Abstract
In this manuscript, the grown and annealed strontium-doped nickel oxide nanoparticles (SrNiONPs) were synthesized using a precipitation method with nickel nitrate and strontium nitrate as precursor agents with trisodium citrate. Various characterization techniques, including X-ray diffraction pattern (XRD), scanning electron microscopy (SEM), transmission electron microscopy (TEM), Fourier transform infrared spectroscopy (FTIR), UV–visible, and zeta sizer, were used to thoroughly examine the samples. The XRD pattern (21 nm) was used to calculate the size, phases, and crystallinity of the material (SrNiONPs). In addition to characterization, the material was tested for cytotoxicity in lung cancer cells (A549). The viability test in A549 cells was performed using [3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyltetrazolium bromide] (MTT) and Neutral Red Uptake (NRU) assay with SrNiONPs concentration ranging from 1 to 100 μg/mL. According to the MTT and NRU data, the toxicity studies are dose-dependent. SrNiONPs also increased reactive oxygen species (ROS) and were involved in apoptosis (A549 cells). Furthermore, quantitative PCR (qPCR) data revealed that the mRNA levels of apoptotic genes marker like p53, bax, and caspase-3 were upregulated, whereas bcl-2, an anti-apoptotic gene, was downregulated. As a result, apoptosis was mediated by the p53, bax, caspase3, and bcl-2 pathways, implying a potential mechanism by which SrNiONPs mediate their toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Strontium is an alkaline earth metal that is extremely reactive in nature. In the periodic table, strontium is represented by the symbol Sr and has the atomic number 38. Because of its high reactivity, strontium turns yellow when it comes into contact with air and produces strontium nitride when burned in air. It has a face center cubic (FCC) structure in its crystal geometry and has paramagnetic properties in nature [1]. Another highly reactive inorganic material that has received a lot of attention is nickel oxide (NiO) nanoparticles, which have unique electrical, magnetic, and catalytic properties when compared to bulk material [2]. NiO is a p-type semiconducting material with wide band gap value (3.6–4.0 eV) that has many applications due to its high band gap and good physicochemical properties, such as electrochromic films, catalysts, solar cells, lithium batteries, light-emitting diodes (LEDs), resistive random access memory, and electrochemical sensors and biosensor gas sensor [3,4,5,6,7]. A variety of methods have been used to prepare NiONPs, including thermal decomposition [8], simple liquid phase process [9], sol–gel method [10], and sonochemical method [11]. Among the various methods, the co-precipitation solution method has clear advantages such as simplicity, low cost, and ease of production of mass collection of samples [11]. The size, morphology, and shape of NPs depend on their synthesis procedures, precursor concentration, surfactant, and calcination temperature [8,9,10,11].
As a result, the increasing use of NiONPs necessitates a better understanding of their potential impact on the environment and human health. Furthermore, the toxic effects of NiONPs have been studied in various human cell systems, with a focus on liver and airway cells [12]. Oxidative stress, defined as the difference between reactive oxygen species production (ROS) and antioxidants that favor a ROS excess, has long been recognized as a common mechanism for cell damage and death. This imbalance can be exacerbated by aging, smoking, diseases, and certain foods [13]. ROS are primarily formed during oxidative stress by the electron transport chain in mitochondria. Furthermore, NPs have been shown to produce more free radicals and ROS than larger particles, most likely due to their larger surface area [13]. The inverse relationship between cell viability and ROS level demonstrated that oxidative stress is most likely a key mechanism by which NPs induce cytotoxicity [13]. In this study, nickel and strontium were co-precipitated to form strontium-doped nickel oxide nanoparticles (termed SrNiONPs) and characterized with various instruments such as an X-ray diffraction pattern to determine the size, phases, and crystallinity of the material. The SEM was used to determine the general morphology of the prepared material, which was equipped with energy dispersive X-ray analysis (EDX), which revealed the elemental proportion of the SrNiO NPs. A structural examination was also performed using TEM. FTIR and UV–visible spectroscopy were used to investigate the material’s chemical functional and optical properties. Microscopy was used to examine the morphology of the cells, while MTT and NRU assays were used to assess cytotoxicity. The study of reactive oxygen species (ROS), which is a key factor in cancer (A549) cells, was also carried out. A possible discussion was explained here based on the obtained results and their analysis.
Materials and Methods
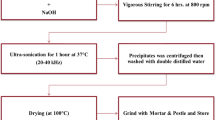
Synthesis of SrNiONPs
The strontium-doped nickel oxide nanoparticles were prepared using strontium nitrate (Sr(NO3)2.4H2O), trisodium citrate dihydrate (C6H5Na3O7.2H2O), and nickel nitrate hexa hydrate (Ni(NO3)2.6H2O) procured from Sigma-Aldrich Chem corporation and used as received. In a typical experiment, about equimolar ratios of Ni(NO3)2.6H2O (0.35 M) and Sr(NO3)2.4H2O (0.35 M) were dissolved in 100 ml of methanol (MeOH) with constant stirring. Trisodium citrate dihydrate (C6H5Na3O7.2H2O) was gently poured into this mixture to reduce metal salts. The pH of the final solution was determined to be 9.73. The solution was transferred to a refluxing pot and refluxed for 3 h at 80 °C. After the refluxing was completed, the semi-aqueous solution was centrifuged (4000 rpm for 3 min) to remove the ionic impurities before being transferred to glass petri dishes and dried in an oven at 60 °C. The oven dried powder was further annealed at 250 °C for 1 h before being cooled at room temperature for further chemical, physical, and biological analysis.
Material Characterization of SrNiONPs
The crystallinity, phases, and size of the grown and annealed powder samples were determined using an X-ray powder diffractometer (XRD) (PANalytical XPert Pro, USA) with CuKα radiation (λ = 1.54178 Å) in the range of 20–80º and a scanning speed of 6º/min. The morphology of the synthesized powder was determined using SEM (JSM-6380, Japan). The prepared powder was squirted on carbon tape and fixed to the sample (SrNiONPs) holder for morphological analysis at room temperature using SEM. The elemental analysis was also completed using an EDX equipped with a SEM. The powder structural examination was also carried out using TEM (JEOL, JSM 2010, Japan). Regardless of the functional groups of the chemicals used, FTIR spectroscopy was used in the 400–4000 cm−1 range. The optical property of the material was also analyzed via UV–visible (UV–visible, Shimazu) spectroscopy with ranges from 200 to 800 nm, at room temperature.
To evaluate the dynamic light scattering (DLS) and zeta (ζ) potential, the SrNiONPs were dissolved in double deionized water (DDW, 10 mg/mL) and further diluted to achieve a concentration of 50 µg/mL. The diluted solution was then sonicated in a bath sonicator (40 W) for 15–20 min to ensure complete NP separation. The hydrodynamic particle size of SrNiONPs and zeta (ζ) potential in an aqueous suspension was examined using dynamic light scattering (DLS) with a ZetaSizer (Malvern, UK).
Reagents and Consumables for the Biological Study
The MTT [3-(4, 5-dimethylthiazol-2-yl)-2, 5 diphenyltetrazolium bromide] was procured from Sigma Chemical Company Pvt. Ltd. St. Louis, MO, USA, and used without any further modification except dilution; additionally, the Dulbecco’s Modified Eagle Medium (DMEM) and MEM culture medium, antibiotic–antimycotic and fetal bovine serum (FBS), were purchased from Invitrogen, USA. Nunc, Denmark, supplied the plastic wares and other consumables for the cell culture.
Cell Culture of Lung (A549) Cancer Cells with Treatment of SrNiONPs
Lung cancer cells (A549) were cultured in a specified medium (DMEM/MEM) with 10–12% fetal bovine serum (FBS), 0.2% sodium bicarbonate, and antibiotic–antimycotic solution (100X, 1 mL/100 mL of medium) with humid atmosphere (5% CO2 & 95% O2) at 37 °C. Previously, for the experiments, the cell viability was evaluated using trypan blue dye according to the protocol [14], and cells with viability greater than 95% were only used in the study. The cells were treated with nano powder over a period of 10 to 12 passages. For the treatment of cancer cells, the material was initially used at high concentrations and then diluted at desired and different concentrations. According to the experiment, the cells were grown in 6-well or 96-well plates.
MTT Assay
The viability of cells was determined using the MTT assay for control and treated samples according to the previously established protocol [15]. In brief, the cells were initially cultured in specialized 96 well plates (at a rate of 1 × 104/well) and incubated for 24 h at 37 °C in a humidified environment. The cells were treated with prepared Sr-NiO-NPs of concentration 2–100 µg/mL, for 24 h. Once the cells were completely mixed in the well plates, stock solution of MTT (5 mg/mL in PBS) was amalgamated at a rate of 10 μL/well in 100 μL of cell suspension, and further incubated for 4 h. After the incubation period, the well solution was washed with PBS, and ~ 200 µL of DMSO was added to aspirate the formazan product and gently mixed. The optical analysis of the solution was performed using multiwall micro-plate reader at 550 nm (Multiskan Ex, Thermo Scientific, Finland). The control cells were used as a reference and were run under the same conditions as the test cells. The maximum absorbance is determined by the solvent used in the sample solution, and the % of cell viability was calculated using the following equations:
NRU Assay
The assessment of toxicity for control and treated SrNiONPs cancer cells was also performed via NRU assay as described previously [16, 17]. Both cancer cells were seeded (10,000/well) in 96 well plates. When the cells had fully grown (after 24 h), they were exposed to the material at the desired concentration (2–100 g/mL) and placed in an incubator for 24 h. Following the exposure, the cells were incubated in NR medium (100 g/mL) for 3 h before being washed and the dye extracted in a 1% acetic acid and 50% ethanol solution. The developed color was analyzed at 540 nm.
Reactive Oxygen Species Generation Assay
The reactive oxygen species (ROS) generation was investigated using a fluorescent agent, 2, 7-dichloro dihydrofluoresce in diacetate dye (DCFH-DA; Sigma-Aldrich, USA). This was analyzed in accordance with the previously described protocol [18]. After being exposed to the processed material for 24 h, the cells were washed thoroughly with PBS and incubated for 30 min in DCFH-DA (20 µM) in dark medium at 37 °C. Once the reaction of DCFH-DA with control and treated cells was completed, it was examined with a fluorescence microscope for intracellular fluorescence.
Isolation of Total RNA and Real-Time PCR
RNA was purified from 3 × 105 cells/well of A549 cells untreated and treated with SrNiONPs (100 μg/ml) for 24 h using Qiagen RNeasy mini kit according to the manufacturer’s protocol. Total RNA purity was verified by using Nanodrop spectrophotometer 8000 (Thermo Scientific, USA) and the integrity of RNA was visualized on 2% agarose gel using the documentation system (Universal Hood II, BioRad, USA). The cDNA was synthesized using the MLV reverse transcriptase (GE Health Care, UK) according to the kit instruction, taking 2 μg of RNA and 100 ng of oligo(dT)12–18 primer. The set of primers for apoptotic and anti-apoptotic genes was quantified using a Roche® LightCycler®480 (96-well block) according to the recommended cycling program [19]. A 20 µl reaction mixture was made up of a combination of 100 ng of the cDNA and 7.5 μM of each primer with 2 × of SYBR Green in I Roche Diagnostics. The real-time PCR (RT-PCR) cycling conditions were 95 °C for 10 min of denaturation, followed by 40 cycles of 95 °C for 15 s of annealing, 60 °C for 20 s of annealing, and 72 °C for 20 s of elongation. GAPDH was used as an internal housekeeping control gene to normalize the data from expressed genes.
Results
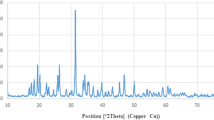
X-Ray Diffraction Pattern
The grown and annealed prepared powders were using an XRD pattern at room temperature (Fig. 1). The XRD pattern depicts several peaks that show the crystalline and phase structure of the material. The spectroscopy shows the presence of indexed and assigned peaks that are very similar to the formation of nickel oxide (NiO) [JCPDS 75–0197] and strontium oxide (SrO) and SrO3 [JCPDS no 00–006-0520], and in accordance with the information available [20]. The XRD defines the phases, peak positions, FWHM, and crystallite size; and the average diameter of the crystallite (nm) of the grown powder was calculated using well-known Scherrer formula as previously described in literature [20]. The particle size is estimated to be 21 nm using the Scherrer equation, as described in previous published work [20]. The annealed powder has a high crystallinity of the prepared material and sharp and intense peaks, indicating that the processed powder particles have a high crystallinity, whereas the FWHM or wide width of the peaks indicates that the size of each crystallite is very small.
Scanning Electron Microscopy Results
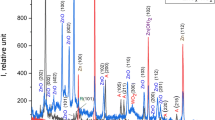
The structural identification of annealed nanocomposite of strontium-doped nickel oxide nanoparticles (SrNiONPs) powder was analyzed with scanning electron microscopy (SEM) and the obtained images are illustrated in Fig. 2. According to the low-resolution images (Fig. 2a–b), perforated sheet-like structures with dense morphology can be seen, with particles arranged and embedded with the accumulation of several minute particles and agglomerated in nature. Furthermore, a more detailed explanation of an individual particle size morphology was confirmed by high magnification images (Fig. 2c–d). The particles appeared to be merged as a result of annealing, and several plate-like structures with nanoscale structures appeared (Fig. 2d). The obtained information from SEM and their material analysis is well justified and in consistent with the XRD analysis (Fig. 1).
SEM images of SrNiO: a–b shows the low magnification images whereas c–d shows the high magnification image with size and surface structures of nanoparticle. e Shows the energy dispersive X-ray analysis (EDX) of SrNiONPs, which shows the elemental compositions rations of Sr, Ni, O, and other respective elements, respectively, and f displayed TEM image and it shows the average size (diameter and length) of each nanoparticle is ~ 21 nm
In addition to this observation, the elemental analysis of the prepared composite material was observed using EDX spectroscopy in conjunction with a SEM micrograph (Fig. 2e). The EDX spectrum reveals that the highest atomic mass ratio percentages of strontium (26.47%), nickel (17.84%), and oxygen (19.39%) appeared in the spectrum, indicating that the formed structures were made with strontium, nickel, and oxygen elements. The carbon value (34.35%) could be due to the annealing conditions of the processed material, whereas the surface impurities could be molybdenum (0.83%) and phosphorous (1.12%). The spectrum and percent mass ratios confirm that the material synthesized is pure SrNiO (Fig. 2e).
Transmission Electron Microscopy Results
The annealed nanocomposite powder was also examined with transmission electron microscopy (TEM) for more structural observations, as described in the materials and methods section, and the obtained result is shown in Fig. 2f. According to the observations recovered from the SEM images, the TEM image is also consistent with the SEM images and shows the structural detail of SrNiONPs. The SrNiONPs are interconnected with the other particles; the average size of each NP is 201 nm; they are very clear, spherical and cubic in shape, with smooth surfaces that are consistent with SEM observations (Fig. 2f).
FTIR Results
The functional properties of the prepared materials are defined by FTIR spectroscopy. Figure 3(a) shows that an O–H stretching mode is represented by a peak range of 3200–3600 cm−1. A very small curved was observed at 3462 cm−1 which is due to the particles annealed at 250 °C for 1 h and it eliminated the O–H contents form the processed material. A sharp and intense peak assigned at 1465 cm−1 is related to the asymmetric stretching mode of C–O molecule (Fig. 3(a)). The peak obtained at 1093 cm−1 and 858 cm−1 is related to the asymmetric and symmetric stretching signal of NO3−1 group [20], whereas the band at ~ 441 cm−1 is related to the metal and oxygen bands (SrNiO), respectively [20]. The spectrum shows that no other band was observed in relation to any other functional group in the spectrum, demonstrating the material’s purity [20, 21].
UV–Visible Spectroscopy
The optical characteristics of the material can be observed via UV–visible spectroscopy and signifies the band gap energy of the formed material. Figure 3(b) shows a flat peak at 298 nm wavelength of the prepared material. The band gap energy of the material increases due to the mixing of other material. The observed band gap energy is analogous to and very similar to values documented in the literature [20, 21].
Hydrodynamic Particle Size and Zeta Potential
The aggregation and merging of particles are a common occurrence in all NPs. Small NPs are aggregated, merged together, densely packed, and form aggregation in this experiment. The dynamic light scattering (DLS) of NPs was investigated, and the size of particulate matter was determined to be approximately 347.94 nm (Fig. 4(a)). The large size of the NPs is due to the high annealing conditions, which aggregated and merged the particles together. The DLS is used to determine particle size in suspension solutions. The particles in suspension solution have a tendency to colloid and form aggregation in an aqueous medium, expressing the primary and secondary sizes of the prepared nanostructures. The size of the NPs grows as they collide/interact with other molecules. Our findings were also consistent with previous published literature [22, 23]. The zeta potential of SrNiONPs in aqueous solution was determined to be − 15.75 mV (Fig. 4(b)).
MTT Assay with SrNiONPs
The cytotoxicity of cancer cells, A549 cells, was investigated using the MTT assay, as previously described, with different concentration ranges of material ranging from 1 to 100 µg/ml for a 24 h incubation. According to the results, the viability of cancer cells was reduced by the prepared SrNiONPs, and the data was concentration/dose-dependent. The viability of A549 cells was reduced by MTT assay at 24 h to 102.7%, 101.3%, 95%, 86%, 73%, 69%, and 62% (Fig. 5(a)) for concentrations of 1, 2, 5, 10, 25, 50, and 100 µg/mL with control (p < 0.05 for each). According to the data obtained, the cytotoxicity of cancer cells was greatly influenced when the concentration of SrNiONPs was increased.
NRU Assay Cancer Cells with SrNiONPs
Similar findings were made in the NRU assay, as described in the materials and methods section. Cancer cells exposed to SrNiONPs were not significantly affected at first, but as concentrations or doses increased, cells were reduced and the effect was dose-dependent. For the A549 cells, NRU assay decreased at 24 h by 101%, 100%, 97%, 93%, 88%, 77%, and 65% for concentrations of 1, 2, 5, 10, 25, 50, and 100 g/mL, respectively (p < 0.05 for each). The NRU assay yielded similar results to the MTT assay, indicating that cytotoxicity is greatly influenced at high concentrations of nanostructures doses.
Induced ROS Generation in Cells with SrNiONPs
When A549 cancer cells were exposed to SrNiONPs at 10, 25, 50, and 100 µg/mL concentrations for 24 h (Fig. 6), a sequential trend in ROS generation was observed (Fig. 6). As shown in the graph (Fig. 6), ROS levels rise as the concentration of SrNiONPs increases when compared to control cells. The ROS levels at 10 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL increased by 120, 145, 160, and 190%, respectively, when compared to the control (100%) (Fig. 6).
Quantitative Real-Time PCR Induced by SrNiONPs
For the study of mRNA level using quantitative RT-PCR, the A549 cells were treated with the SrNiONPs for 24 h at a concentration of 100 μg/ml, and quantitative qPCR was performed to evaluate the mRNA level of fold changes for genes bax (1.9), p53 (2.9), casp3 (2.8), and bcl-2 (0.45), with control (1.0), respectively. The regulation of apoptotic genes A549 cells (p < 0.05 for each gene) was significantly altered. The qPCR results show that the tumor suppressor gene p53 and the pro-apoptotic gene bax were both upregulated. In SrNiONPs-treated cells, we also found increased expression of the caspase-3 gene. The expression of bcl-2 (0.45), an anti-apoptotic gene, was downregulated in cells treated with SrNiONPs (Fig. 7).
For mRNA quantification levels, fold change of apoptotic genes (p53, bax, bcl2, and casp3) was analyzed. Cells treated 100 μg/mL of SrNiONPs for 24 h. For the normalization, an internal control of GAPDH was used. The values are mean ± SE of three independent experiments. *Statistically significant difference compared to control (p < 0.05)
Discussion
In the current study, the material was first synthesized via solution process using strontium, nickel salt, and trisodium citrate dihydrate. Various techniques, including XRD, SEM, and TEM, were used to classify the crystallinity, size (̴ 20 ± 1 nm), and structure of the synthesized SrNiONPs, as well as EDX elemental study. The functional identification was confirmed via FTIR spectroscopy whereas the optical characteristic was examined via UV–visible spectroscopy. The hydrodynamic dynamics and zeta potential were confirmed and defined the size and potential of the processed material in liquid medium. In addition to this, the cytotoxic study was also investigated with lung A549 cancer cells at different doses (1, 2, 5, 10, 25, 50, and 100 μg/mL) of nanostructure material. The MTT assay was employed with and without SrNiONPs to check the cytotoxicity against cancer cells for 24 h. The viability of cancer cells against the processed material was accessed and it was dose reliant and the recovered results are in accordance with the previously published literature on cytotoxic activity [24, 25]. A similar pattern was also observed in the NRU assay. To learn more about the cytotoxicity of cancer cells and the role of SrNiONPs in their mechanism, we measured the ROS, which played a key role in the cytotoxicity in A549 cells after 24 h of exposure. The NPs have a proclivity to generate ROS in the culture solution and are responsible for the toxicological effects [26]. Various components of cells, such as DNA, protein, and lipids, can be disrupted in their functions by ROS generation and are responsible for cell death [27]. It is also evident from gene expression, which reveals apoptosis in cells and has a significant influence on the processed nanostructures. The study recapitulates that cell death was induced by SrNiONP interaction, which resulted in increased ROS generation when exposed to SrNiONPs. Based on the findings and possible explanations, we can conclude that cytotoxicity is dose-dependent and is greatly influenced at higher SrNiONP concentrations. In comparison to our previous research, NiO is more toxic [28].
Conclusion
The current work summarizes that the nanostructured material was prepared using a solution process and was thoroughly characterized. The average diameter of the crystallite was found to be ~ 21 nm (as observed from SEM and TEM). The % elemental ratios of strontium (26.47%), nickel (17.84%), and oxygen (19.39%) in the prepared material were revealed by EDX. The FTIR analysis also reveals that the formed material is pure SrNiONPs devoid of any chemical scum. The hydrodynamic size and zeta potential reveal that the particles in the liquid medium are colloidal in nature. The SrNiONPs were tested for cytotoxicity against lung (A549) cancer cells using the MTT and NRU assays, and it was discovered that there was initially no effect, but that as doses were increased, the cytotoxicity was greatly influenced. Cell cytotoxicity is also confirmed by RT-PCR and ROS generation in A549 cells treated with SrNiONPs. Because of the unique structural organization of nanostructures, it enables an outstanding improvement in the eradication of cancer cells. When compared to other existing drugs, the tiny and unique geometry allows a passage to enter easily and quickly to the cancer cell organelles, allowing for the possibility of cancer cell reduction.
Data Availability
Available whenever requested.
References
Colarusso P, Guo B, Zhang KQ, Bernath PF (1996) High-resolution infrared emission spectrum of strontium monofluoride. J Mol Spectrosc 175:158–171
Teoh LG, Li KD (2012) Synthesis and characterization of NiO nanoparticles by sol gel method. Mater Trans 53:2135–2140
Garcia-Miquel JL, Zhang Q, Allen SJ, Rougier A, Blyr A, Davies HO (2003) Nickel oxide sol–gel films from nickel diacetate for electrochromic applications. Thin Solid Films 424:165–170
Ganachari SV, Bhat R, Deshpande R, Abbaraju V (2012) Synthesis and characte- rization of nickel oxide nanoparticles by self-propagating low temperature combustion method. Recent Res Sci Technol 4:50–53
Zorkipli NNM, Kaus NHM, Mohamad AA (2016) Synthesis of NiO nanoparticles through sol-gel method. Proc Chem 19:626–631
Dooley KM, Chen SY, Ross JRH (1994) Stable Nickel-containing catalysts for the oxidative coupling of methane. J Catal 145:402–408
Rao KV, Sunandana CS (2008) Co3O4 nanoparticles by chemical combustion: effect of fuel to oxidizer ratio on structure, microstructure and EPR. Solid State Commun 148:32–37
Hosny NM (2011) Synthesis, characterization and optical band gap of NiO nano parti cles derived from anthranilic acid precursors via a thermal decomposition route. Polyhedron 30:470–476
Rahdar A, Aliahmad M, Azizi Y (2015) NiO nanoparticles: synthesis and charac terization. J Nanosci 5:145–151
Xiang L, Deng XY, Jin Y (2002) Experimental study on synthesis of NiO nano particles. Scr Mater 47:219–224
Mohammadijoo M, Naderi Khorshidi Z, Sadrnezhaad SK, Mazinani V (2014) Synthesis and characterization of nickel oxide nanoparticle with wide band gap energy prepared via thermochemical processing. Nanosci Nanotechnol Int J 4:6–9
Capasso L, Camatini M, Gualtieri M (2014) Nickel oxide nanoparticles induce infla mmation and genotoxic effect in lung epithelial cells. Toxicol Lett 226:28–34
Halliwell B (2003) Oxidative stress in cell culture: an under-appreciated problem? FEBS Lett 540:3–6
Siddiqui MA, Kashyap MP, Kumar V, Khedhairy AAA, Musarrat J, Pant AB (2010) Protective potential of trans-resveratrol against 4-hydroxynonenal induced damage in PC12 cells. Toxicol In Vitro 24:1592–1598
Siddiqui MA, Singh G, Kashyap MP, Khanna VK, Yadav S, Chandra D, Pant AB (2008) Influence of cytotoxic doses of 4-hydroxynonenal on selected neuro transmitter receptors in PC-12 cells. Toxicol In Vitro 22:1681–1688
Borenfreund E, Puerner JA (1985) Toxicity determined in vitro by morphological alterations and neutral red absorption. Toxicol Lett 24(2–3):119–124
Fotakis G, Timbrell JA (2006) In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett 160(2):171–177
Zhao J, Riediker M (2014) Detecting the oxidative reactivity of nanoparticles: a new protocol for reducing artifacts. J Nanopart Res 16(7):2493
Ahmad J, Wahab R, Siddiqui MA, Saquib Q, Khedhairy AAA (2020) Cytotoxicity and cell death induced by engineered nanostructures (quantum dots and nano particles) in human cell lines. J Biol Inorg Chem 2:325–338
Apsana G, George PP, Devanna N, Yuvasravana R (2018) Biomimetic synthesis and antibacterial properties of strontium oxide nanoparticles ocimum sanctum leaf extract. Asian J Pharm Clin Res 11(3):384–389
Anand GT, Nithiyavathi R, Ramesh R, Sundaram SJ, Kaviyarasu K (2020) Structural and optical properties of nickel oxide nanoparticles: investigation of antimicrobial applications. Surf Interface 18:100460
Iqbal J, Abbasi BA, Mahmood T, Hameed S, Munir A, Kanwal S (2019) Green synthesis and characterizations of nickel oxide nanoparticles using leaf extract of Rhamnus virgata and their potential biological applications. Appl Organomet Chem 33(8):e4950
Kim SH, Lee DH, Lee JH, Yang JY, Seok JH, Jung K, Lee JK (2021) Evaluation of the skin sensitization potential of metal oxide nanoparticles using the ARE-Nrf2 Luciferase KeratinoSensTM assay. Toxicol Res 37:277–284
Sioutas C, Delfino RJ, Singh M (2005) Exposure assessment for atmospheric Ultrafine Particles (UFPs) and implications in epidemiologic research. Environ Health Perspect 113:947–955
Kim YS, Lim CH, Shin SH, Kim JC (2017) Twenty-eight-day repeated inhalation toxicity study of nano-sized neodymium oxide in male Sprague-Dawley rats. Toxicol Res 33(3):239–253
Wang FS, Fan JG, Zhang Z, Gao B, Wang HY (2020) The global burden of liver disease: the major impact of China. Hepatol Rev 60:2099–2108
Ahmad J, Alhadlaq HA, Alshamsanc A, Siddiquia MA, Saquib Q, Khan ST, Wahab R, Khedhairy AAAl, Musarrat J, Akhtar MJ, Ahamed M, (2016) Differential cytotoxicity of copper ferrite nanoparticles in different human cells. J Appl Toxicol 36(10):1284–1293
Ahmad J, Alhadlaq HA, Siddiqui MA, Saquib Q, Khedhairy AAA, Musarrat J, Ahamed M (2015) Concentration-dependent induction of reactive oxygen species, cell cycle arrest and apoptosis in human liver cells after nickel nanoparticles exposure. Environ Toxicol 30(2):137–148
Funding
The authors are grateful to the Deanship of Scientific Research, King Saud University for funding through Vice Deanship of Scientific Research Chairs.
Author information
Authors and Affiliations
Contributions
J Ahmad: conceptualization, methodology, project administration writing–review and editing, and writing–original draft. R Wahab: methodology, data curation, conceptualization, and software. MA Siddiqui: conceptualization, methodology, investigation, and formal analysis. Q Saquib: methodology, validation, and formal analysis. N Ahmad: software and formal analysis: AA Al-Khedhairy: conceptualization and funding acquisition.
Corresponding author
Ethics declarations
Ethics Approval
We are using human cancer cell lines purchased from ATCC. In our case, no ethical approval is required.
Consent to Participate
Not applicable.
Consent for Publication
The authors approve processing this manuscript for publication.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmad, J., Wahab, R., Siddiqui, M.A. et al. Strontium-Doped Nickel Oxide Nanoparticles: Synthesis, Characterization, and Cytotoxicity Study in Human Lung Cancer A549 Cells. Biol Trace Elem Res 200, 1598–1607 (2022). https://doi.org/10.1007/s12011-021-02780-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-021-02780-5