Abstract
Analysis of DNA polymorphisms are the primary technique used for personal identification in forensic cases. However, DNA samples collected as evidence from crime scenes are usually degraded by environmental, physical, and chemical factors, which may interfere with the PCR analysis and, consequently, personal identification. Whole genome amplification (WGA) is a useful method to amplify genomic DNA from samples containing low quantity and poor quality of DNA, and it approach that shows promise to overcome the limited small fragments based upon random fragmentation by universal priming sites. In this study, we describe the use of WGA to genotype 15 short tandem repeat (STR) loci from dried blood samples irradiated with different types of ultraviolet (UV) light (UVA, UVB, and UVC). The result showed that UVC was the most damaging to DNA, followed by UVB and UVA. Samples exposed to UVA could be genotyped for all STR loci with or without WGA. For UVB and UVC irradiated blood samples, a greater number of STR loci could be genotyped after WGA. Although it hard to amplified a few higher molecular weight alleles, overall, the WGA method was useful in genotyping template DNA of poor quality but low quantity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA polymorphism analysis is frequently used in forensic studies to identify potential suspects. Short tandem repeats (STRs) are the most commonly used genetic markers. Each individual carries a unique combination of alleles across predefined set of STR loci, thus providing an exclusive genetic profile with a high power of discrimination. However, DNA samples collected as evidence from the crime scene are frequently degraded because of environmental, physical, and chemical factors [1, 2] and cannot be used for PCR analysis. Additionally, the technical capacity to analyze samples containing very few cells is often limited by the quality and quantity of DNA. Therefore the most important things is how many STR loci could be detected from the DNA of the crime scene. This may be due to a number of factors, the most common being the presence of PCR inhibitors and low copy number and/or damaged DNA template.
Ultraviolet (UV) light is a major factor affecting DNA quality [3, 4]. UV radiation is a part of the electromagnetic spectrum that reaches the earth from the sun and consists of three types of light with different wavelengths UVA, UVB, and UVC. The UVB and UVC are of shorter wavelengths and therefore more damaging. The predominant types of UV-induced DNA damage include base modifications, double-strand breaks, and photoproducts. The mechanisms of DNA damage have largely been elucidated in living cells and cell-free systems [5]. Despite damage caused by UV irradiation, DNA remains a forensically relevant sample. DNA quality of a physiologically dried stain could be damaged in the same manner biochemically, although the kinetics and extent of the damage may vary with the nature of the sample.
Several methods have been developed to overcome problematic STR amplification from low copy number samples [6, 7]. Whole genome amplification (WGA) is one such method, which involves random fragmentation of genomic DNA and the conversion of these fragments into PCR-amplifiable sequences flanked by universal priming sites [8, 9]. Thus, WGA facilitates the amplification of genomic DNA from samples containing insufficient and fragmented template. This method was specifically designed to amplify the DNA template in clinical studies, such as single cell analysis, pre-implantation genetic diagnosis, and forensic DNA genotyping. WGA will improve the ability to generate full profile and / or increase the number of genotyped loci in partial profiles from DNA samples exposed to UV irradiation.
Materials and methods
Collection and UV irradiation of blood samples
Blood samples were collected from 20 Japanese subjects by venipuncture, spotted onto sterilized cotton gauze, and allowed to dry at room temperature, thus mimicking bloodstains recovered from a crime scene. The dried blood samples were exposed to UVA (365 nm), UVB (302 nm), or UVC (254 nm) light in a UV cross-linker (Ultra Violet Products, Upland, CA, USA) at an intensity of 8.7 mJ/cm2 under a 16 h light/8 h dark cycle. The irradiation cycles were repeated until no STR loci of the 15 analyzed were detectable. All samples were stored in the dark until PCR amplification.
DNA isolation
DNA from the dried blood samples was extracted using QIAamp DNA Blood Investigator Kit (Qiagen Inc., Valencia, CA, USA). The isolated DNA samples were prepared for quantification using Qubit dsDNA HS Assay Kit (Life Technologies) and quantified using the Qubit 3.0 Fluorometer (Life Technologies), according to the manufacturer’s instructions.
STR analysis
All the samples were processed using AmpFISTR® Identifiler® Plus PCR Amplification Kit (Applied Biosystems) which contains 15 autosomal STR loci (D8S1179, D2S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818 and FGA) and Amelogenin in accordance with the manufacturer’s instructions. PCR reactions contained 1 ng genomic DNA. Amplified fragments were detected using ABI Prism 310 capillary electrophoresis system (Applied Biosystems). STR analysis was conducted on DNA isolated from dried blood samples before and after UV irradiation for comparison of STR profiles.
Whole genome amplification
To genotype the STR loci from UV irradiated samples, DNA was subjected to WGA using GenomePlex® Complete WGA Kit (Sigma). The process of WGA involves random fragmentation of DNA followed by PCR amplification using universal primers. The DNA sample supplied in the kit as a positive control was used in all experiments. Serial dilutions of K562 DNA standard (Promega) (0.001, 0.01, 0.1, 1.0, and 10 ng) were used to determine the sensitivity of the kit.
STR analysis from WGA products
Products of WGA were purified using the GenElute PCR Clean-Up Kit (Sigma) to remove excess reagents, including primers, nucleotides, and DNA polymerase, according to the manufacturer’s instructions. Purified WGA products were then subjected to STR genotyping as described above.
Results and discussion
STR genotyping of UV irradiated blood samples
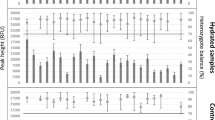
The objective of this study was to determine the ability to generate useful STR profile from DNA exposed to different types of UV irradiation. Incomplete STR profiles were detected in samples irradiated with 60 cycles of UVC, 75 cycles of UVB, and 90 cycles of UVA (Fig. 1a), indicating that UVC was the most damaging, followed by UVB and UVA. Although more than 10 STR loci were detectable after 90 cycles of UVA irradiation, none of the loci were detectable in UVC irradiated samples and less than half of the STR loci (7 of 15 loci) were detectable in UVB irradiated samples after 90 cycles. The number of detected STR loci gradually decreased with an increase in exposure number of exposure cycles.
STR genotyping after WGA
Because UV irradiation damages DNA making it unusable for genotyping, we investigated whether DNA from UV irradiated samples could be successfully genotyped following WGA. Recently, several approaches and commercial kits have been developed for WGA, enabling DNA amplification from a small amount of template, such as that from a single cell [10, 11]. In this study, GenomePlex® Complete WGA Kit was used to amplify DNA from UV irradiated blood samples for STR genotyping. Testing the kit with serial dilutions of the standard K562 DNA indicated that the kit was able to generate amplicons from as little as 0.1 ng of DNA template (Fig. 2), and the resulting products could be used to genotype all 15 autosomal STR loci.
For UVA irradiated samples, all 15 STR loci could be PCR amplified and genotyped from their WGA products, regardless of the number of cycles of irradiation (Fig. 1b). However, fewer STR profiles could be detected in the WGA products of UVB and UVC irradiated blood samples. In the case of UVB irradiated samples, although the complete STR profile could be obtained from the WGA products of samples treated with 60 cycles of UVB, only 12 and 10 STR loci could be genotyped from samples exposed to 105 and 120 cycles of UVB irradiation, respectively (Fig. 1b). In the case of UVC irradiated samples, no more than six STR loci could be genotyped from the WGA products of samples treated with 120 cycles of UVC (Fig. 1b).
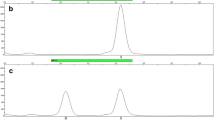
In forensic cases, genotyping of STR loci is key to unambiguous identification of potential suspects. Accurate genotyping of at least 10 STR loci can assist in the identification of suspects in some forensic cases. Thus, WGA may assist in STR analysis of samples exposed to UVA and UVB irradiation. Further, we noted that loci with higher molecular weight (MW) are more prone to UV damage than those with lower MW. In the case of heterozygous loci, the peak representing higher MW allele was lost earlier than the peak representing the lower MW allele of the same locus, which could withstand a few additional cycles of UVB irradiation. On the other hand, after damaged by UVB, some small peaks which were totally different from the control sample allele was observed. As showing in Fig. 3, allele 13 and 19 peaks were detected in D13S317 and the D18S51 locus, it was analyzed at the very limit from an RFU (relative fluorescence units) level for a peak, but those peaks were not detected in the WGA.
Several factors affect DNA quality. In forensic cases, samples collected from a crime scene are commonly exposed to damaging factors, such as humidity, microbial and chemical contamination, and UV irradiation. A variety of lesions in the primary structure of DNA are indicative of damage, such as oxidation products, single or double strand breaks, UV-induced photoproducts, DNA or protein crosslinks, and chemically induced covalent adducts [12]. Several reports have described methods to repair the damaged DNA to ensure successful genotyping [13, 14]. However, the potential of DNA repair and genotyping varies with the type and degree of DNA damage [15]. Although repaired DNA can be used for STR genotyping, the success of genotyping depends on the quantity of the template obtained [16].
The method of WGA, unlike DNA repair, allows for the amplification of genomic DNA and has been successfully used for genetic profiling of samples containing fragmented DNA template in trace amounts [17, 18]. In this study, we were able to accurately genotype STR loci from the WGA products of K562 DNA standard at a concentration of 0.1 ng or higher. Although DNA quality may not be of concern in the case of standard DNA exposed to UV light, as high as 3 ng of DNA might still not be sufficient for STR genotyping of blood samples subjected to prolonged UV exposure. This may be because of repeated lesions in DNA structure and/or insufficient DNA template because of UV irradiation, which might not be satisfactory for WGA.
Overall, WGA was not used to determine STR profiles but to increase probability of obtaining STR genotype from UV damaged DNA. Results showed that among samples exposed to UVA, the lost STR alleles can be recovered for all specimens after comparison with direct STR assay during full experiment period. In the case of UVB and UVC irradiated blood samples, except for a few high MW alleles, WGA improved the success of genotyping, thus proving to be a useful tool for genotyping DNA samples with poor quality but low quantity, such as those recovered from most crime scenes.
References
Kayser M (2015) Forensic DNA phenotyping: predicting human appearance from crime scene material for investigative purposes. Forensic Sci Int Genet 18:33–48
Friedberg EC (2003) DNA damage and repair. Nature 421:436–440
Hall A, Ballantyne J (2004) Characterization of UVC-induced DNA damage in bloodstains: forensic implications. Anal Bioanal Chem 380:72–83
Poepping C, Beck SE, Wright H, Linden KG (2014) Evaluation of DNA damage reversal during medium-pressure UV disinfection. Water Res 56:181–189
Vaisman A, Woodgate R (2017) Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 52(3):274–303
Diegoli TM, Farr M, Cromartie C, Coble MD, Bille TW (2012) An optimized protocol for forensic application of the PreCR™ Repair Mix to multiplex STR amplification of UV-damaged DNA. Forensic Sci Int Genet 6(4):498–503
Hall A, Sims LM, Ballantyne J (2014) Assessment of DNA damage induced by terrestrial UV irradiation of dried bloodstains: forensic implications. Forensic Sci Int Genet 8(1):24–32
Rinke J, Schäfer V, Schmidt M, Ziermann J, Kohlmann A, Hochhaus A, Ernst T (2013) Genotyping of 25 leukemia-associated genes in a single work flow by next-generation sequencing technology with low amounts of input template DNA. Clin Chem 59(8):1238–1250
Han T, Chang CW, Kwekel JC, Chen Y, Ge Y, Martinez-Murillo F, Roscoe D, Težak Z, Philip R, Bijwaard K, Fuscoe JC (2012) Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development. BMC Genom 13(217):1–15
Huang L, Ma F, Chapman A, Lu S, Xie XS (2015) Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet 16:79–102
Borgström E, Paterlini M, Mold JE, Frisen J, Lundeberg J (2017) Comparison of whole genome amplification techniques for human single cell exome sequencing. PLoS ONE 12(2):e0171566
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Robertson JM, Dineen SM, Scott KA, Lucyshyn J, Saeed M, Murphy DL, Schweighardt AJ, Meiklejohn KA (2014) Assessing PreCR™ repair enzymes for restoration of STR profiles from artificially degraded DNA for human identification. Forensic Sci Int Genet 12:168–180
Mitchell D, Willerslev E, Hansen A (2005) Damage and repair of ancient DNA. Mutat Res 571:265–276
Tate CM, Nuñez AN, Goldstein CA, Gomes I, Robertson JM, Kavlick MF, Budowle B (2012) Evaluation of circular DNA substrates for whole genome amplification prior to forensic analysis. Forensic Sci Int Genet 6(2):185–190
Kroneis T, El-Heliebi A (2015) Quality control of isothermal amplified DNA based on short tandem repeat analysis. Methods Mol Biol 1347:129–140
Lee JC, Tsai LC, Lai PY, Lee CC, Lin CY, Huang TY, Linacre A, Hsieh HM (2012) Evaluating the performance of whole genome amplification for use in low template DNA typing. Med Sci Law 52(4):223–228
Nishikawa Y, Hosokawa M, Maruyama T, Yamagishi K, Mori T, Takeyama H (2015) Monodisperse picoliter droplets for low-bias and contamination-free reactions in single-cell whole genome amplification. PLoS ONE 10(9):e0138733
Acknowledgements
We thank Mr. Eiji Isobe for assistance with the samples irradiation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Approval for this study was granted by the Ethics Committee of Nihon University School of Medicine and National Research Committee. All procedures performed in studies involving human participants were in accordance with the ethical standards of this Ethics Committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all participants included in the study.
Rights and permissions
About this article
Cite this article
Uchigasaki, S., Tie, J., Sobashima, E. et al. Genotyping DNA isolated from UV irradiated human bloodstains using whole genome amplification. Mol Biol Rep 45, 925–929 (2018). https://doi.org/10.1007/s11033-018-4240-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4240-6