Abstract
The ideal condition of earthworm gut promotes growth and multiplication of beneficial soil microorganisms eliminating pathogens and converts organic wastes into nutrients rich compost. The present study has been carried out to determine the population dynamics of earthworm gut bacteria and to find out relative abundance of different functional bacterial groups in the foregut, midgut, and hindgut of earthworm Perionyx excavatus. To assess bacterial diversity, a viable plate count method was adopted. In the different gut region of earthworm, aerobic heterotrophic, amylolytic, Bacillus, Gram-negative, proteolytic, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing bacterial populations ranged from 22.2 to 241.6 × 106, 8.0 to 171.60 × 106, 1.83 to 2.79 × 106, 10.68 to 23.04 × 104, 3.70 to 5.52 × 104, 59.60 to 208.40 × 104, 1.86 to 7.34 × 104, 10.94 to 19.78 × 104, 0.80 to 3.42 × 104, 7.83 to 13.70 × 104, 1.31 to 2.67 × 104 cfu/ml gut suspension, respectively. The results of the one-way ANOVA revealed that the bacterial load of most of the bacterial groups was significantly higher (p < 0.05) in the hindgut region, followed by midgut and foregut. Only the density of the proteolytic group was significantly higher (p < 0.05) in the midgut region followed by foregut and hindgut. Starch hydrolyzing bacteria constitute the largest group of bacteria in the gut content. From principal component analysis, two components were extracted with the eigenvalues of 8.485 and 1.132. Agglomerative hierarchical cluster analysis revealed that the bacterial populations were clustered into four different groups. Quantitative variation among bacterial groups in earthworm’s gut seems to determine the soil health and composting efficiency; from this point of view, the present study will provide a better understanding about different functional bacterial groups of earthworm’s guts and might be helpful in sustainable agriculture and waste management.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Earthworms are considered as ecosystem engineers as they play imperative responsibility in the modification of physicochemical and biological characteristics of the soil [1]. They break down bulky soil particles and organic wastes, making them accessible for microbial degradation and ultimately convert the debris to precious vermicompost with the help of microbes [2]. Indiscriminate use of chemical fertilizers and pesticides causes serious threats to natural water bodies and living organisms. In 2017, agrochemicals are projected at around 7.55 billion and by 2050 are anticipated to hit 9.8 billion to provide enough aliment for the global population [3]. The toxicity of chemical fertilizer and pesticides on physiology, oxidative stress biomarkers, development, and growth of different aquatic non-target organisms had been reported previously [4]. On the other hand, earthworm’s involvement improves disintegration and biodegradation of organic wastes 60–80% by face lifting the growth and multiplication of beneficial decomposer bacteria [5]. Soil microorganisms, i.e., fungi, protozoa, algae, and bacteria are considered to be the major portion of diet for earthworms [6]. Earthworm gut is a tubular structure consisting of a mouth, muscular pharynx, gizzard, intestine, and associated digestive glands [7]. The intestine is further divided into foregut, midgut, and hindgut [8]. Earthworm gut provides ideal physico-chemical conditions like neutral pH, high moisture, optimum temperature, organic and mineral-rich mucus for the growth, and development of microorganisms [9, 10]. Earthworm gut is considered as a natural filter as it promotes growth and multiplication of beneficial soil microorganisms, elimination of soil pathogens, and conversion of organic wastes into nutrient rich compost [11, 12]. Microbial load in the earthworm gut is higher than that in the surrounding soil, and there is immense bacterial diversity in earthworm gut [13]. This bacterial community plays a crucial role in the degradation processes [14]. Several factors in the earthworm gut may regulate the bacterial community [15]. Enzymes like cellulose, amylase, lipase, chitinase, protease, lichenase, urease, nitrate reductase, invertase, acid phosphatase, and alkaline phosphatase have been reported from earthworm gut content [16, 17]. Some of them were produced by the earthworm itself [18, 19] while others were secreted by the ingested micro-organisms [20,21,22]. Those enzymes along with mucus and antibiotics in earthworm’s gastrointestinal tract break down the organic macromolecules [17]. Thus, the microorganisms and earthworms act symbiotically and synergistically to speed up and improve the organic matter decomposition [23, 24].
Though earthworm gut bacteria play an important role in soil fertility, scanty literature is available on bacterial diversity in the gut region of earthworm [25]. In this context, the present study has been carried out to determine the population dynamics of earthworm gut bacteria and to find out their relative abundance in the foregut, midgut, and hindgut of earthworm Perionyx excavatus (Perrier, 1872).
Material and Methods
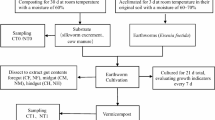
Collection of Earthworms
Live mature earthworms (Perionyx excavatus) were collected in a sterile plastic container from 30 days old composting beds of Kulti vermicomposting farm (23°12′ N, 88°30′ E) of Purba Bardhaman district of West Bengal, India. After collection, earthworms were brought to the laboratory for further microbial analysis. Specimen was submitted to the Zoological Survey of India, Kolkata, and identified as Perionyx excavatus (Perrier) based on external morphological features such as number of segments, clitellum characteristics, and color.
Dissection of Earthworm and Collection of Gut Content
Live adult earthworms were superficially sterilized with 50% alcohol and rapidly transferred to the dissection tray. The digestive tract was cut open and the content from foregut, midgut, and hindgut was collected with a sterile loop and kept separately into sterile Eppendorf tubes.
Analysis of Earthworm Gut Content
To assess the bacterial diversity in the earthworm gut content, viable plate count method was adopted during the present study. Enumeration of the bacterial populations belonging to different groups was performed following standard methodologies [26, 27]. The gut content was diluted with sterile distilled water up to 10−4. To determine the aerobic heterotrophic bacterial population, 50 μl gut suspension from 10−4 dilutions was added to 100 ml of nutrient agar (peptone 5.0, yeast extract 1.5, HM peptone 1.5, NaCl 5.0, agar 15 g l−1, pH-7.4). Then the inoculated medium was distributed on five sterile petri plates and incubated at 30 ± 1 °C in the BOD incubator for 48 h. To determine other bacterial populations, 50 μl of gut suspension of different dilutions was added to 100 ml of each bacterial group specific culture medium and distributed in five plates and incubated at 30 ± 1 °C. The Gram-negative bacterial population was enumerated by incubating the gut suspension into MacConkey agar (peptone 3.0, pancreatic digest of gelatin 17.0, lactose monohydrate 10.0, crystal violet 0.001, NaCl 5.0, neutral red 0.030, agar 13.5, and bile salts 1.5 g l−1, pH-7.1) for 24 h. To determine the amylolytic bacterial population, the sample was incubated in starch agar medium (soluble starch 2.0, meat extract 3.0, peptic digest 5.0, agar 15.0 g l−1, pH 7.2 ± 0.1) for 24 h and only the colonies producing clear zone after flooding with Gram’s iodine were counted as starch hydrolyzer. The total Bacillus population was recorded after 48 h incubation of gut suspension in the HiCrome Bacillus agar (peptone 10.0, HM extract 1.0, NaCl 10.0, D-mannitol 10.0, chromogenic mixture 3.2, Phenol red 0.025, agar 15 g l−1, pH-7.1). To determine the nitrifying bacterial population, the sample was incubated in Winogradsky’s medium (ammonium sulfate 1.0, dipotassium hydrogen phosphate 1.0, manganese sulfate heptahydrate 0.5, NaCl 2.0, ferrous sulfate heptahydrate trace, calcium chloride dihydrate 0.02, agar 10 g l−1, pH-8.5), and the pink colonies were visualized by flooding the plates with α-naphthylamine and sulfanilic acid (1:1). The colony number of this particular group of bacteria was recorded at 5 days intervals from the date of incubation up to 30 days. But all other groups of bacterial populations were recorded after 72 h of incubation. Inorganic phosphate solubilizing bacterial population was determined by incubating the gut suspension on Pikovskaya’s agar media (yeast extract 5.0, dextrose 10.0, calcium phosphate 5.0, ammonium sulfate 0.5, KCl 0.2, MgSO4 0.1, MnSO4 trace, FeSO4 trace, agar 15 g l−1, pH-7.0) and counting the number of bacterial colonies producing a clear zone after 72 h incubation. Nitrate reducing bacterial population was recorded on nitrate reducing agar medium (potassium nitrate 1.0, peptone 5.0, HM peptone 3.0, agar powder 12.0, pH-6.8) by visualizing pink colonies after flooding the plates with α-naphthylamine and sulfanilic acid (1:1). The same procedure was followed to count asymbiotic nitrogen-fixing bacterial population on nitrogen-free medium (mannitol 10.00, dipotassium hydrogen phosphate 0.50, magnesium sulfate 0.20, sodium chloride 0.20, manganese sulfate trace, ferric chloride trace, agar powder 18.00 g l−1, pH 7.20 ± 0.2). Protein (gelatine) hydrolyzing bacterial population was determined by the presence of the halo zone around the colonies by flooding with HgCl2 on nutrient agar medium with 2% gelatine. Spirit blue agar medium (casein enzymic hydrolysate 10.00, yeast extract 5.00, spirit blue 0.15, agar 17.00 g l−1, pH 6.8 ± 0.2) with TWEEN 80 were used to determine fat hydrolyzing groups. Azotobacter population was enumerated by incubating the gut suspension on Azotobacter agar medium (mannitol 20.00, dipotassium phosphate 1.0, magnesium sulfate 0.20, sodium chloride 0.20, FeSO4 trace, agar 15 g l−1, pH-8.3) for 24 h. Incubation period varied among certain bacterial groups because off the growth rate of different bacterial groups varied in different culture media.
Statistical Analysis
The data obtained from the study were further subjected to statistical analysis using XLSTAT software to draw a more specific conclusion. After performing the normality check utilizing the Shapiro Wilk test, the difference in the abundance of bacterial groups in the foregut, midgut, and hindgut of earthworm was evaluated by one-way ANOVA followed by the Tukey test. The data obtained on the different groups of bacteria present in the earthworm gut were subjected to heat mapping and agglomerative hierarchical cluster analysis. Furthermore, the relation between various bacterial groups of earthworm gut was evaluated by principal component analysis (PCA) [28]. The principal components were chosen as per Gniazdowski [29].
Result
Bacterial population isolated from different gut regions of earthworm Perionyx excavatus showed a normal distribution in QQ plot (Fig. 1).
In the different gut regions of earthworm P. excavatus, bacterial loads of aerobic heterotrophic, starch-hydrolyzing, total Bacillus, Gram-negative, gelatin hydrolyzing, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing populations ranged from 22.2 to 241.6 × 106, 8.0 to 171.60 × 106, 1.83 to 2.79 × 106, 10.68 to 23.04 × 104, 3.70 to 5.52 × 104, 59.60 to 208.40 × 104, 1.86 to 7.34 × 104, 10.94 to 19.78 × 104, 0.80 to 3.42 × 104, 7.83 to 13.70 × 104, 1.31 to 2.67 × 104 cfu/ml gut suspension, respectively (Fig. 2). The results of the one-way ANOVA (Table 1) revealed that bacterial load of aerobic heterotrophic, starch-hydrolyzing, total Bacillus, Gram-negative, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing population were significantly higher (p < 0.05) in the hindgut region, followed by midgut and foregut. Only the density of the gelatin hydrolyzing group was significantly higher (p < 0.05) in the midgut region followed by foregut and hindgut.
The result from heatmap also confirms that the aerobic heterotrophic, starch-hydrolyzing, total Bacillus, Gram-negative, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing population exhibited higher density in the hindgut region and gelatin hydrolyzing bacterial population was higher in the midgut region (Fig. 3).
From principal component analysis (PCA), a total of 10-factor components were observed in the scree plot (Fig. 4c). Based on eigenvalues, factor 1 (8.485) and factor 2 (1.132) were considered that explained 96.171% (component 1: 84.847% and component 2: 11.324%) of the variance on the bacterial population in the different gut regions of the earthworm Perionyx excavatus (Table 2). The score plot of the observed variables (Fig. 4b) showed that component 1 successfully separated the bacterial population of hindgut from fore- and midgut whereas the component 2 separated the data obtained from bacterial population of midgut from fore- and hindgut regions.
a Ordination diagram of quadrant distribution of different bacterial groups from earthworm gut via PCA. b Score plot of observed variable showing the distribution and clustered of the obtaining data among two major components. c Scree plot showing the Eigenvalues, principal components and cumulative variability (%) derived from PCA regarding the bacterial groups from earthworm gut. d Pearson correlation matrix (n) between different bacterial groups of earthworm gut. Crossed mark represents insignificant correlation; blue color indicates positive and red color indicates negative correlation. (psb: phosphate solubilizing bacteria, fhb: fat hydrolyzing bacteria, gnb: Gram-negative bacteria, anfb: asymbiotic nitrogen-fixing bacteria, nfb: nitrifying bacteria, azo: Azotobacter, bac: total Bacillus, shb: starch-hydrolyzing bacteria, nrb: nitrate-reducing bacteria and ghb: gelatine hydrolyzing bacteria)
The factor loading table showed that the 1st principal component is strongly correlated with aerobic heterotrophic, starch hydrolyzing, total Bacillus, Gram-negative, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing population and 2nd principal component is strongly correlated with gelatin hydrolyzing population (Table 3).
The bacterial groups were plotted on the quadrant plot, and it showed a projection of the initial variable in the four quadrants. The first quadrant was comprised of starch hydrolyzing, total Bacillus, nitrate-reducing, and asymbiotic nitrogen-fixing bacterial group; the second quadrant contained Azotobacter, nitrogen-fixing, fat hydrolyzing, Gram-negative and phosphate-solubilizing bacteria, and the fourth quadrant was comprised of a single group of gelatine hydrolyzing bacteria (Fig. 4a).
The result of the Pearson correlation matrix (Fig. 4d) revealed significant (p < 0.05) positive correlation among aerobic heterotrophic, starch-hydrolyzing, total Bacillus, Gram-negative, fat hydrolyzing, nitrate-reducing, nitrifying, asymbiotic nitrogen-fixing, Azotobacter, and phosphate solubilizing population, and all these bacterial groups were negatively correlated with the gelatin hydrolyzing bacterial group.
Agglomerative hierarchical cluster analysis (AHC) in respect of the number of various bacterial groups in different gut regions revealed that the bacterial populations were clustered into four different groups below the automatic truncation line. Phosphate solubilizing, fat hydrolyzing, Gram-negative, asymbiotic nitrogen-fixing, and nitrifying bacterial group created a single cluster in respect of their distribution pattern while the Azotobacter group diverged from them forming another cluster. Bacillus and starch-hydrolyzing and the nitrate-reducing bacterial group formed a different cluster and gelatine hydrolyzing bacteria further differed from these three groups forming single cluster (Fig. 5).
Agglomerative hierarchical clustering (AHC) of different bacterial groups from earthworm gut; the dotted line indicates the automatic truncation, leading to four groups. (psb: phosphate solubilizing bacteria, fhb: fat hydrolyzing bacteria, gnb: Gram-negative bacteria, anfb: asymbiotic nitrogen-fixing bacteria, nfb: nitrifying bacteria, azo: Azotobacter, bac: total Bacillus, shb: starch-hydrolyzing bacteria, nrb: nitrate-reducing bacteria and ghb: gelatine hydrolyzing bacteria)
Discussion
The earthworm gut holds enormous bacterial diversity but only a little portion is explored [25]. Parle, one of the pioneer researchers in the field of on microbial presence in the earthworm gut, reported fungal, bacterial, and actinomycetes populations in three different earthworm species in 1963 [18]. Thereafter, several studies have been made regarding the total aerobic bacterial population and changes in total bacterial loads while passing through the gut. But the detailed study on the qualitative and quantitative estimation of different bacterial groups from the different regions of the earthworm gut probably is not reported yet from these regions. Present study evinces that most of the bacterial groups exhibited higher density in the hindgut region of the earthworm whereas bacterial load was comparatively lower in the foregut region. Both the bacterial heat map and AHC analysis reflect the presence of a higher number of bacterial populations in the hindgut region. The highest aerobic heterotrophic bacterial load was found in the hindgut region followed by midgut and foregut. The gradual rise of the number of aerobic heterotrophs from the foregut to hind gut in Perionyx excavatus may be due to the epigeic nature of this earthworm species. When the organic debris passes through the digestive tract, it accumulates in hindgut which offers an ideal microclimatic environment for the profuse growth of the bacterial population. The earthworm gut acts like selective filter as well as fermentation vessel which provides favorable condition for the growth and activity of bacterial community. Several factors such as anoxic environment, neutral pH, high moisture content, ideal temperature conditions [30], and mucus-containing nutrients and easily metabolizable compounds collectively contribute making the earthworm gut ideal habitats for bacteria [31]. However, the total heterotrophic bacterial load may vary through the different regions of the earthworm digestive tract, and it may vary among different species of earthworms. Earlier reports support these findings as they stated that the population of soil microorganisms ingested by earthworm increases while passing through the gut [32]. The microbial population in the fresh vermicasts was higher than the midgut content of earthworms. Birundha et al. [33] reported if the bacterial population of midgut content was considered as 1, it increased to 1.20 in the fresh vermicastings of Perionyx excavatus. This result indicates that further bacterial replication occurs in the hindgut region, and the overall bacterial population increases. One of the earliest studies stated that the highest number of total bacterial population was found on hindgut followed by midgut and then foregut in L. terrestris [34]. Chowdhury et al. [35] studied total microbial communities in the gut content of Perionyx excavatus and found the highest abundance of bacteria-actinomycetes in hindgut followed by midgut and minimum in foregut. Though the maximum generic variation of bacteria-actinomycetes was detected in foregut followed by midgut and hindgut, Kristufek et al. [36] reported that cfu of total heterotrophic bacteria in the foregut region was 7 × 106/g gut content, but it amplified to 16 × 106 and 29 × 106 in the midgut and hindgut region, respectively in Lumbricus rubellus. But an opposite observation was also reported where bacterial population decrease towards the posterior region in the case of earthworm Onycochaeta borincana [37] and Aporrectodea caliginosa [36]. From another study, it was found that the midgut fluid taken from three earthworm species namely Aporrectodea caliginosa, Lumbricus terrestris, and Eisenia fetida could selectively suppress bacterial growth. But the hindgut fluid did not show such suppressive activity; moreover, the growth of most of the bacterial strains increased under the influence of hindgut fluid [6].
During the present study, most of the bacterial group exhibited higher density in the hindgut region but the proteolytic bacterial population was found maximum in the midgut region, followed by foregut and then hindgut. Another experiment was conducted by Mishra and Dash [38], and they reported higher proteolytic activity in the midgut region, followed by foregut and hindgut in Lampito mauritii. This enzymatic activity may be achieved by the proteolytic bacteria present in the midgut content.
From the present study, it was observed that the Bacillus population constituted a large portion of total bacterial load, and it was found higher in the posterior region. Kim et al. [39] also described the genus Bacillus as the leading group in the gut region of earthworm. An increase in the number of Bacillus cereus was observed as the food shifts backward along the intestinal length of earthworm [40]. Among heterotrophos, starch-hydrolyzing bacteria constitute the largest group of bacteria in the gut content. They produce enzyme amylase which breaks down starch into simple sugars. Amylase-producing Bacillus tequilensis was isolated from the gut content of Perionyx excavatus [41]. There are many pieces of evidence of the presence of amylase in the different regions of the earthworm gut [38].
Microbial load in earthworms is an important parameter to determine the efficiency of composting. Benitez et al. [42] reported that earthworms and microorganisms act mutually in the degradation of organic matter. It was found that the bacterial load of 37 × 105 of cfu/g in the waste was amplified to 87 × 105 of cfu/g when treated with the earthworm Eudrilus eugeniae. Similarly, during the present study, it is observed that the total bacterial load significantly increases as the food passes through the different regions of the earthworm gut. The total bacterial load is increased from 22.2 × 106 to 241.6 × 106 while passing through the foregut to the hindgut. Other beneficial bacterial groups like nitrifying, denitrifying, phosphate solubilizing, lipolytic, asymbiotic N2 fixing, but Azotobacter population were also significantly amplified during their journey in the earthworm gut.
Earthworm had been considered as ecological engineers for a long time; however, the importance of the earthworm gut microbial community in regulation of earthworm’s metabolism and thereby in nutrient transformation had been studied in recent past years [43]. The anaerobic condition of the gut region of earthworm facilitates the colonization of different types of anaerobic bacteria [44]. This huge bacterial population in earthworm digestive tract facilitates the biotransformation of several soil pollutants including metals, microplastics, inorganic and organic chemicals, and antibiotics [45]. These bacterial groups are immensely important for soil fertility, plant growth, and recycling of nutrients, and the earthworms accelerate all these processes by facilitating growth of these bacterial groups [44].
Conclusion
Although the study of earthworm gut microbiota has been studied in the past few years, information regarding different functional bacterial groups remains limited. The present study shows the population dynamics of the different bacterial groups in the different gut regions of earthworm Perionyx excavatus for the very first time in these regions. From the observation of the present study, it may be concluded that most of the bacterial groups exhibited higher density in the hindgut region of the earthworm and bacterial load was comparatively lower in the foregut region. As bacterial load in earthworms seems to determine the soil health and efficiency of composting, the present piece of work will certainly illuminate the judicious and scientific exploitation of hindgut bacterial diversity of earthworm Perionyx excavatus as bioresource bacteria in sustainable agriculture and waste management program.
Data availability
The data that support the findings of this study are available on request from the corresponding author.
References
Zhang, W., Dima, C., & Cancan, Z. (2007). Functions of earthworm in ecosystem. Biodiversity Science, 15(2), 142.
Horn, M. A., Drake, H. L., & Schramm, A. (2006). Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Applied and Environment Microbiology, 72(2), 1019–1026.
Nataraj, B., Hemalatha, D., Rangasamy, B., Maharajan, K., & Ramesh, M. (2017). Hepatic oxidative stress, genotoxicity and histopathological alteration in fresh water fish Labeo rohita exposed to organophosphorus pesticide profenofos. Biocatalysis and Agricultural Biotechnology, 12, 185–190. https://doi.org/10.1016/j.bcab.2017.09.006
Majumder, R., & Kaviraj, A. (2019). Acute and sublethal effects of organophosphate insecticide chlorpyrifos on freshwater fish Oreochromis niloticus. Drug and Chemical Toxicology, 42, 487–495. https://doi.org/10.1080/01480545.2018.1425425
Sinha, R. K., Agarwal, S., Chauhan, K., & Valani, D. (2010). The wonders of earthworms & its vermicompost in farm production: Charles Darwin’s ‘friends of farmers’, with potential to replace destructive chemical fertilizers. Agricultural Science, 01(02), 76–94. https://doi.org/10.4236/as.2010.12011
Ravindran, B., Contreras-Ramos, S. M., & Sekaran, G. (2015). Changes in earthworm gut associated enzymes and microbial diversity on the treatment of fermented tannery waste using epigeic earthworm Eudrilus eugeniae. Ecological Engineering, 74, 394–401.
Chao, H. L., Kong, H., Zhang, M., Sun, M., Ye, D., Huang, Z., Zhang, D., Sun, S., Zhang, Y., Yuan, M., Liu, M., Hu, F., & Jiang, X. (2019). Metaphire guillelmi gut as hospitable micro-environment for the potential transmission of antibiotic resistance genes. Science of The Total Environment, 669, 353–361.
Drake, H. L., & Horn, M. A. (2007). As the worm turns: The earthworm gut as a transient habitat for soil microbial biomes. Annual Review of Microbiology, 61, 169–189.
Edwards, C. A. , Domínguez, J. & Arancon, N. Q. (2004). The influence of vermicomposts on plant growth and pest incidence. In: S. H., Shakir & W. Z. A., Mikhail (Eds.), Soil Zoology for Sustainable Development in the 21st Century, Cairo, pp 397–420. http://jdguez.webs.uvigo.es/wp-content/uploads/2011/10/The-influence-of-vermicompost-on-plant-growth-and-pest-incidence.pdf
Munnoli, M. P. (2007). Management of industrial organic solid waste through vermi-culture biotechnology with special reference to microorganisms. PhD diss., Goa University, India. http://hdl.handle.net/10603/12461. Accessed 19 Nov 2021.
Govindarajan, V. & Prabaharan. (2014). Gut micro-floral of earthworms: a review. American Journal of Biological and Pharmaceutical Research, 1(3), 125–130. https://www.researchgate.net/publication/265595977_GUT_MICRO-FLORA_OF_EARTHWORMS_A_REVIEW
Lv, B., Xing, M., & Yang, J. (2018). Exploring the effects of earthworms on bacterial profiles during vermicomposting process of sewage sludge and cattle dung with high-throughput sequencing. Environmental Science and Pollution Research, 25, 12528–12537. https://doi.org/10.1007/s11356-018-1520-6
Horn, A. M., Andreas, S., & Harold, L. D. (2003). The earthworm gut: An ideal habitat for ingested N2O-producing microorganisms. Applied and Environment Microbiology, 69(3), 1662–1669.
Villar, D., Alves, D., Pérez-Díaz, S., & Mato,. (2016). Changes in microbial dynamics during vermicomposting of fresh and composted sewage sludge. Waste Management, 48, 409–417. https://doi.org/10.1016/j.wasman.2015.10.011
Liu, D. F., Lian, B., & Wang, B. (2016). Solubilization of potassium containing minerals by high temperature resistant Streptomyces sp isolated from earthworm’s gut. Acta Geochimica, 35(3), 262–270.
Kiyasudeen, K., Ibrahim, M. H., Quaik, S., & Ismail, S. A. (2015). Prospects of organic waste management and the significance of earthworms. Springer.
Pathma, J., & Sakthivel, N. (2013). Molecular and functional characterization of bacteria isolated from straw and goat manure based vermicompost. Applied Soil Ecology, 70, 33–47. https://doi.org/10.1016/j.apsoil.2013.03.011
Parle, J. N. (1963). Micro-organisms in the intestines of earthworms. Microbiology, 31(1), 1–11.
Ranganathan, L. S., & Vinotha, S. P. (1998). Influence of pressmud on the enzymatic variations in the different reproductive stages of Eudrilus eugeniae (Kinberg). Current Science, 74(7), 634–635.
Tracey, M. V. (1951). Cellulase and chitinase of earthworms. Nature, 167(4254), 776–777.
Edwards, C. A., & Fletcher, K. E. (1988). Interactions between earthworms and microorganisms in organic-matter breakdown. Agriculture, Ecosystems & Environment, 24(1–3), 235–247.
Pramanik, P., Ghosh, G. K., Ghosal, P. K., & Banik, P. (2007). Changes in organic–C, N, P and K and enzyme activities in vermicompost of biodegradable organic wastes under liming and microbial inoculants. Bioresource Technology, 98(13), 2485–2494.
Koubová, A., Chroňáková, A., Pižl, V., Sánchez-Monedero, M. A., & Elhottová, D. (2015). The effects of earthworms Eisenia spp. on microbial community are habitat dependent. European Journal of Soil Biology, 68, 42–55.
Zhu, B. K., Fang, Y. M., Zhu, D., Christie, P., Ke, X., & Zhu, Y. G. (2018). Exposure to nanoplastics disturbs the gut microbiome in the soil oligochaete Enchytraeus crypticus. Environmental Pollution, 239, 408–415.
Liu, D. F., Lian, B., Wu, C., & Guo, P. (2018). A comparative study of gut microbiota profiles of earthworms fed in three different substrates. Symbiosis, 74(1), 21–29.
Pelczar, M. J., Bard, R. C., Burnett, G. W., Conn, H. J., Demoss, R. D., Euans, E. E., Weiss, F. A., Jennison, M. W., Meckee, A. P., Riker, A. J., Warren, J. & Weeks, O. B. (1957). Manual of microbiological methods, Society of American Bacteriology. McGraw Hill Book Company Inc.
Lacey, L. A. ed. (1997). Manual of techniques in insect pathology. Academic Press.
Zar, J. H. (1999). Biostatistical analysis. Pearson Education India.
Gniazdowski, Z. (2017). New interpretation of principal components analysis. arXiv preprint arXiv, 1711, 10420.
Martin, A., Cortez, J., Barosis, I., & Lavelle, P. (1987). Les mucus intestinaux de Var de Terre, moteur de leurs interactions avec la microflore. Revue d’Écologie et de Biologie du Sol, 24, 549–558.
Barois, I., & Lavelle, P. (1986). Changes in respiration rate and some physico-chemical properties of a tropical soil during transit through Pontoscolex corethrurus (Glossoscolecidae, Oligochaeta). Soil Biology & Biochemistry, 18, 539–541.
Idowu, A. B., Edema, M. O., & Adeyi, A. O. (2006). Distribution of bacteria and fungi in the earthworm Libyodrillus violaceous (Annelida: Oligochaeta), a native earthworm from Nigeria. Revista de Biologia Tropical, 54(1), 49–58.
Birundha, M., Paul, J. J., & Mariappan, P. (2013). Growth and reproduction of Perionyx excavatus in different organic wastes. International Journal of Current Microbiology and Applied Sciences, 2(2), 28–35.
Stockli, A. (1928). Studien tiber den Einfluss der Regenwtirmer auf die Beschaffenheit des Bodens. Landwirtsch Jahrb Schweiz, 42, 1–121.
Chowdhury, A., Hazra, A. K., Mahajan, S. & Choudhury, J. (2007). Microbial communities of earthworm (Perionyx excavatus Perrier) gut, cast and adjacent Soil in two different fields of west Bengal. Rec. Zoo. Surv. India. l07(Part-4), 101–113. http://faunaofindia.nic.in/PDFVolumes/records/107/04/0101-0113.pdf
Kristufek, V., Ravasz, K., & Pizl, V. (1992). Changes in densities of bacteria and microfungi during gut transit in Lumbricus rubellus and Aporrectodea caliginosa (Oligochaeta: Lumbricidae). Soil Biology & Biochemistry, 24, 1499–1500.
Valle-Molinares, R., Borges, S., & Rios-Velazquez, C. (2007). Characterization of possible symbionts in Onychochaeta borincana (Annelida: Glossoscolecidae). European Journal of Soil Biology, 43, S14–S18.
Mishra, P. C., & Dash, M. C. (1980). Digestive enzymes of some earthworms. Experientia, 36(10), 1156–1157.
Kim, H. J., Shin, K. H., Cha, J. C., & Hur, H. G. (2004). Analysis of aerobic and culturable bacterial community structures in earthworm (Eisenia fetida) intestine. Journal of Applied Biological Chemistry, 47(3), 137–142.
Ponomareva, S. I. (1953). The influence of the activity of earthworms on the creation of a stable structure in a sod-podzolized soil. Tr Pochv Inst Dokuchaeva., 41, 304–378.
Ghosh, S., & Chatterjee, S. (2020). Isolation and characterization of Bacillus tequilensis from gut content of Perionyx excavatus and evaluation of its starch hydrolyzing property. Biosc. Biotech. Res. Comm., 13, 670–675. https://doi.org/10.21786/bbrc/13.2/45
Benitez, E., Nogales, R., Elvira, C., Masciandaro, G. & Ceccanti, B. (1999). Enzyme and earthworm activities during vermicomposting of carbaryl-treated sewage sludge. American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America, 28(4), 1099–1104. https://doi.org/10.2134/jeq1999.00472425002800040006x
Zhou, G. W., Yang, X. R., Sun, A. Q., Li, H., Lassen, S. B., Zheng, B. X., & Zhu, Y. G. (2019). Mobile incubator for iron (III) reduction in the gut of the soil-feeding earthworm Pheretima guillelmi and interaction with denitrification. Environmental Science and Technology, 53(8), 4215–4223.
Sun, M., Chao, H., Zheng, X., Deng, S., Ye, M. & Hu, F. (2020). Ecological role of earthworm intestinal bacteria in terrestrial environments: a review. Science Total Environment, p.140008. https://doi.org/10.1016/j.scitotenv.2020.140008
Wang, H. T., Zhu, D., Li, G., Zheng, F., Ding, J., O’Connor, P. J., Zhu, Y. G., & Xue, X. M. (2019). Effects of arsenic on gut microbiota and its biotransformation genes in earthworm Metaphire sieboldi. Environmental Science and Technology, 53(7), 3841–3849.
Acknowledgements
The authors are grateful to WBDST, DST-FIST, and DST-PURSE for providing instrumental facilities. The authors are thankful to the Head, Department of Zoology, and the authority of The University of Burdwan for giving all sorts of laboratory facilities to conduct this research.
Author information
Authors and Affiliations
Contributions
Sucharita Ghosh: conceptualization, validation, statistical analysis, data curation, methodology, and writing original draft. Soumendranath Chatterjee: study design, conceptualization, data curation, writing, review, editing, visualization, and supervision. Dipanwita Sarkar Paria: conceptualization, writing, review, editing, and visualization.
Corresponding author
Ethics declarations
Ethics approval
Proper approval from Institutional Biosafety Committee was obtained at starting of the research work.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ghosh, S., Sarkar Paria, D. & Chatterjee, S. Comparative Study on Bacterial Population Dynamics of Foregut, Midgut, and Hindgut Content of Perionyx excavatus (Perrier) Isolated from Eco-friendly, Non-hazardous Vermicompost. Appl Biochem Biotechnol 194, 6126–6139 (2022). https://doi.org/10.1007/s12010-022-03970-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-03970-0