Abstract
Earthworms and their casts have been widely used for organic waste degradation and plant growth promotion. The microbial communities that reside in the guts and casts of earthworms markedly influence both applications. In the present study, next-generation sequencing was applied to identify the microbial communities in the guts and casts of three epigeic earthworm species, Eudrilus eugeniae, Perionyx excavatus, and Eisenia fetida, reared under two different feeding conditions. A total of 580 genera belonging to 43 phyla were identified. By comparing bacterial diversity among samples divided into groups based on the earthworm species, sample types, and conditions, the beta diversity analysis supported the impact of the sample type and suggested that there was significant dissimilarity of the bacterial diversity between the gut and cast. Besides, bacterial Phylum compositions within the group were compared. The result showed that the top three high relative frequency phyla found in the casts were the same regardless of earthworm species, while those found in the gut depended on both the condition and earthworm species. Focusing on the cellulolytic and plant growth-promoting bacteria, certain cellulolytic bacteria, Paenibacillus, Comamonas, and Cytophaga, were found only in the cast. Citrobacter and Streptomyces aculeolatus were detected only in the guts of earthworms reared in the bedding containing vegetables and bedding alone, respectively. Besides, Actinomadura and Burkholderia were detected only in the gut of E. eugeniae and E. fetida, respectively. The results proved that the microbial composition was affected by sample type, condition, and earthworm species. In addition, the proportion of these beneficial bacteria was also influenced by these factors. Hence, the information from this study can be used as a guide for selecting earthworm species or their casts for more efficient organic waste decomposition and plant growth promotion.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Earthworms are very important invertebrates that have been used in agriculture, animal feed, and medicine (Grdiša et al. 2013). They are taxonomically classified in the phylum Annelida and can be separated into three groups, anecic, endogeic, and epigeic, based on their ecological and trophic functions. Epigeic worms live in the soil surface and mainly feed on fresh organic matter (Pathma and Sakthivel 2012). They can rapidly convert organic matter into rich humus and have been used for vermicomposting organic, infected biomedical, and industrial wastes (Natarajan and Gajendran 2014).
Eudrilus eugeniae, Perionyx excavatus, and Eisenia fetida have been extensively used in vermicomposting because they have the ability to colonize in organic waste, consume at a high rate, tolerate a wide range of environmental conditions, and have short life cycles and high reproductive rates (Dominguez and Gomez-Brandon 2012). Earthworms can break down plant materials and improve soil fertility via processes that occur in their digestive tracts and continue in their casts (Katheem et al. 2014). Inside the earthworm gut, there is mucus that contains proteins, polysaccharides, organic compounds, amino acids, enzymes, and microorganisms. Bacteria, fungi, protozoa, and yeast have all been detected in the guts and casts of several earthworm species, and many reports indicate that certain microorganisms can increase in number inside the earthworm gut (Parthasarathi et al. 2007). Certain bacteria, such as Serratia, Klebsiella, and Paenibacillus, live in symbiosis with earthworms, producing enzymes for digesting organic compounds, such as cellulose, chitin, and pectin (Parthasarathi et al. 2007; Pathma and Sakthivel 2012).
Cellulose is the most abundant organic waste. It can be digested by cellulase and cellobiase, which convert cellulose into D-glucose. Cellulase can be produced by certain earthworm species, such as Pheretima hilgendorfi, Eisenia fetida, and Eisenia andarei, and by several microorganisms, such as Pseudomonas, Flavobacterium, and Klebsiella (Shweta 2012). Although some earthworm species can produce cellulolytic enzymes by themselves, there is a significant correlation between cellulase activity and microorganisms living in the earthworm gut (Chauhan et al. 2010). In addition, Shweta (2012) reported that the cellulolytic activities involved in vermicomposting mainly come from microbial communities. These findings indicate that microbial communities in the guts of earthworms play a major role in cellulose decomposition.
Besides cellulolytic microorganisms, there are several plant-growth-promoting microbes in the guts and casts of earthworms. These plant growth promoters can produce substances that can inhibit or kill phytopathogens, mineralize inorganic compounds, and fix nitrogen (N2) (Parthasarathi et al. 2007; Pathma and Sakthivel 2012). In addition, microorganisms in earthworm casts can release available macronutrients and micronutrients at a slow rate, and can increase the concentration of phenolic compounds in plants, which helps to lower pest attack, better than the use of inorganic fertilizers (Pathma and Sakthivel 2012). Hence, the use of earthworm casts, which contain beneficial microbes, may help to improve both the quality and yield of plants.
Because the microorganisms in the guts and casts of earthworms have numerous benefits, researchers have been identifying them, using various techniques, including culturing on synthetic media, polymerase-chain-reaction (PCR)-denaturing gradient gel electrophoresis, and single-strand conformation polymorphism analysis (Byzov et al. 2009; Nechitaylo et al. 2010; Huang et al. 2013). However, using these techniques, only a small number of microbes can be examined, with a number of microfloral taxa potentially remaining undiscovered. Interestingly, another alterative, next-generation sequencing has been used to identify the majority of microbiome taxa in several organisms, including in gut samples (Wu et al. 2010; Surat et al. 2016). Using this technique, the majority of the taxa in the microbiotas, including minor and unculturable microfloral taxa, can be detected.
The present study explored the microbial diversity in the guts and casts of three earthworm species, E. eugeniae (the African night crawler), P. excavatus (the blue worm), and E. fetida (the tiger worm), using Illumina sequencing. The microbial communities in the guts and casts of the three earthworm species reared under two different conditions, bedding material only and bedding containing vegetables, were compared. The profiles and proportions of the microbiota, especially cellulolytic microorganisms and plant growth promoters in the guts and casts of the earthworms, were presented in this study.
Materials and methods
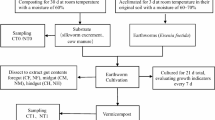
Earthworm culture and DNA extraction
The three earthworm species, E. eugeniae, P. excavatus, and E. fetida, were provided by Associate Prof. Somchai Chantsavang, Department of Animal Science, Faculty of Agriculture, Kasetsart University, Bangkok, Thailand. The earthworm bedding was composed of cow manure and coconut husks in a proportion of 3:1, respectively (Yuvaraj et al. 2019). The 20-kg bedding mixture was soaked with water and subsequently stored in a manure bag in the shade at 29 ± 3 °C for 1 month. Three-hundred grams of each earthworm species was cultured in the 3-kg prepared bedding in separate containers. The earthworm containers (20 cm × 30 cm × 10 cm) were kept under shade at 29 ± 3 °C with 70% moisture. After 2 days, 10 earthworms, both juvenile and adult stage, of each species were surface sterilized with 70% ethanol. The earthworm guts were dissected and kept in sterilized microtubes at − 20 °C. Then, the remaining earthworms were reared in the cow manure and coconut husk mixture, supplemented with 100 g of finely chopped vegetables every 2 days. Fourteen days later, 10 earthworms of each species were sterilized with 70% ethanol. The earthworm guts were dissected and kept in sterilized microtubes at − 20 °C. The bedding and casts of the three earthworm species were collected in sterilized tubes and kept at − 20 °C until analyzed.
DNA was isolated from the guts of the earthworms, both before and after feeding with vegetables, as well as from the bedding and casts, using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Germany), following the manufacturer’s protocol.
16S rRNA amplification
DNA samples from the bedding material, guts, and casts of the earthworms were used to amplify 16S rRNA fragments. A primer pair comprising a forward primer, 341F (CCTACGGGNGGCWGCAG), and a reverse primer, 802R (TACNVGGGTATCTAATC), was used to amplify a partial fragment (V4) of bacterial 16S rRNA gene (Klindworth et al. 2013). The total of 25 µL of PCR reaction was consisting of 2 µL of DNA template; 1 U of HotStar Hifidelity Polymerase (QIAGEN, Germany); 1 × HotStar Hifidelity PCR buffer, containing 1.5 mM MgSO4 and 0.3 mM dNTPs; and 500 nM of each primer. The amplification cycles involved pre-incubation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 1 min, with a final extension step at 72 °C for 5 min.
The PCR product was purified using a MinElute PCR Purification Kit (QIAGEN, Germany). The concentrations of the purified products were determined using a Qubit Fluorometer. Then, the DNA integrity was checked using agarose gel electrophoresis, with the quantified DNA samples being used to construct libraries. Finally, a paired-end-read library was generated, with the sequences being read by the Illumina Miseq platform. The sequencing was carried out according to the manufacturer’s protocol.
Bacterial taxonomy classification
The short-reads cleaned by removing adapters and index sequences were obtained from the BGI sequencing center. The demultiplexed and cleaned paired-end reads with the inserted size of approximately 500 bp of ten samples were imported into QIIME2 (Bolyen et al. 2019). DADA2 (Callahan et al. 2016) was applied for denoising, error-correcting, chimeric removing, singleton removing, and joining the qualified paired-end reads. The dereplicated non-chimeric sequences were applied for phylogenetic and diversity analysis using MAFFT (Katoh and Standley 2013), FastTree2 (Price et al. 2010), and core-metric-phylogenetic method in QIIME2 (Bolyen et al. 2019). The observed OTUs and Shannon’s diversity index were calculated for alpha diversity analysis. The Bray–Curtis distance was calculated for beta diversity analysis. The OTU sequences were classified by comparing them to the pre-trained Greengene database (Bokulich et al. 2018). The relative frequency of bacterial phyla of each sample was presented as a stacked bar graph. The 16S rRNA sequences from the earthworm gut, the bedding, and the cast samples are available in the Sequence Read Archive (SRA) on NCBI with the accession number SAMN20346337-46.
Statistical analysis
For alpha diversity values, the rarefaction plot was generated to present Shannon’s diversity index distribution. Then, the Kruskal–Wallis test was applied to compare bacterial diversity among groups of samples to evaluate whether the factor determining the group would cause differences in bacterial diversity among groups. Finally, the pairwise Kruskal–Wallis analysis was applied to determine which pair of groups was significantly different.
The principal coordinate analysis (PCoA) was applied to Bray–Curtis distances to explore whether the factor defined by the Kruskal–Wallis test generated adequate dissimilarity among groups. The PERMANOVA analysis (Anderson 2001) was further applied to examine whether the within-group distances differed significantly from between-group distances.
The comparison of bacterial richness among groups was visualized by a Venn diagram. The abundance of cellulolytic, nitrogen-fixing, and phosphate-mineralizing bacteria in each sample was compared using gniess (Morton et al. 2017). The dendrogram generated based on the correlation clustering method and the heatmap presenting bacterial abundances were built using gneiss (Morton et al. 2017).
These statistical analyses and graphs were performed using QIIME2 (Bolyen et al. 2019) and R (Team 2020).
Results
Quality of sequence data and within-group bacterial diversity comparison
This study aimed to explore the microbial diversity in the guts and casts of three earthworm species: Eudrilus eugeniae (African nightcrawler), Perionyx excavatus (blue worm), and Eisenia fetida (tiger worm). Ten samples, one bedding control, six gut samples, and three casts, were obtained for this study. As shown in Table 1, the number of paired-end reads per sample was between 99,145 and 196,088 sequences; however, the number of non-chimeric sequences per sample was between 32,061 and 63,766 sequences. The length of non-chimeric sequences was between 291 and 487, with an average of 451.18 bp. The number of observed OTUs was 7439, and the numbers of observed OTUs per sample were between 470 and 1741 (Table 1). Shannon’s diversity index was between 6.240 and 9.929 (Table 1). The samples presenting the lowest and the highest bacterial richness (observed OTUs) were the gut sample of P. excavatus reared in the bedding and the cast of E. fetida, respectively. These samples were the same for bacterial diversity (Shannon’s diversity index). Therefore, Shannon’s diversity index was chosen for within-sample bacterial comparison (Fig. 1). Because the lowest number of cleaned sequences per sample was 32,061, this number was applied to normalize every sample’s sampling depth for bacterial diversity analysis (Fig. 1).
The distribution of bacterial diversity (Shannon’s diversity index) presented as rarefaction curves (a) and the comparison of diversity between groups of samples (b–d). The ten samples were divided into groups based on the species of earthworm (b), type of samples (c), and condition (d). The gut samples (c) were separated based on the feeding substrate, the bedding, and bedding containing vegetables (d). The pairwise Kruskal–Wallis test provided the p-value and q-value (adjusted p-value based on Benjamini and Hochberg correction)
The rarefaction plot (Fig. 1a) showed the saturation of bacterial diversity in each sample. Thus, these normalized data were qualified for bacterial diversity comparison. The ten samples were grouped based on three criteria: the earthworm species (Fig. 1b), the type of samples (Fig. 1c), and the condition of samples (Fig. 1d). Four groups of samples were shown when the samples were divided based on the condition of samples as the gut samples were separated based on feeding conditions. The overall statistical comparison presented a non-significant difference among groups of samples divided by the earthworm species or the condition of samples. The statistically significant difference was shown among groups of samples divided by their types, in which the gut and the cast samples presented the highest difference of average Shannon’s diversity index (Fig. 1c). Even though the statistical value (q-value) showed non-significant, the graph (Fig. 1c) clearly showed that the cast samples carried bacterial diversity higher than the gut samples. Because of the single sample of the bedding control, the diversity difference between the control and gut samples shown in Fig. 1c could not be statistically confirmed. These results suggested that the sample type would be an essential factor determining the bacterial diversity of the samples included in this study.
Comparison of between-group bacterial diversity and bacterial richness
The impact of the sample type on bacterial diversity was further examined by evaluating whether this factor could generate adequate dissimilarity of bacterial diversity between samples belonging to different groups. As shown in Fig. 2a, the three clusters of ten samples corresponded well to classify samples based on their types. The significant difference in the Bray–Curtis distances shown by the PERMANOVA test (Fig. 2b) with an overall p-value of 0.002 indicated that the bacterial composition of samples belonging to the same group was similar to each other than that belonging to different groups. The pairwise comparison further showed that the bacterial composition in the gut sample significantly differed from that in the cast. These results supported Shannon’s diversity index analysis that the sample type influenced the bacterial diversity and suggested that this factor distinguished the gut from the cast samples.
The statistical analyses on Bray–Curtis distance (a and b), the relative frequency of bacterial phyla in each sample (c), and the Venn diagram that presented the comparison of bacterial genera richness among different samples’ types: bedding, gut, and cast (d). The principal coordinate analysis on Bray–Curtis distance (a) and the pairwise PERMANOVA result (b) presented bacterial community dissimilarity among groups of samples separated by samples’ type
All 7439 OTUs were taxonomic classified and 641 species (Table S1) of 580 genera (Fig. 2d) belonging to 43 phyla (Fig. 2c) were identified. Approximately 20.81% of sequences per sample were classified as belonging to the kingdom Bacteria without lower taxonomic level information (Fig. 2c). Every sample’s highest relative frequency phylum was Proteobacteria, and OD1 was the second-high relative frequency phylum of every sample except the gut sample of E. eugeniae collected after feeding with vegetables. The second and third most abundance phyla found in the gut sample of E. eugeniae collected after feeding with vegetables were Fusobacteria and OD-1, respectively. The third most relative frequency phylum was varied among different samples: Actinobacteria for the gut sample of E. eugeniae reared in the bedding material, Firmicutes for the gut sample of E. fetida, and Bacterioidetes for the rest. Therefore, the top three high relative frequency phyla found in the casts were the same regardless of earthworm species, while those phyla found in the gut depended on both the condition and earthworm species. These results corresponded with the relatively high variation of within-group bacterial diversity of the gut samples compared to the variation found in the casts (Fig. 1c).
Both the alpha (Fig. 1c) and beta (Fig. 2a, b) diversity analysis indicated that the bacterial composition in the gut samples differed from that in the casts. Therefore, bacterial richness was compared among groups of samples separated by sample types (Fig. 2d). The 270, 372, and 399 genera were found in the bedding, gut, and cast samples. Among 580 genera, 148 genera, including Flavobacterium, Cellvibrio, Acinetobacter, and Lysobacter, were commonly found in every group, and 165 were found in two out of three groups of samples. Among 267 genera found uniquely in each group, Actinomadura, Arthrobacter, Burkholderia, Cellulomonas, Citrobacter, Flavisobacter, Streptomyces aculeolatus, and Xylanimicrobium were observed only in the guts. In addition, Citrobacter and Streptomyces aculeolatus were detected only in the guts of earthworms reared in the bedding containing vegetables and bedding alone, respectively. Besides, Actinomadura, Burkholderia, and Xylanimicrobium were detected only in the gut of E. eugeniae, E. fetida, and P. excavatus. These results contributed that bacterial community in the gut was influence by the condition and earthworm species.
The abundance of cellulolytic and plant growth-promoting bacteria in the samples
Twenty genera found in samples included in this study have been reported as cellulolytic and plant growth-promoting bacteria. Among these genera, 12 of them were cellulolytic bacteria: Actinomadura, Aeromonas, Bacillus, Cellulomonas, Cellvibrio, Citrobacter, Comamonas, Cytophaga, Erwinia, Lysobacter, Paenibacillus, and Streptomyces. Four genera, Acinetobacter, Brachybacterium, Burkholderia, and Clostridium, were N2-fixing bacteria. Klebsiella and Pseudomonas were cellulolytic bacteria, N2 fixer, and phosphate solubilizer. Flavobacterium was cellulolytic and phosphate-solubilizing bacteria while Rhodobacter was cellulolytic bacteria and N2 fixer.
By comparing the list of cellulolytic and plant growth-promoting bacterial genera among different sample types (Fig. 3a), four genera, Flavobacterium, Cellvibrio, Lysobacter, and Acinetobacter, were found across all types. Bedding samples and the guts contained these genera and Streptomyces. Five genera, Aeromonas, Bacillus, Klebsiella, Pseudomonas, and Rhodobacter, were observed in both gut and cast samples. Six (Actinomadura, Brachybacterium, Burkholderia, Cellulomonas, Citrobacter, and Clostridium) and four (Comamonas, Cytophaga, Erwinia, and Paenibacillus) genera were found only in gut and cast, respectively. These results showed that the guts contained higher number of cellulolytic and plant growth-promoting bacteria than the casts.
The Venn diagram presents the cellulolytic and plant growth-promoting bacterial genera richness among different sample types (a) and the heatmaps presenting abundances of these bacteria in each sample (b). The samples were grouped based on their types: bedding, gut, and cast. The cladogram tips’ labels showed four genera: Flavobacterium, Cellvibrio, Acinetobacter, and Lysobacter, found in all types in grey letters. Streptomyces, commonly found in bedding and gut samples, was shown in green. Aeromonas, Bacillus, Klebsiella, Pseudomonas, and Rhodobacter commonly found in gut and cast were shown in purple. Six genera, Actinomadura, Brachybacterium, Burkholderia, Cellulomonas, Citrobacter, and Clostridium, shown in red, were found only in the gut, and four genera, Comamonas, Cytophaga, Erwinia, and Paenibacillus, shown in blue, were found only in the cast. The red color presented high abundance for the heatmap, while the blue presented low to no abundance
However, the criteria for selecting either the earthworms or their casts for organic waste decomposition or plant-growth promotion should include the abundance of each bacterial genera as shown in Fig. 3b. The number of OTUs belonging to the bacteria genera found in the cast were also relatively high when compared to those found in the gut as shown in red color. However, several genera of these bacteria in the cast were detected at very low amount as shown in dark blue color. Moreover, certain genera, Aeromonas, commonly found in the gut and cast had higher proportion in the gut than that in the cast, especially the gut of E. fetida.
The number of genera and proportion of cellulolytic and plant growth-promoting bacteria varied among all samples. The highest OTUs of cellulolytic bacteria were observed in the gut of E. fetida reared in the bedding containing vegetables (17.06%, 11 genera), followed by that in the cast of E. eugeniae (12.37%, 12 genera), P. excavatus (8.49%, 9 genera), and E. fetida (8.42%, 11 genera) (Table S1). For N2 fixers, the highest proportion of the bacteria was found in the guts of E. fetida reared in the bedding containing vegetables (2.49%), followed by that in the cast of E. fetida (2.04%), P. excavatus (1.72%), and E. eugeniae (1.66%). For phosphate-solubilizing bacteria, the casts contained more phosphate solubilizers than the gut samples. The highest proportion of phosphate-solubilizing bacteria was detected in the cast of P. excavatus (2.79%), followed by the gut of E. fetida reared in the bedding containing vegetables (2.38%) and the cast of E. fetida (2.21%).
Discussion
Epigeic earthworms mainly feed on fresh organic matter and, consequently, have been used for organic waste management and plant growth promotion (Domínguez and Gómez-Brandón 2012). The guts and casts of earthworms contain very high microbial diversity, with many taxa involved in carbon- and N2-cycle processes, micronutrient mineralization, and the inhibition of phytopathogens, arthropod pests, and nematodes (Pathma and Sakthivel 2012). To maximize the efficiency of using earthworms and their casts in waste management and agriculture, identification of the microbes in their guts and casts needs to be undertaken. The three epigeic earthworms examined in this study produced a total of 641 species, belonging to 580 genera belonging to 43 bacterial phyla.
Effect of earthworm species, sample type, and condition on the microbial communities
Several factors, including sample type, feed substrate, and earthworm species, can affect the microbial compositions in the guts and casts of earthworms (Parthasarathi et al. 2007; Nechitaylo et al. 2010; Gómez-Brandón et al. 2012). In this study, the PCoA analysis (Fig. 2a) clearly showed three clusters corresponding to the sample type: the bedding, gut, and cast. Moreover, the statistical analyses supported the difference of bacterial community between the gut and cast. In addition, by comparing the list of bacteria between these two groups, certain microbes were present only in the gut or cast samples. For instance, the genera Arthrobacter, Flavisobacter, and Xylanimicrobium were observed only in the guts while Paenibacillus, Comamonas, and Cytophaga were presented only in the cast. Therefore, it is clear that sample type had an influence on the bacterial communities.
Besides, feed substrate and earthworm species also affected bacterial diversity and composition in the present study. Citrobacter and Streptomyces aculeolatus were detected only in the guts of earthworms reared in the bedding containing vegetables and bedding alone, respectively. Similar to the previous study, different feeding substrates have different effects on such microbial communities, including in E. eugeniae, E. fetida, and P. excavatus (Parthasarathi et al. 2007). In addition, Actinomadura, Burkholderia, and Xylanimicrobium were detected only in the gut of E. eugeniae, E. fetida, and P. excavatus, respectively. This specificity might be due to the selection of bacterial endosymbionts by the gut fluid in the earthworm’s digestive tract (Byzov et al. 2009; Gómez-Brandón et al. 2012). This selection may stimulate or inhibit particular microbes in the digestive tract, thereby shaping the microbial composition in the gut and casts (Monroy et al. 2009; Gomez-Brandon et al. 2012).
Potential of using earthworms and their casts in organic waste management and plant growth promotion
An increase in organic waste, such as leftover vegetable matter, has prompted the need to develop an efficient waste management process (Parthasarathi et al. 2014). Cellulose is a significant component in the plant cell wall, which can be degraded by cellulolytic enzymes (Shweta 2012). Although some earthworms, such as P. hilgendorfi, can degrade cellulose by themselves, earthworm gut microbes still play a significant role in cellulose decomposition (Nozaki et al. 2009; Chauhan et al. 2010). Sixteen bacterial genera, Actinomadura, Aeromonas, Bacillus, Cellulomonas, Cellvibrio, Citrobacter, Comamonas, Cytophaga, Erwinia, Flavobacterium, Klebsiella, Lysobacter, Paenibacillus, Pseudomonas, Rhodobacter, and Streptomyces, found in the guts and casts of the earthworms investigated in this study have been previously reported as cellulolytic bacteria (Wang et al. 2008; Vijayakumar et al. 2009; Shweta, 2012; Bamidele et al. 2014). The cast of E. eugeniae contained the highest cellulolytic genera (12 genera, 12.37%) but the gut of E. fetida reared in the bedding containing vegetables presented the highest proportion of these bacteria (11 genera, 17.06%). The result suggested that E. fetida reared in bedding containing vegetables may have good potential as a cellulose degrader in organic waste management.
Several studies have reported that earthworms can improve plant yields (shoot and root length, flowering, fruiting, and germination rate) and reduce root diseases (Chauhan et al. 2010; Munnoli et al. 2010; Pathma and Sakthivel 2012). Furthermore, the addition of worm casts helps to increase the available nutrients (N, P, K, Ca, and Mg) in soil, as a result of their plant-growth-promoting activities (Parthasarathi et al. 2007). In this study, several N2-fixing and phosphate-solubilizing bacteria were found in the gut and cast samples.
Nitrogen is one of the essential nutrients that can limit plant growth. Shamseldin (2013) showed that several N2 fixers, including Klebsiella, Clostridium, Acetobacter, Azotobacter, Bacillus, Serratia, and Pseudomonas, were isolated from the intestine wall in earthworms (Brito-Vega and Espinosa-Victoria 2009). Seven N2-fixing bacteria, Acinetobacter, Brachybacterium, Burkholderia, Clostridium, Klebsiella, Pseudomonas, and Rhodobacter, were observed in this study. Interestingly, the cast of E. fetida contained the highest number of N2-fixing bacteria (5 genera, 2.04%) while the gut of E. fetida reared in the bedding containing vegetables (3 genera, 2.49%) had the highest proportion of this group of bacteria. Hence, both E. fetida and its cast may have a greater potential to increase the nitrogen content in soil than the other earthworms examined in this study.
After nitrogen, phosphorous (P) is the second key element for promoting root growth and enhancing disease resistance in plants (Sharma et al. 2013). Plants can take up phosphate only in solubilized form, and phosphate-mineralizing microbes markedly influence the available P content in soils (Sharma et al. 2013). The Pseudomonas, Klebsiella, and Flavobacterium found in the present study have been reported as having the potential to solubilize insoluble phosphates (Parthasarathi et al. 2007; Sharma et al. 2013). The highest proportion of the phosphate-mineralizing microbes was detected in the cast of P. excavatus (2.79%). Therefore, the cast of this earthworm species has the highest potential to increase the available P content in soil.
Conclusions
In this study, a total of 580 genera belonging to 43 phyla were identified from the guts and casts of three earthworm species: E. eugeniae, E. fetida, and P. excavatus. The bacterial diversity and composition in the guts and casts of the earthworms was affected by sample type, earthworm species, and feed substrate. Several beneficial microbes, particularly cellulolytic microbes and plant growth promoters, were detected. The highest proportion of cellulolytic and N2-fixing microbes was detected in the gut of E. fetida reared in the bedding containing vegetables; consequently, it should be selected for cellulose degradation and increasing nitrogen in the soil. The highest proportion of phosphate-mineralizing bacteria was observed in the cast of P. excavatus; hence, the use of this earthworm’s cast could efficiently increase the available phosphorus contents in soil.
References
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Bamidele JA, Idowu AB, Ademolu KO, Atayese AO (2014) Microbial diversity and digestive enzyme activities in the gut of earthworms found in sawmill industries in Abeokuta, Nigeria. Rev Biol Trop 62:1241–1249
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Knight R, Huttley GA, Caporaso JG (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. https://doi.org/10.1186/s40168-018-0470-z
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Brito-Vega H, Espinosa-Victoria D (2009) Bacterial diversity in the digestive tract of earthworms (Oligochaeta). J Biol Sci 9:192–199
Byzov BA, Nechitaylo T, Bumazhkin BK, Kurakov AV, Golyshin PN, Zvyagintsev DG (2009) Culturable microorganisms from the earthworm digestive tract. Microbiology 78:360–368
Callahan B, McMurdie P, Rosen M et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Chauhan A, Kumar S, Singh AP, Gupta M (2010) Vermicomposting of vegetable wastes with cowdung using three earthworm species Eisenia foetida, Eudrilus eugeniae and Perionyx excavatus. Nat Sci 8:33–43
Domínguez J, Gómez-Brandón M (2012) Vermicomposting: composting with earthworms to recycle organic wastes In: Dr. Sunil Kumar (Ed) Management of Organic Waste, InTech, London, pp 2–20
Gómez-Brandón M, Lores M, Domĭnguez J (2012) Species-specific effects of epigeic earthworms on microbial community structure during first stages of decomposition of organic matter. PLoS ONE 7:e31895. https://doi.org/10.1371/journal.pone.0031895
Grdiša M, Grsic K, Grdisa M (2013) Earthworms — role in soil fertility to the use in medicine and as a food. Invertebr Surviv J 10:38–45
Huang K, Li F, Wei Y, Chen X, Fu X (2013) Changes of bacterial and fungal community compositions during vermicomposting of vegetable wastes by Eisenia fetida. Bioresour Technol 150:235–241
Katheem KS, Jessy RS, Ibrahim MH (2014) Earthworm’s gut as reactor in vermicomposting process: a mini review. Int J Sci Res Publ 4:1–6
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Monroy F, Aira M, Domínguez J (2009) Reduction of total coliform numbers during vermicomposting is caused by short-term direct effects of earthworms on microorganisms and depend on the dose of application of pig slurry. Sci Total Environ 407:5411–5416
Morton JT, Sanders J, Quinn RA et al (2017) Balance trees reveal microbial niche differentiation. Msystems 2:e00162-e1116. https://doi.org/10.1128/mSystems.00162-16
Munnoli PM, Teixeira da Silva JA, Bhosle S (2010) Dynamics of the soil-earthworm-plant relationship: a review. Dyn Soil Dyn Plant 4:1–21
Natarajan N, Gajendran M (2014) Vermiconversion of paper mill sludge for recycling the nutrients using earthworm Eudrilus eugeniae. IOSR J Environ Sci Toxicol Food Technol 8:6–11
Nechitaylo TY, Yakimov MM, Godinho M, Timmis KN, Belogolova E, Byzov BA, Kurakov AV, Jones DL, Golyshin PN (2010) Effect of the earthworms Lumbricus terrestris and Aporrectodea caliginosa on bacterial diversity in soil. Microb Ecol 59:574–587
Nozaki M, Miura C, Tozawa Y, Miura T (2009) The contribution of endogenous cellulase to the cellulose digestion in the gut of earthworm (Pheretima hilgendorfi: Megascolecidae). Soil Biol Biochem 41:762–769
Parthasarathi K, Jayanthi L, Soniya MA, Sekar J, Basha SA (2014) Efficiency of Perionyx excavatus (perrier) in litter (Anacardium occidium L.) decomposition and nutrient mineralization. Int J Mod Res Rev 2:453–458
Parthasarathi K, Ranganathan LS, Anandi V, Zeyer J (2007) Diversity of microflora in the gut and casts of tropical composting earthworms reared on different substrates. J Environ Biol 28:87–97
Pathma J, Sakthivel N (2012) Microbial diversity of vermicompost bacteria that exhibit useful agricultural traits and waste management potential. Springer plus 1:26. https://doi.org/10.1186/2193-1801-1-26
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. https://doi.org/10.1371/journal.pone.0009490
R Core Team (2020) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: www.R-project.org
Shamseldin A (2013) The role of different genes involved in symbiotic nitrogen fixation — review. Global J Biotechnol Biochem 8:84–94
Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springer plus 2:587. https://doi.org/10.1186/2193-1801-2-587
Shweta M (2012) Cellulolysis: a transient property of earthworm or symbiotic/ingested microorganisms? Int J Sci Res Publ 2:1–8
Surat W, Mhuantong W, Sangsrakru D, Chareonviriyaphap T, Arunyawat U, Kubera A, Sittivicharpinyo T, Siripan O, Pootakham W (2016) Gut bacterial diversity in Plasmodium-infected and Plasmodium-uninfected Anopheles minimus. Chiang Mai J Sci 43:427–440
Vijayakumar B, Shanmugasundaram P, Menaga SV, Sujatha R, Anandhi VA (2009) Study on isolation and characterization of earthworm gut flora of Perionyx excavatus. Biomed Pharmacol J 2:403–406
Wang CM, Shyu CL, Ho SP, Chiou SH (2008) Characterization of a novel thermophilic, cellulose-degrading bacterium Paenibacillus sp. strain B39. Lett App Microbiol 47:46–53
Wu G, Lewis J, Hoffmann C, Chen Y-Y, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, Li H, Bushman FD (2010) Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol 10:206. https://doi.org/10.1186/1471-2180-10-206
Yuvaraj A, Thangaraj R, Maheswaran R (2019) Decomposition of poultry litter through vermicomposting using earthworm Drawida sulcata and its effect on plant growth. Int J Environ Sci Technol 16:7241–7254
Acknowledgements
We would like to thank Assoc. Professor Somchai Chantsavang for providing all earthworm samples and suggesting how to rear and identify these earthworm species.
Funding
This study was financially supported by Kasetsart University Research and Development Institute (KURDI), Kasetsart University, Bangkok, Thailand (ว-ท(ด)112.58).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wonnapinij, P., Sriboonlert, A. & Surat, W. Exploration of microbial communities in the guts and casts of Eudrilus eugeniae, Perionyx excavatus, and Eisenia fetida. Folia Microbiol 67, 329–337 (2022). https://doi.org/10.1007/s12223-022-00948-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-00948-7