Abstract
Intervertebral disc degeneration (IDD) is characterised by nucleus pulposus (NP) loss and extracellular matrix (ECM) degeneration. Circular RNAs (circRNAs) have been reported to be dysregulated during IDD progression. Recently, reports showed that hsa_circ_0040039 was increased in degenerated lumbar disc samples. The aim of this study was to explore the specific role and underlying mechanisms of hsa_circ_0040039 in IDD. The expression of hsa_circ_0040039 was investigated in NP tissues of IDD patients. IL-1β was used to treat NP cells to construct an IDD in vitro model. Overexpression and loss-of-function assays and bioinformatic analysis were performed to evaluate the role and potential mechanism of hsa_circ_0040039 during IDD progression. Hsa_circ_0040039 expression was increased about 2 folds in NP tissues compared with normal tissues and IL-1β-stimulated NP cells also presented hsa_circ_0040039 upregulation, and its overexpression promoted cell proliferation and ECM degeneration. The depletion of hsa_circ_0040039 had the opposite effects. Based on bioinformatics prediction, Luciferase assay, PCR and Western blot, our study verified that hsa_circ_0040039 directly bond to miR-146b-3p, then mediated its targeted MMP2 and PCNA. Moreover, the overexpression of miR-146b-3p and the silence of MMP2 or PCNA, partially abolished the effect of hsa_circ_0040039 on IL-1β-stimulated NPs. Hsa_circ_0040039 may participate in IDD development by mediating the repair and regeneration of NPs through upregulation MMP2 and PCNA mediated by miR-146b-3p.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intervertebral disc degeneration (IDD) is characterised by aging and degeneration of nucleus pulposus (NP), annulus fibrosus and cartilaginous endplate, etc. [1], which is a crucial pathological basis of a series of spinal degenerative diseases. The pathogenesis of IDD is complex, including genetics, aging, apoptosis, inflammation, biomechanical and environmental factors [1,2,3]. IDD imposes a huge burden on the global healthcare system, economic sociology and patients’ lives [4]. Hence, it is necessary to deeply illustrate the mechanism of NP loss and ECM degeneration and to find new therapy targets of IDD.
It is well known that cell apoptosis, phenotype alteration and immune dysregulation are main pathological features of IDD [5]. Moreover, the increased degradation of extracellular matrix, with matrix metalloproteinases (MMPs) as major proteases, is strikingly correlated with the pathological progression of IDD [6]. In another side, interleukin-1β (IL-1β) is the predominant pro-inflammatory cytokine that hyper-secreted in degenerated intervertebral discs and one of the promotors to disc degeneration [7]. Evidence indicated that IL-1β activation triggered the production of MMPs and inhibited the expressions of type II collagen and aggrecan in NP cells [7]. Therefore, IL-1β-stimulated NP is commonly used to construct a cellular model for IDD development in vitro [8, 9].
Circular RNAs (circRNAs) have been reported be dysregulated during IDD progression. Wang et al. discovered 7294 circRNAs differentially expressed in degenerated human NP cells, compared with the non-degenerated ones [10]. The important function of circRNAs in IDD progression has been validated [11]. Moreover, accumulated evidence indicated that circRNA functions as miRNA ‘sponge’ to regulate downstream gene expression [12, 13]. The interaction axis of circRNA-miRNA-mRNA has been verified to play a crucial role in the initiation and development of IDDs [14]. For examples, circ-4099 suppressed TNF-α-induced ECM degradation and inflammatory responses in NPs through miR-616-5p/Sox9 axis, which participated in the regulation of IDD [13]. Xie et al. found that circERCC2 ameliorated IDD by sponging to miR-182-5p, thereby mediating SIRT1 and activating mitophagy and inhibiting apoptosis in NPs [15]. Recently, hsa_circ_0040039 (chr16: 69,279,504–69,318,147), also called hsa_circRNA_101852, is derived from the host gene SNTB2 and found to be upregulated in the lumbar disc samples of IDD patients [16]. However, there lacks evidence in which the specific role and potential mechanism of hsa_circ_0040039 in IDD development.
In this study, we verified the alteration of hsa_circ_0040039 in IDD tissue samples. Besides, an IL-1β-stimulated NP cell model was established to explore the specific role of hsa_circ_0040039 in NP cell proliferation and ECM metabolism by performing gain-of-function and loss-of-function experiments. Furthermore, the underlying mechanisms of hsa_circ_0040039 in NP cells were explored by investigating the ‘sponge’ of hsa_circ_0040039 and downstream genes. This research might provide a novel target for IDD diagnosis and therapy.
Materials and Methods
Patient Tissues and Ethic Approval
The human intervertebral disc nucleus pulposus tissues included in this study were all collected from patients who were diagnosed as IDD and received intervertebral disc resection at Honghui Hospital with no IDD history before. As for the non-degenerated tissue samples recruited in our study, the derived subjects were confirmed as scoliosis disease or suffered vertebral fracture, and they all received discectomy or spinal fusion. Radiotherapy, chemotherapy and surgery were not included in the history of all subjects in this study. And all the patients and subjects provided written informed consents. The experimental procedures were conducted in accordance with the Declaration of Helsinki and got approval by the Ethics Committee of Honghui Hospital. The NP tissues were obtained from the intervertebral disc tissues of the included subjects, according to a previous study [17]. The tissues were immediately cryopreserved by using liquid nitrogen and finally stored at -80℃ condition.
Cell Culture
Human nucleus pulposus cells (HNPCs) were provided by the American Type Culture Collection (Rockville, Md.). Cells were maintained in high glucose DMEM (Invitrogen, Carlsbad, CA, USA) which contained 10% fetal bovine serum (FBS, Gibco, Rockville, MD) and supplemented with 1% penicillin-streptomycin. HNPCs were grown in an incubator with 5% CO2 under a humidified atmosphere at 37℃.
Cell Transfection and Treatment
The lentivirus overexpression vector of hsa_circ_0040039 (ov-circ) and control lentivirus vector (ov-NC) were constructed by Hanbio Technology (Shanghai, China). SiRNAs targeting to the back splice junction of hsa_circ_0040039, eukaryotic translation initiation factor 4A3 (eIF4A3), MMP2, proliferating cell nuclear antigen (PCNA), and siRNA-NC, as well as the mimic of miR-146b-3p and scramble mimic were synthesised by Geenseed Biotech (Guangzhou, China). The HNPCs cells were planted in 24-well plates with 250 µL medium and grown to a confluence at 80%. Subsequently, all transfections were performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for 48 h following the manufacturer’s instructions. The doses of overexpression vector and siRNAs for the transfection assays were 0.5 µg and 50 nM, respectively. The concentration of mimics used in this study was also at 50 nM. Then, IL1β (10 ng/mL) was utilized to stimulate HNPCs cells for 24 h. The cells were finally collected for the next experiments.
Extraction RNA and Quantitative Real-Time PCR (qRT-PCR)
For the detection of target genes’ expressions, total RNA was extracted from HNPCs using Trizol reagent (Invitrogen). After the concentration of RNA samples were detected, the reverse transcription was performed using a PrimeScript RT Reagent Kit (Takara, Otsu, Shiga, Japan). SYBR Premix Ex Taq II (Takara) was used to conduct qPCR amplification on an ABI 7500 StepOnePlus system (Applied Biosystems, CA, USA). All the manipulation procedures were performed in line with the protocols of manufacturers. GAPDH was adopted as the internal control for mRNAs and circRNAs, while U6 was regarded as the internal control for miRNAs. Triplicates of each experiment were performed. The final results were quantified by using the 2−ΔΔCT method. The primers used in this study are shown in Table 1. Especially, total RNA was incubated with 3 U/µg RNase R (Epicentre Biotechnologies, Madison, WI, USA) for 20 min at 37 °C before reverse transcription during detecting circRNA expression. Besides, miRNA was isolated by using an miRNeasy Mini Kit (Qiagen, Duesseldorf, Germany).
CCK-8 Assay
Cell proliferation was measured using a CCK-8 assay kit (Dojindo Corp, Japan) according to its illustration. Briefly, the treated HNPCs were plated into a 96-microwell plate. CCK-8 reagent (10 µL) was added into each well and incubated for 2 h at 37 °C. The optional density values were recorded at 450 nm on a plate reader (Thermo Fisher Scientific).
Western Blot Assay
The cells after treatments were lysed for 30 min using RIPA lysis buffer (Beyotime, Shanghai, China) on ice, supplemented with a protease inhibitor. After centrifugation and quantification with the BCA™ Protein Assay kit (Beyotime, Shanghai, China), the protein samples were separated on 10% SDS-PAGE gels (80 V, 90 min) and transferred onto nitrocellulose (NC) membranes (2.2 μm). Then, the blots were blocked with 5% skim milk at room temperature for 2 h and followed with primary antibody incubation (overnight at 4 °C). The antibodies included anti-eIF4A3, anti-collagen II, anti-aggrecan, anti-ADAMTS5 and anti-MMP2 (Proteintech, IL, USA). Subsequently, HRP rabbit IgG secondary antibodies were used to incubate proteins for 1 h at room temperature. Finally, the immunoreactive bands were visualised using an ECL Chemiluminescence kit (Thermo, MA) and quantified with an Image Lab™ Software (Bio-Rad, Shanghai, China).
Luciferase Reporter Assay
The reporter plasmids containing wild type hsa_circ_0040039 sequence (WT-hsa_circ_0040039), mutant hsa_circ_0040039 sequence (MUT-hsa_circ_0040039), wild type MMP2 sequence (WT-MMP2), mutant MMP2 sequence (MUT-MMP2), wild type PCNA sequence (WT-PCNA), and mutant PCNA sequence (MUT-PCNA) were designed by Geenseed Biotech (Guangzhou, China). The plasmids, miR-146b-3p mimics and NC mimic were co-transfected into HNPCs using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). After 48 h-transfection, the final luciferase activities were measured using a dual luciferase reporter gene assay kit (BioAssay Systems, Hayward, CA, USA).
RNA Immunoprecipitation (RIP) Assay
The RIP assay was performed using a Magna RIP™ RNA-Binding Protein Immunoprecipitation kit (Millipore, USA). The magnetic beads were incubated with anti-EIF4A3 antibodies or negative control IgG antibody (Millipore, USA). HNPCs were harvested and lysed by RIP lysis buffer. Next, the cell lysis solution was incubated with the corresponding antibody-coated beads. The coprecipitated RNA was isolated using Trizol reagent (Invitrogen) and the enrichments of measured by qRT-PCR.
Statistical Analysis
Experimental data are presented as mean ± standard error of means (SEM). Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software Inc., CA, USA). The differences between groups were analysed using Student’s t-test. A p value < 0.05 was regarded as demonstrating statistical significance.
Results
Hsa_circ_0040039 was Upregulated During IDD Progression
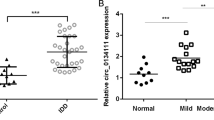
First, we evaluated the expression of hsa_circ_0040039 in clinical samples of IDD patients and IL-1β-induced in vitro model of IDD. As the results presented in Fig. 1A, hsa_circ_0040039 expression in IDD group was significantly upregulated (P = 0.00681) compared with the normal subject group. The cell viability of HNPCs was dramatically decreased (6 h, P = 0.0351; 12 h, P = 0.0164; 24 h P = 0.0281; 5 ng/mL, P = 0.0312; 10 ng/mL, P = 0.00812; 20 ng/mL, P = 0.0751) by the incubation of IL-1β in both time- and dose-dependent manners; with the greatest decreases firstly occurred at 24 h and 10 ng/mL, respectively (Fig. 1B C). Next, according to results of in vitro IDD cell model, the expression of hsa_circ_0040039 in HNPCs was dramatically increased (6 h, P = 0.0293; 12 h, P = 0.00614; 24 h P = 0.0177; 5 ng/mL, P = 0.00803; 10 ng/mL, P = 0.0412; 20 ng/mL, P = 0.0451) after IL-1β incubation also in both time- and dose-dependent manners; with the highest hsa_circ_0040039 expression at 24 h and 20 ng/mL (Fig. 1D and E). In general, the evidence presented that hsa_circ_0040039 had an increased expression level in both IDD tissues and in vitro cell model.
Hsa_circ_0040039 was upregulated in NP tissues and IL-1β-stimulated HNPCs during IDD progression. HNPCs were stimulated with IL-1β (10 ng/mL) for different times or at different concentrations for 24 h. (A, D and E) The expression of hsa_circ_0040039 in NP tissues of IDD and normal subjects, as well as IL-1β-stimulated HNPCs, was measured using qRT-PCR. (B and C) CCK-8 assay was used to detect cell viability under different treatments. In A, ** p < 0.01; in B and D, * p < 0.05 and ** p < 0.01 compared with 0 h group, # p < 0.05 and ## p < 0.01 compared with 6 h group, $ p < 0.05 compared with 12 h group; in C and E, * p < 0.05 and ** p < 0.01 compared with 0 ng/mL group, # p < 0.05 and ## p < 0.01 compared with 5 ng/mL group, $ p < 0.05 compared with 10 ng/mL group
Effect of Overexpression and Inhibition of hsa_circ_0040039 on the Proliferation and ECM Metabolism IL-1β-stimulated HNPCs
To investigate the role of hsa_circ_0040039 in IDD progression, loss-of- and gain-of-function assays of hsa_circ_0040039 were performed based on an IL-1β-stimulated HNPCs model. The results showed that ov-circ transfection dramatically increased the expression of hsa_circ_0040039 (P = 0.00633), and si-circ decreased it (P = 0.00519) in IL-1β-stimulated HNPCs (Fig. 2A). However, the expression of SNTB2, the host gene of hsa_circ_0040039, was not influenced by ov-circ (P = 0.0830) and si-circ (P = 0.0791) transfections (Fig. 2B). CCK-8 assays indicated that the overexpression of hsa_circ_0040039 suppressed IL-1β-induced decline on the cell proliferation of HNPCs (P = 0.0226), while silencing hsa_circ_0040039 aggravated the effect of IL-1β (P = 0.0189) (Fig. 2C). Meanwhile, the mRNA levels of PCNA and Ki67 were increased in ov-circ group (PCNA, P = 0.00538; Ki67, P = 0.0316) and decreased in si-circ group (PCNA, P = 0.0228; Ki67, P = 0.00713) compared with ov-NC and si-NC groups, respectively (Fig. 2D and E). In addition, the overexpression of hsa_circ_0040039 downregulated collagen II (mRNA, P = 0.0191; protein, P = 0.0132) and aggrecan (mRNA, P = 0.0308; protein, P = 0.0403) in IL-1β-treated HNPCs, and promoted ADAMTS5 (mRNA, P = 0.0231; protein, P = 0.00632) and MMP2 expressions (mRNA, P = 0.0158; protein, P = 0.00720), while silencing hsa_circ_0040039 had the opposite effects (collagen II: mRNA, P = 0.00632; protein, P = 0.0119. aggrecan: mRNA, P = 0.00813; protein, P = 0.00732. ADAMTS5: mRNA, P = 0.00772; protein, P = 0.00812. MMP2: mRNA, P = 0.00681; protein, P = 0.00591) (Fig. 2F-2H).
The effect of hsa_circ_0040039 overexpression or inhibition on the proliferation and ECM metabolism of IL-1β-stimulated HNPCs. (A) The transfection efficiency of hsa_circ_0040039 overexpression vectors (ov-circ) and siRNA of hsa_circ_0040039 (si-circ) in IL-1β-stimulated HNPCs was measured. ** p < 0.01 compared with IL-1β + ov-NC (si-NC) treated group. (B) The effects of ov-circ and si-circ on SNTB2 expression in IL-1β-stimulated HNPCs. (C) Cell proliferation of IL-1β-stimulated HNPCs was evaluated after overexpression or silence of hsa_circ_0040039. (D and E) The relative mRNA expression of PCNA and Ki67 in HNPCs was detected through RT-PCR. (F-H) Protein and mRNA expression of Collagen II, aggrecan, ADAMTS5 and MMP2 in HNPCs were detected. ** p < 0.01 compared with control group; # p < 0.05 and ## p < 0.01 compared with IL-1β + ov-NC group; $ p < 0.05 and $$ p < 0.01 compared with IL-1β + si-circ group
eIF4A3 Induced hsa_circ_0040039 Expression in IL-1β-stimulated HNPCs
eIF4A3 is an RNA binding protein and involved in the post-transcriptional regulation processes of RNA, such as mRNA splicing and the biogenesis of circRNAs [18]. Based on that, we investigated the factor that induced hsa_circ_0040039 alteration during IDD progression. First, with the help of the Circ interactome platform (https://circinteractome.nia.nih.gov/index.html), two putative binding sites of eIF4A3 were found in the upstream (site ‘a’) and downstream (site ‘b’) regions of hsa_circ_0040039 pre-mRNA transcript (Fig. 3A). The transfection of si-eIF4A3 significantly suppressed the expression of eIF4A3 in HNPCs, at both mRNA (P = 0.00613) and protein (P = 0.00238) levels (Fig. 3B C). Furthermore, eIF4A3 silence suppressed the increased expression of hsa_circ_0040039 (P = 0.00536) in IL-1β-stimulated HNPCs (Fig. 3D). The RIP assay was utilized to validate the bindings between eIF4A3 hsa_circ_0040039 whose results verified that eIF4A3 anti-body dramatically enriched the wild type transcripts of hsa_circ_0040039 pre-mRNA, SNTB2, but not the mutant types (MUT-SNTB2-a, P = 0.00708; MUT-SNTB2-a, P = 0.00593) (Fig. 3E).
eIF4A3 induced hsa_circ_0040039 expression in IL-1β-stimulated HNPCs. (A) The binding sites for eIF4A3 on the SNTB2 mRNA transcript at the upstream/downstream region of hsa_circ_0040039 was obtained from circ interactome. (B and C) The transfection efficiency of si-EIF4A3 in HNPCs. ** p < 0.01 and *** p < 0.005 compared with si-NC transfected group. (D) Silence of EIF4A3 suppressed the production of hsa_circ_0040039 in HNPCs. ** p < 0.01 compared with control group; ## p < 0.01, compared with IL-1β + si-NC treated group. (E) The RIP assay verified eIF4A3 could directly bind to the SNTB2 mRNA in HNPCs. ** p < 0.01 compared with WT-SNTB2 group
Hsa_circ_0040039 bond with miR-146b-3p and Promoted MMP2 and PCNA Expression
To further explore the potential regulatory mechanism of hsa_circ_0040039 in IDD development, we analysed the possible targeted miRNAs of hsa_circ_0040039 based on circBase (http://www.circbase.org/) and circ interactome predictions. There were 9 miRNAs which had binding sites with hsa_circ_0040039 were predicated by both circBANK and Circ interactome, including hsa-miR-1203, hsa-miR-136, hsa-miR-146b-3p, hsa-miR-330-3p, hsa-miR-370, hsa-miR-545, hsa-miR-574-5p, hsa-miR-638 and hsa-miR-665 (Fig. 4A). Next, we found that hsa_circ_0040039 overexpression significantly suppressed the expressions of hsa-miR-136 (P = 0.00662), hsa-miR-146b-3p (P = 0.00243), hsa-miR-330-3p (P = 0.0416), hsa-miR-370 (P = 0.0440), and hsa-miR-638 (P = 0.00758), and the expression of hsa-miR-146b-3p was decreased by hsa_circ_0040039 overexpression most (P = 0.00243) (Fig. 4B). Hence, we furtherly targeted to validate the relationship between miR-146b-3p and hsa_circ_0040039. IL-1β significantly suppressed the expression of miR-146b-3p in HNPCs (P = 0.00131), which was promoted by miR-146b-3p mimic (P = 0.00507) (Fig. 4C). The sequences of WT and MUT putative miR-146b-3p binding sites of hsa_circ_0040039 were shown in Fig. 4D. Besides, miR-146b-3p mimics weakened the luciferase activity of WT-hsa_circ_0040039 compared with the NC mimic group (P = 0.00182), while no significant difference was found between NC mimic and miR-146b-3p mimic transfected groups under MUT-hsa_circ_0040039 treatment (P = 0.394) (Fig. 4D). These results revealed that hsa_circ_0040039 directly bond to and mediated miR-146b-3p in HNPCs.
Hsa_circ_0040039 binds with miR-146b-3p and promotes MMP2 and PCNA expression. (A) Venn diagram showed 9 miRNAs that predicted interacting with hsa_circ_0040039 by both circBANK and circ interactome. (B) The expression of 9 miRNAs in HNPCs after overexpression of hsa_circ_0040039. * p < 0.05, ** p < 0.01 and *** p < 0.005 compared with ov-NC treated group. (C) The transfection efficiency of miR-146b-3p mimics in IL-1β-stimulated HNPCs was measured. (D) Luciferase reporter assay was performed to detect the activity of hsa_circ_0040039 in HNPCs cells after transfected with miR-146b-3p mimic or NC mimic. *** p < 0.005 compared with NC mimic group. (E) The putative binding sites of miR-146b-3p on the MMP2 and PCNA wild-type (WT) or mutated sequence (MUT). (F and G) The relative luciferase activities were measured in HNPCs co-transfected with WT-MMP2 or MUT-MMP2 (WT-PCNA or MUT-PCNA) and miR-146b-3p mimic or NC mimic. *** p < 0.005 compared with NC mimic group. (H-J) Protein expression of MMP2 and PCNA in HNPCs was measured. ** p < 0.01 compared with control group; # p < 0.05 and ## p < 0.01 compared with IL-1β + ov-NC + NC mimic treated group; $$ p < 0.01 compared with IL-1β + ov-circ treated group
Subsequently, Targetscan (http://www.targetscan.org) was adopted to analyse the direct targets of miR-146b-3p. We found that MMP2 contains three conserved target sites and PCNA contains one conserved target site of miR-146b-3p (Fig. 4E). The results of dual-luciferase reporter gene assay indicated that only miR-146b-3p mimic suppressed the luciferase activity of WT-MMP2 (P = 0.00233) and WT-PCNA reporters (P = 0.00181) compared with the NC mimic group, but had no obvious effects on MUT-MMP2 (P = 0.362) and MUT-PCNA reporters (P = 0.651) (Fig. 4F and 4G). Additionally, we found that the overexpression of hsa_circ_0040039 significantly promoted the expressions of MMP2 (P = 0.00630) and PCNA (P = 0.00823) in IL-1β-treated HNPCs, while the transfection of miR-146b-3p mimic dramatically suppressed the expressions of MMP2 (P = 0.00716) and PCNA (P = 0.0409), and reversed the promoted effects of hsa_circ_0040039 overexpression (MMP2, P = 0.00506; PCNA, P = 0.00638) (Fig. 4H-4J). These results suggested that MMP2 and PCNA were downstream factors of the hsa_circ_0040039/miR-146b-3p axis.
Hsa_circ_0040039 Promoted Cell Proliferation and ECM Degeneration Through Regulating the Expression of MMP2 and PCNA Mediated by miR-146b-3p
Furthermore, our study validated the roles of miR-146b-3p, MMP2 and PCNA in hsa_circ_0040039-mediated HNPC proliferation and ECM degeneration. PCNA expression was significantly inhibited by si-PCNA in IL-1β-treated HNPCs (mRNA, P = 0.00613; protein, P = 0.00734) (Fig. 5A-5C). The transfections of miR-146b-3p mimic and si-PCNA both dramatically depressed the cell proliferation of IL-1β-stimulated HNPCs (miR-146b-3p, P = 0.0154; si-PCNA, P = 0.00651), and partially reversed the promoting effect of hsa_circ_0040039 on HNPC proliferation (miR-146b-3p, P = 0.00598; si-PCNA, P = 0.00633) (Fig. 5D). In addition, the expression of MMP2 was significantly inhibited by si-MMP2 in IL-1β-treated HNPCs, both at mRNA and protein levels (mRNA, P = 0.00604; protein, P = 0.00685) (Fig. 5E and G). The overexpression of miR-146b-3p and silence of MMP2 both promoted collagen II (miR-146b-3p: mRNA, P = 0.00710; protein, P = 0.00505. si-PCNA: mRNA, P = 0.00531; protein, P = 0.00593) and aggrecan (miR-146b-3p: mRNA, P = 0.00682; protein, P = 0.00801. si-PCNA: mRNA, P = 0.00726; protein, P = 0.00611.) expressions, and suppressed ADAMTS5 (miR-146b-3p: mRNA, P = 0.0315; protein, P = 0.0208. si-PCNA: mRNA, P = 0.00631; protein, P = 0.00839) and MMP2 expressions (miR-146b-3p: mRNA, P = 0.00912; protein, P = 0.0152. si-PCNA: mRNA, P = 0.00694; protein, P = 0.00760) in IL-1β-treated HNPCs (Fig. 5H-5J). Meanwhile, the results presented that miR-146b-3p mimic and si-MMP2 both reversed the inhibitory effects of hsa_circ_0040039 overexpression on collagen II (miR-146b-3p: mRNA, P = 0.0396; protein, P = 0.0218. si-PCNA: mRNA, P = 0.0309; protein, P = 0.00851) and aggrecan (miR-146b-3p: mRNA, P = 0.0403; protein, P = 0.00711. si-PCNA: mRNA, P = 0.0632; protein, P = 0.00583) in IL-1β-treated HNPCs, as well as the promoted effects on ADAMTS5 (miR-146b-3p: mRNA, P = 0.00776; protein, P = 0.0173. si-PCNA: mRNA, P = 0.00833; protein, P = 0.0214) and MMP2 (miR-146b-3p: mRNA, P = 0.00601; protein, P = 0.00735. si-PCNA: mRNA, P = 0.00537; protein, P = 0.00516) (Fig. 5H-5J).
Hsa_circ_0040039 promotes cell proliferation and ECM degeneration through regulating the expression of MMP2 and PCNA mediated by miR-146b-3p. (A-C) The expression of PCNA mRNA and protein in IL-1β-stimulated HNPCs was measured after transfection with siRNA of PCNA. ** p < 0.01 compared with IL-1β + si-NC treated group. (D) Cell proliferation of IL-1β-stimulated HNPCs was evaluated. * p < 0.05 and ** p < 0.01 compared with IL-1β treated group; ## p < 0.05 compared with IL-1β + ov-circ treated group. (E-G) The expression of MMP2 mRNA and protein in IL-1β-stimulated HNPCs was measured after transfection with siRNA of MMP2. ** p < 0.01 compared with IL-1β + si-NC treated group. (H-J) Expression of Collagen II, aggrecan, ADAMTS5 and MMP2 mRNA and protein in HNPCs were detected. * p < 0.05 and ** p < 0.01 compared with IL-1β treated group; # p < 0.05 and ## p < 0.01 compared with IL-1β + ov-circ treated group
Discussion
CircRNA has been emerging as a crucial regulator of various cellular processes and takes a fundamental part in multiple physiological functions and pathological progression. According to the current evidence, the involvement of circRNA in the precise molecular mechanism of IDD has been demonstrated which mediates IDD development [19]. For examples, the downregulation of circERCC2 was found in the degenerative NP tissues of IDD patients and impacted the apoptosis of NP cells and ECM degradation [15]. In the study of Cheng and colleagues, circVMA21 attenuated the NP cell apoptosis and ECM imbalance, both anabolism and catabolism, which contributed to alleviating IDD progression [20]. Previously, Lan et al. discovered that hsa_circ_0040039 was upregulated by about four-folds in degenerated intervertebral disc tissue samples compared with the normal ones [16]. Our research consistently found hsa_circ_0040039 upregulation in NP tissues of IDD patients and in IL-1β-stimulated HNPCs, an in vitro cell model of IDD. Also, by overexpressing hsa_circ_0040039, the IL-1β-induced NP cell injury and ECM degradation, with decreased collagen II and aggrecan expression and increased MMP2 and ADAMTS5 expressions, were depressed in this study which suggested that hsa_circ_0040039 attenuated IDD development. Previous studies indicated that loss of NPs and ECM degradation in the intervertebral discs are the major causes of IDD. The increased degradations of collagen II and aggrecan have been found in the early stages of IDD [21]. The expressions of MMPs and ADAMTSs are also promoted as the severity of the IDD degeneration worsens while non-degenerated discs present low levels of MMPs and ADAMTSs, even undetectable [22]. Meanwhile, MMPs and ADAMTSs possessed the ability to degrade collagens and aggrecan, leading to structural damage and IVD [22]. Consistent with our findings, Tan et al. discovered that miR-665 alteration-induced elevation on NP cell proliferation and decreases on collagen II and aggrecan suppressed the progression of IDD [23]. The therapeutic effect of aucubin on IDD was relied on ameliorating ECM degradation via depressing the expressions of ADAMTS5 and MMP3 and activating collagen II in HNPCs [24].
Moreover, the present study also discovered the binding between eIF4A3 and hsa_circ_0040039 mRNA transcript, and silencing eIF4A3 suppressed the production of hsa_circ_0040039 in IL-1β-stimulated HNPCs. We indicated that eIF4A3 might be the factor that induced the abnormal accumulation of hsa_circ_0040039. Previous studies regarded that eIF4A3 is an important core compound of the exon junction complex, and plays a key role in pre-mRNA splicing [25]. Besides, eIF4A3 participates in the mediation of circRNA biogenesis, e.g. circMMP9, circBNIP3 and circSEPT9, thus to regulate the carcinogenesis and development of various tumour diseases [18, 25, 26]. Jiang et al. demonstrated that eIF4A3 stimulated the cyclization of circARHGAP29 by binding its back-spliced junction region and promoted circARHGAP29 transcription which led to the enhanced aerobic glycolysis in prostate cancer cells [27]. However, few evidence has confirmed the role of eIF4A3 in IDD progression. Further investigations are required to perform to verify whether eIF4A3 could mediate IDD pathogenesis by inducing the biogenesis of circRNAs.
In terms of the regulatory mechanism, most circRNAs could function as a competitive RNA (ceRNA) to regulate gene expression by sponging miRNAs[28]. For example, circ-4099 played a protective role in IDD by exerting as a sponge of miR-616-5p to regulate SOX9 expression [13]. Huang and colleagues indicated that circSPG21/miR-1197/ATP1B3 ceRNA network was validated in both NP cell model and in vivo mice model of IDD [29]. The therapeutic value of circRNA-CIDN was proved to mediate NP cell damage by sponging to miR-34a-5p and regulating SIRT1 [30]. To further determine the underlying molecular mechanisms of hsa_circ_0040039 in HNPCs, we explored the potential miRNAs that bind to hsa_circ_0040039. The results presented that miR-146b-3p was a pivotal target of hsa_circ_0040039 and depressed by hsa_circ_0040039 overexpression. Previously, miR-146b-3p has been reported to play roles in tumours, thrombosis and acute respiratory distress syndrome [31,32,33]. Our research filled the vacancy that the role of miR-146b-3p in IDD remains unclear: miR-146b-3p was decreased in IL-1β-induced in vitro cell model of IDD and the overexpression of miR-146b-3p aggravated cell proliferation loss and ECM degradation in IL-1β-stimulated HNPCs. Meanwhile, miR-146b-3p reversed the promoting effect of hsa_circ_0040039 on HNPCs cell growth and inhibitory effect on ECM degradation.
It is well known that PCNA plays a critical role in DNA replication and repairment, and is regarded as a marker of cell proliferation [34, 35]. Additionally, MMP2, a member of the matrix metalloproteinases family, is upregulated in human degenerative intervertebral disc tissue and contributes to the degeneration of the intervertebral disc [21]. Evidence showed that SNHG1 promoted cell proliferation of NP cells via elevating PCNA expression [36]. Gao and colleagues illustrated that MMP2 was dysregulated in NP tissues of IDD patients and PART1 mediated NP cell degeneration by depressing MMP2 via sponging to miR-93 [37]. Consistently, our findings suggested that hsa_circ_0040039 promoted PCNA and MMP2 expressions to enhance cell proliferation and ECM degradation of IL-1β-stimulated HNPCs, and silencing PCNA and MMP2 both reversed the promoting effects of hsa_circ_0040039/miR-146b-3p axis on IDD progression.
Conclusion
In summary, this study indicated that hsa_circ_0040039 was upregulated in IDD disc tissues and IL-1β-stimulated HNPCs. Hsa_circ_0040039 showed a positive effect on cell proliferation and an inhibitory effect on ECM degradation in IL-1β-stimulated HNPCs. Besides, eIF4A3 impacted hsa_circ_0040039 transcription. Furthermore, the inhibition of hsa_circ_0040039 mediated IDD progression by exerting as a sponge of miR-146b-3p to regulate PCNA and MMP2 expressions. The evidence illustrated the influence of abnormal hsa_circ_0040039 expression on the development of IDD and its corresponding regulatory mechanism for the first time, which may provide a novel biomarker for IDD diagnosis and therapy.
Data Availability
The data supporting the conclusions of this article are included within the article.
Abbreviations
- IDD :
-
Intervertebral disc degeneration
- NP :
-
Nucleus pulposus
- ECM :
-
Extracellular matrix
- IL-1β :
-
Interleukin-1β
- MMPs :
-
Matrix metalloproteinases
- HNPCs :
-
Human nucleus pulposus cells
- ov-circ :
-
Overexpression vector of hsa_circ_0040039
- eIF4A3 :
-
Eukaryotic translation initiation factor 4A3
- PCNA :
-
Proliferating cell nuclear antigen
References
Kos, N., Gradisnik, L., & Velnar, T. (2019). A Brief Review of the Degenerative Intervertebral Disc Disease. Medical archives (Sarajevo, Bosnia and Herzegovina), 73,421–424.
Ding, F., Shao, Z. W., Yang, S. H., Wu, Q., Gao, F., & Xiong, L. M. (2012). Role of mitochondrial pathway in compression-induced apoptosis of nucleus pulposus cells. Apoptosis: an international journal on programmed cell death, 17, 579–590.
Sambrook, P. N., MacGregor, A. J., & Spector, T. D. (1999). Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis and rheumatism, 42, 366–372.
Sakai, D., & Grad, S. (2014). Advancing the Cellular and Molecular Therapy for Intervertebral Disc Disease. Advanced Drug Delivery Reviews, 84, 159–171.
Yu, P., Mao, F., Chen, J., Ma, X., Dai, Y., Liu, G., Dai, F., & Liu, J. (2022). Characteristics and mechanisms of resorption in lumbar disc herniation. Arthritis Res Ther, 24, 205.
Cui, N., Hu, M., & Khalil, R. A. (2017). Biochemical and Biological Attributes of Matrix Metalloproteinases. Progress in molecular biology and translational science, 147,1–73.
Johnson, Z. I., Schoepflin, Z. R., Choi, H., Shapiro, I. M., & Risbud, M. V. (2015). Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. European cells & materials, 30, 104–116. discussion 116 – 107.
Huang, J. F., Zheng, X. Q., Lin, J. L., Zhang, K., Tian, H. J., Zhou, W. X., Wang, H., Gao, Z., Jin, H. M., & Wu, A. M. (2020). Sinapic acid inhibits IL-1β-Induced apoptosis and catabolism in Nucleus Pulposus cells and ameliorates intervertebral Disk Degeneration. Journal of inflammation research, 13, 883–895.
Tu, J., Li, W., Zhang, Y., Wu, X., Song, Y., Kang, L., Liu, W., Wang, K., Li, S., Hua, W., & Yang, C. (2017). Simvastatin Inhibits IL-1β-Induced Apoptosis and Extracellular Matrix Degradation by Suppressing the NF-kB and MAPK Pathways in Nucleus Pulposus Cells. Inflammation, 40, 725–734.
Wang, S., Sun, J., Yang, H., Zou, W., Zheng, B., Chen, Y., Guo, Y., & Shi, J. (2019). Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta biochimica et biophysica Sinica, 51, 571–579.
Li, Z., Chen, X., Xu, D., Li, S., Chan, M. T. V., & Wu, W. K. K. (2019). Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration.Cell Prolif, 52, e12704.
Sang, Y., Chen, B., Song, X., Li, Y., Liang, Y., Han, D., Zhang, N., Zhang, H., Liu, Y., Chen, T., Li, C., Wang, L., Zhao, W., & Yang, Q. (2019). circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a Axis in breast Cancer. Molecular therapy: the journal of the American Society of Gene Therapy, 27, 1638–1652.
Wang, H., He, P., Pan, H., Long, J., Wang, J., Li, Z., Liu, H., Jiang, W., & Zheng, Z. (2018). Circular RNA circ-4099 is induced by TNF-α and regulates ECM synthesis by blocking mir-616-5p inhibition of Sox9 in intervertebral disc degeneration. Experimental & molecular medicine, 50, 27.
Zhu, J., Zhang, X., Gao, W., Hu, H., Wang, X., & Hao, D. (2019). lncRNA/circRNA–miRNA–mRNA ceRNA network in lumbar intervertebral disc degeneration. Molecular Medicine Reports, 20, 3160–3174.
Xie, L., Huang, W., Fang, Z., Ding, F., Zou, F., Ma, X., Tao, J., Guo, J., Xia, X., Wang, H., Yu, Z., Lu, F., & Jiang, J. (2019). CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death And Disease, 10, 751.
Lan, P. H., Liu, Z. H., Pei, Y. J., Wu, Z. G., Yu, Y., Yang, Y. F., Liu, X., Che, L., Ma, C. J., Xie, Y. K., Hu, Q. J., Wan, Z. Y., & Wang, H. Q. (2016). Landscape of RNAs in human lumbar disc degeneration. Oncotarget, 7, 63166–63176.
Jian, S., Wang, H. L., Song, K. H., Ding, Z. W., Wang, H. L., Ma, X. S., Lu, F. Z., Xia, X. L., Wang, Y. W., & Fei-Zou. (2018). CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA (50 vol., p. 94). Experimental & Molecular Medicine.
Wang, R., Zhang, S., Chen, X., Li, N., Li, J., Jia, R., Pan, Y., & Liang, H. (2018). EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Molecular Cancer, 17, 166.
Mitra, A., Pfeifer, K., & Park, K. S. (2018). Circular RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer Res, 7, S624–S628.
Cheng, X., Zhang, L., Zhang, K., Zhang, G., Hu, Y., Sun, X., Zhao, C., Li, H., Li, Y. M., & Zhao, J. (2018). Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Annals Of The Rheumatic Diseases, 77, 770–779.
Wang, W. J., Yu, X. H., Wang, C., Yang, W., He, W. S., Zhang, S. J., Yan, Y. G., & Zhang, J. (2015). MMPs and ADAMTSs in intervertebral disc degeneration. Clinica chimica acta; international journal of clinical chemistry, 448,238–246.
Dowdell, J., Erwin, M., Choma, T., Vaccaro, A., Iatridis, J., & Cho, S. K. (2017). Intervertebral Disk Degeneration and Repair. Neurosurgery, 80, S46-s54.
Tan, H., Zhao, L., Song, R., Liu, Y., & Wang, L. (2018). microRNA-665 promotes the proliferation and matrix degradation of nucleus pulposus through targeting GDF5 in intervertebral disc degeneration. Journal Of Cellular Biochemistry, 119, 7218–7225.
Yang, S., Li, L., Zhu, L., Zhang, C., Li, Z., Guo, Y., Nie, Y., & Luo, Z. (2019). Aucubin inhibits IL-1beta- or TNF-alpha-induced extracellular matrix degradation in nucleus pulposus cell through blocking the miR-140-5p/CREB1 axis. Journal Of Cellular Physiology, 234, 13639–13648.
Zheng, X., Huang, M., Xing, L., Yang, R., Wang, X., Jiang, R., Zhang, L., & Chen, J. (2020). The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Molecular Cancer, 19, 73.
Li, Y., Ren, S., Xia, J., Wei, Y., & Xi, Y. (2020). EIF4A3-Induced circ-BNIP3 aggravated Hypoxia-Induced Injury of H9c2 cells by targeting miR-27a-3p/BNIP3. Molecular therapy. Nucleic acids, 19, 533–545.
Jiang, X., Guo, S., Wang, S., Zhang, Y., Chen, H., Wang, Y., Liu, R., Niu, Y., & Xu, Y. (2022). EIF4A3-Induced circARHGAP29 promotes aerobic glycolysis in Docetaxel-Resistant prostate Cancer through IGF2BP2/c-Myc/LDHA signaling. Cancer Research, 82, 831–845.
Huo, Z., Li, H., Tian, L., Li, J., Zhang, K., Li, Z., Li, G., Du, L., Xu, H., & Xu, B. (2021). Construction of a Potentially Functional circRNA-miRNA-mRNA Network in Intervertebral Disc Degeneration by Bioinformatics Analysis. Biomed Res Int, 2021, 8352683.
Huang, Y., Zhang, Z., Wang, J., Shen, S., Yao, T., Xu, Y., Chen, Z., Fang, B., & Ma, J. (2021). circSPG21 protects against intervertebral disc disease by targeting miR-1197/ATP1B3. Experimental & Molecular Medicine, 53, 1547–1558.
Xiang, Q., Kang, L., Wang, J., Liao, Z., Song, Y., Zhao, K., Wang, K., Yang, C., & Zhang, Y. (2020). CircRNA-CIDN mitigated compression loading-induced damage in human nucleus pulposus cells via miR-34a-5p/SIRT1 axis. EBioMedicine, 53, 102679.
Riesco-Eizaguirre, G., Wert-Lamas, L., Perales-Patón, J., Sastre-Perona, A., Fernández, L. P., & Santisteban, P. (2015). The miR-146b-3p/PAX8/NIS Regulatory Circuit modulates the differentiation phenotype and function of thyroid cells during carcinogenesis. Cancer Research, 75, 4119–4130.
Liu, Y., Zhu, J. Q., Jin, X. H., Dong, M. P., & Zheng, J. F. (2020). Up-regulation of miR-146b-3p protects septic mice with acute respiratory distress syndrome by inhibiting PI3K/AKT signaling pathway. Journal of bioenergetics and biomembranes, 52, 229–236.
Su, Z., & Wu, F. (2020). Inflammatory Factors Induce Thrombosis through the miR-146b-3p/p38MAPK/COX-2 Pathway. BioMed research international, 2020, 8718321.
Juríková, M., Danihel, Ľ., Polák, Š., & Varga, I. (2016). Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta histochemica, 118, 544–552.
Wang, X., Peng, L., Gong, X., Zhang, X., Sun, R., & Du, J. (2018). LncRNA-RMRP promotes nucleus pulposus cell proliferation through regulating miR-206 expression. Journal of cellular and molecular medicine, 22, 5468–5476.
Tan, H., Zhao, L., Song, R., Liu, Y., & Wang, L. (2018). The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. American Journal of Physiology-Cell Physiology, 315, C21–C27.
Gao, D., Hao, L., & Zhao, Z. Long non-coding RNA PART1 promotes intervertebral disc degeneration through regulating the miR-93/MMP2 pathway in nucleus pulposus cells.International Journal of Molecular Medicine,46.
Funding
This work was supported by Key Research and Development Program of Shaanxi (2023-YBSF-617).
Author information
Authors and Affiliations
Contributions
Xiaochen Fan contributed to the conception of the study; Yi Cao and Haidong Deng performed the experiment; Jing Jia contributed significantly to analysis and manuscript preparation; Liang Li performed the data analyses and wrote the manuscript; Xiaochen Fan helped perform the analysis with constructive discussions.
Corresponding author
Ethics declarations
Ethic Approval
All patients involved provided written informed consents. The experimental procedures were conducted in accordance with the Declaration of Helsinki and got approval by the Ethics Committee of Honghui Hospital.
Consent to Participate
None.
Consent to Publish
All authors were agree with the publication of the article.
Conflict of Interest
All authors have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liang Li and Yi Cao contribute equally to this study and regarded as co-first author.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, L., Cao, Y., Deng, H. et al. EIF4A3-Induced Upregulation of hsa_circ_0040039 is a Biomarker and Aggravates IL-1β-Stimulated Intervertebral Disc Degeneration. Appl Biochem Biotechnol 195, 4292–4307 (2023). https://doi.org/10.1007/s12010-023-04334-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04334-y