irradiation. Consequently, a radioprotective effect of sample dehydration, as is commonplace for forensic biological specimens, is apparent. However, disparity between nuDNA and mtDNA integrity in dried samples suggests additional radioprotection is afforded to nuDNA. Future investigation should focus on the HVRs as direct targets for degradation in conjunction with broader integrity indicators, such as those applied in this study.
Key points
-
1.
γ-irradiation up to 50 kGy did not greatly impact forensic genotyping success.
-
2.
Heterozygote imbalance was the primary contributor to subthreshold alleles.
-
3.
Cell desiccation protected DNA, while cell hydration exacerbated DNA damage.
-
4.
Damage to mitochondrial DNA was greater than nuclear DNA at equivalent doses.
Data availability
Research data is available online: Goodwin, C; Wotherspoon, A; Gahan, M; McNevin, D (2019), “Degradation of nuclear and mitochondrial DNA after γ-irradiation and its effect on forensic genotyping”, Mendeley Data, v1, https://doi.org/10.17632/hytsjn9zbv.1
References
Ferguson CD, Kazi T, Perera J. Commercial radioactive sources: Surveying the security risks. Occassional Paper No. 11. Monterey, CA: Center for Nonproliferation Studies, Monterey Institute of International Studies; 2003. Report No.: 1885350066.
Ackerman GA. Chemical, biological, radiological and nuclear (CBRN) terrorism. In: Silke A, editor. Routledge handbook of terrorism and counterterrorism. Abingdon, United Kingdom: Routledge; 2019.
Koehler D, Popella P. Mapping far-right chemical, biological, radiological, and nuclear (CBRN) terrorism efforts in the west: characteristics of plots and perpetrators for future threat assessment. Terrorism and Political Violence. 2018. https://doi.org/10.1080/09546553.2018.1500365.
IAEA. Categorization of radioactive sources. IAEA safety standard series guide no. RS-G-1.9. International Atomic Energy Agency: Austria; 2005.
Dutra MP, Aleixo GC, Ramos ALS, Silva MHL, Pereira MT, Piccoli RH, et al. Use of gamma radiation on control of Clostridium botulinum in mortadella formulated with different nitrite levels. Rad Phys Chem. 2016;119:125–9.
Elliott LH, McCormick JB, Johnson KM. Inactivation of Lassa, Marburg, and Ebola viruses by gamma irradiation. J Clin Microbiol. 1982;16:704–8.
Ortatatli M, Canitez K, Sezigen S, Eyison RK, Kenar L. Evaluation of gamma-radiation inactivation of a bioterrorism agent, Bacillus anthracis spores, on different materials. Indian J Microbiol. 2018;58:76–80.
Jeffreys AJ. Genetic fingerprinting. Nature Med. 2005;11:1035–9.
Kimpton C, Fisher D, Watson S, Adams M, Urquhart A, Lygo J, et al. Evaluation of an automated DNA profiling system employing multiplex amplification of four tetrameric STR loci. Int J Legal Med. 1994;106:302–11.
Kimpton CP, Gill P, Walton A, Urquhart A, Millican ES, Adams M. Automated DNA profiling employing multiplex amplification of short tandem repeat loci. Genome Res. 1993;3:13–22.
Hutchinson F. Chemical changes induced in DNA by ionising radiation. Prog Nucleic Acid Res Mol Biol. 1985;32:115–54.
Dextraze ME, Gantchev T, Girouard S, Hunting D. DNA interstrand cross-links induced by ionizing radiation: an unsung lesion. Mutat Res. 2010;704:101–7.
Téoule R. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51:573–89.
Matuo Y, Izumi Y, Sato N, Yamamoto T, Shimizu K. Evaluation of DNA lesions caused by high-LET radiation using the polymerase chain reaction. Radiat Meas. 2013;55:93–5.
Sikorsky JA, Primerano DA, Fenger TW, Denvir J. DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochem Biophys Res Comm. 2007;355:431–7.
Takahashi M, Kato Y, Mukoyama H, Kanaya H, Kamiyama S. Evaluation of five polymorphic microsatellite markers for typing DNA from decomposed human tissues: correlation between the size of the alleles and that of the template DNA. Forensic Sci Int. 1997;90:1–9.
Hodgson A, Baxter A. Preliminary studies into profiling DNA recovered from a radiation or radioactivity incident. J Radioanalyt Nuc Chem. 2013;296:1149–54.
Hoile R, Banos C, Colella M, Walsh SJ, Roux C. Gamma irradiation as a biological decontaminant and its effect on common fingermark detection techniques and DNA profiling. J Forensic Sci. 2010;55:171–7.
Das S. Critical review of water radiolysis processes, dissociation products, and possible impacts on the local environment: a geochemist’s perspective. Aust J Chem. 2013;66:522–9.
Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32:1102–15.
Leach JK, Van Tuyle G, Lin P-S, Schmidt-Ullrich R, Mikkelsen RB. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer Res. 2001;61:3894–901.
Yamamori T, Yasui H, Yamazumi M, Wada Y, Nakamura Y, Nakamura H, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53:260–70.
Budowle B, Chakraborty R, Carmody G, Monson KL. Source attribution of a forensic DNA profile. Forensic Sci Comm. 2000;2:1–6.
Mabuchi T, Susukida R, Kido A, Oya M. Typing the 1.1 kb control region of human mitochondrial DNA in Japanese individuals. J Forensic Sci. 2007;52:355–63.
Morales A, Miranda M, Sánchez-Reyes A, Biete A, Fernández-Checa JC. Oxidative damage of mitochondrial and nuclear DNA induced by ionizing radiation in human hepatoblastoma cells. Int J Radiat Oncol. 1998;42:191–203.
Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Nat Acad Sci. 1988;85:6465–7.
Yakes FM, van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Nat Acad Sci. 1997;94:514–9.
Kam WWY, McNamara AL, Lake V, Banos C, Davies JB, Kuncic Z, et al. Predicted ionisation in mitochondria and observed acute changes in the mitochondrial transcriptome after gamma irradiation: a Monte Carlo simulation and quantitative PCR study. Mitochondrion. 2013;13:736–42.
Qiagen. QIAamp® DNA Mini and Blood Mini handbook. User Guide. Hilden, Germany; 2016.
Biosystems A. Quantifiler™ human and Y human male DNA quantification kits. Cheshire, UK: User Guide; 2018.
Goodwin C, Higgins D, Tobe SS, Austin J, Wotherspoon A, Gahan ME, et al. Singleplex quantitative real-time PCR for the assessment of human mitochondrial DNA quantity and quality. Forensic Sci Med Pathol. 2018;14:70–5.
Biosystems A. GlobalFiler™ PCR amplification kit. Carlsbad, USA: User Guide; 2016.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300.
Kantidze OL, Velichko AK, Luzhin AV, Razin SV. Heat stress-induced DNA damage. Acta Nat. 2016;8:75–8.
Hu S, Gao Y, Zhou H, Kong F, Xiao F, Zhou P, et al. New insight into mitochondrial changes in vascular endothelial cells irradiated by gamma ray. Int J Radiat Biol. 2017;93:470–6.
Joseph P, Bhat NN, Copplestone D, Narayana Y. Production of gamma induced reactive oxygen species and damage of DNA molecule in HaCaT cells under euoxic and hypoxic condition. J Radioanalyt Nuc Chem. 2014;302:983–8.
Yamaguchi M, Kashiwakura I. Role of reactive oxygen species in the radiation response of human hematopoietic stem/progenitor cells. PLoS One. 2013;8:e70503.
Abbondante SF. The effect of radioactive materials on forensic DNA evidence: procedures and interpretation [dissertation]: University of Canberra; 2009.
Neureuther K, Rohmann E, Hilken M, Sonntag ML, Herdt S, Koennecke T, et al. Reduction of PCR-amplifiable DNA by ethylene oxide treatment of forensic consumables. Forensic Sci Int. 2014;12:185–91.
Monson KL, Ali S, Brandhagen MD, Duff MC, Fisher CL, Lowe KK, et al. Potential effects of ionizing radiation on the evidentiary value of DNA, latent fingerprints, hair, and fibers: a comprehensive review and new results. Forensic Sci Int. 2018;284:204–18.
Withrow AG, Sikorsky J, Downs JCU, Fenger T. Extraction and analysis of human nuclear and mitochondrial DNA from electron beam irradiated envelopes. J Forensic Sci. 2003;48:1302–8.
Shaw K, Sesardić I, Bristol N, Ames C, Dagnall K, Ellis C, et al. Comparison of the effects of sterilisation techniques on subsequent DNA profiling. Int J Legal Med. 2008;122:29–33.
Hansson O, Egeland T, Gill P. Characterization of degradation and heterozygote balance by simulation of the forensic DNA analysis process. Int J Legal Med. 2017;131:303–17.
Thompson RE, Duncan G, McCord BR. An investigation of PCR inhibition using Plexor®-based quantitative PCR and short tandem repeat amplification. J Forensic Sci. 2014;59:1517–29.
Desouky O, Ding N, Zhou G. Targeted and non-targeted effects of ionizing radiation. J Radiat Res Appl Sci. 2015;8:247–54.
Mitchel REJ. The dose window for radiation-induced protective adaptive responses. Dose Response. 2010;8:192–208.
Tubiana M, Feinendegen LE, Yang C, Kaminski JM. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiol. 2009;251:13–22.
Rafiei J, Yavari K, Moosavi-Movahedi AA. Preferential role of iron in heme degradation of hemoglobin upon gamma irradiation. Int J Biol Macromol. 2017;103:1087.
Jung J, Jo HJ, Lee SM, Ok YS, Kim JG. Enhancement of biodegradability of EDTA by gamma-ray treatment. J Radioanalyt Nuc Chem. 2004;262:371–4.
Hall AT, Zovanyi AM, Christensen DR, Koehler JW, Minogue TD. Evaluation of inhibitor-resistant real-time PCR methods for diagnostics in clinical and environmental samples. PLoS One. 2013;8:e73845.
Huggett JF, Novak T, Garson JA, Green C, Morris-Jones SD, Miller RF, et al. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008;1:70.
Opel KL, Chung D, McCord BR. A study of PCR inhibition mechanisms using real time PCR. J Forensic Sci. 2010;55:25–33.
Wang DY, Mulero JJ, Hennessy LK. Different effects of PCR inhibitors on multiplex STR assays. Applied Biosystems: Foster City; 2008.
Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci. 2006;32:37–43.
Kim EM, Yang HS, Kang SW, Ho JN, Lee SB, Um HD. Amplification of the γ-irradiation-induced cell death pathway by reactive oxygen species in human U937 cells. Cell Signal. 2008;20:916–24.
Falk M, Lukášová E, Kozubek S. Chromatin structure influences the sensitivity of DNA to γ-radiation. Biochim Biophys Acta. 1783;2008:2398–414.
Takata H, Hanafusa T, Mori T, Shimura M, Iida Y, Ishikawa K, et al. Chromatin compaction protects genomic DNA from radiation damage. PLoS One. 2013;8:e75622.
Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5:a012641.
Hall A, Sims LM, Ballantyne J. Assessment of DNA damage induced by terrestrial UV irradiation of dried bloodstains: forensic implications. Forensic Sci Int. 2014;8:24–32.
Zhou X, Liu X, Zhang X, Zhou R, He Y, Li Q, et al. Non-randomized mtDNA damage after ionizing radiation via charge transport. Sci Rep. 2012;2:780.
Acknowledgements
The authors would like to thank Connie Banos and Justin Davies of ANSTO gamma-irradiation services for assistance in the planning and implementation of sample irradiations, as well as Australian Federal Police Forensics for the use of their genotyping facilities and Timothy Shaw for assistance with genotyping and profile analysis.
Funding
This research was supported by an Australian Institute of Nuclear Science and Engineering research award (ALNSTU11896) and an Australian Government Research Training Program Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All procedures involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments. Approval for the collection and use of human biological material was granted by the University of Canberra Committee for Ethics in Human Research (Project Number 14–70). This article does not contain any studies with animals performed by any of the authors.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Goodwin, C., Wotherspoon, A., Gahan, M.E. et al. Degradation of nuclear and mitochondrial DNA after γ-irradiation and its effect on forensic genotyping. Forensic Sci Med Pathol 16, 395–405 (2020). https://doi.org/10.1007/s12024-020-00251-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-020-00251-2