Abstract
ω-Transaminase (ω-TA) is an attractive biocatalyst for stereospecific preparation of amino acids and derivatives, but low catalytic efficiency and unfavorable substrate specificity hamper their industrial application. In this work, to obtain applicable (R)-ω-TA responsible for amination of α-keto acids substrates, the reactivities of eight previously synthesized ω-TAs toward pyruvate using (R)-α-methylbenzylamine ((R)-α-MBA) as amine donor were investigated, and Gibberella zeae TA (GzTA) with the highest (R)-TA activity and stereoselectivity was selected as starting scaffold for engineering. Site-directed mutagenesis around enzymatic active pocket and access tunnel identified three positive mutation sites, S214A, F113L, and V60A. Kinetic analysis synchronously with molecular docking revealed that these mutations afforded desirable alleviation of steric hindrance for pyruvate and α-MBA. Furthermore, the constructed single-, double-, and triple-mutant exhibited varying degrees of improved specificities toward bulkier α-keto acids. Using 2-oxo-2-phenylacetic acid (1d) as substrate, the conversion rate of triple-mutant F113L/V60A/S214A increased by 3.8-fold relative to that of wide-type GzTA. This study provided a practical engineering strategy for improving catalytic efficiency and substrate specificity of (R)-ω-TA. The obtained experience shed light on creating more industrial ω-TAs mutants that can accommodate structurally diverse substrates.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amino acids and derivatives are important compounds for synthesis of pharmaceuticals, chemicals, and agricultural products. Amino acids and derivatives can be usually synthesized via chemical and enzymatic approaches, including asymmetric synthesis, deracemization, fractional crystallization, and kinetic resolution [1]. Among them, asymmetric synthesis approach employing transaminase (TA) is considered as an attractive choice, due to efficient one-step transformation, stringent stereoselectivity, and no requirement for cofactor regeneration [2].
TA (EC 2.6.1.X) is pyridoxal-5’-phosphate (PLP)-dependent enzyme that catalyzes the transfer of amine groups between amine donors and acceptors. TAs are grouped into α-TAs and ω-TAs. Compared with α-TAs, ω-TAs enable to transfer amine groups that are located away from carboxylic moiety; thus, they are more applicable to aminate structurally diverse substrates (e.g., keto acids, aldehydes, and ketones) [3]. ω-TAs have either (R)- or (S)-enantiopreference. There has been a series of literature reported regarding screening, identification, and application of (S)-ω-TAs, such as enzymes from Klebsiella pneumonia JS2F [4], Vibrio fluvialis JS17 [5], Alcaligenes denitrificans Y2k-2 [6], Bacillus megaterium SC6394 [7], Moraxella lacunata WZ34 [8], Rhodobacter sphaeroides [9], and Mesorhizobium sp. LUK [10]. The commercial ω-TAs are usually (S)-enantiopreference [3]. In contrast, the search for (R)-ω-TAs has been a major objective. Arthrobacter sp. KNK168 was the first reported (R)-TA with ability to aminate 3,4-dimethoxyphenylacetone to yield 3,4-dimethoxyamphetamine with 99% e.e [11]. In order to enrich (R)-TAs library, Höhne et al. [12] revealed the structural characteristics and key residues of (R)-ω-TAs and developed an in silico strategy to identify twenty-one potential (R)-ω-TAs from NCBI database. Jiang et al. [13] and Iglesias et al. [14] exploited a simplified procedure, using a short motif sequence blast method, to identify other different (R)-ω-TAs. Nonetheless, the requirement for applicable (R)-ω-TAs remains urgent with aim to expand the application for asymmetric synthesis of valuable amino acids and derivatives.
For natural ω-TAs, low catalytic efficiency and unfavorable substrate specificity hamper their practical application [15]. Taking α-keto acids as substrate, (R)-ω-TAs are usually more accessible to accept a range of small α-keto acids, such as pyruvate, glyoxylate and 2-oxobutyric acid [15], whereas showed little or no reactivities toward α-keto acids bearing larger substituents. For example, Aspergillus terreus and Aspergillus fumigatus TAs showed inert activities toward 4-methyl-2-oxopentanoate, phenylglyoxylate and α-ketoglutarate [16]. Arthrobacter sp. KNK168 TA showed no detectable activity toward 3-phenylpyruvic acid [17]. Protein engineering has been successfully applied to overcome enzymatic biochemical limitations. As compared to abundant literature regarding engineering (S)-ω-TAs via reconstructing either pocket or tunnel [18,19,20], the instances regarding molecular modification of (R)-ω-TA are very few, the famous model relates to Arthrobacter sp. KNK168 that was successfully engineered to create a twenty-seven amino acids substitutions mutant by Codexis®, designating (R)-ATA117, with good ability to convert 200 g/L prositagliptin ketone to sitagliptin with 92% yield and > 99.9% ee value [21]. To date, the improvement of (R)-ω-TA for converting bulkier α-keto acids still need to be lucubrated.
In this work, the reactivities of eight previously synthesized ω-TAs toward pyruvate were firstly investigated using (R)-α-methylbenzylamine ((R)-α-MBA) as amine donor. Then, the candidate enzyme was subjected to molecular modification to improve the catalysis performance. Finally, the enzymatic properties including original enzyme and mutants were systematically investigated, and the obtained mutants exhibited better specificity toward different α-keto acids particularly those bearing bulkier substituents. The discovered engineering regularity provided reference for creating more industrially important (R)-ω-TAs that can accommodate structurally diverse substrates.
Material and Methods
Materials and Strains
The ω-TAs used in this study were previously synthesized and preserved in our lab (Table S1); their GenBank accession numbers were shown in the “Results” section. Pyruvate, PLP, (L)- and (D)-Ala were purchased from J&K Scientific Ltd. (Beijing, China). O-phthalaldehyde (OPA) and N-acetyl-L-cysteine (NAC) were purchased from Sangon Biotech (Shanghai, China). α-MBA, acetophenone, and different α-keto acids were purchased from Aladdin Reagent Inc. (Shanghai, China). All other reagents were of analytical grade unless otherwise stated.
Expression of Recombinant ω-TAs
Eight ω-TAs fused with 6 × His-tag at C-terminus have been previously ligated into pET28b( +) vector and transformed into E. coli BL21 (DE3) for expression. The fermentation was performed in Luria–Bertani (LB) medium containing 50 μg/mL kanamycin [22]. The recombinant strain was cultivated at 37 °C for 12 h and transferred to 100 mL fresh LB medium supplemented with 50 μg/mL kanamycin. The cultivation continued to be performed at 37 °C until OD600 reaching 0.6, the fermentation temperature decreased to 28 °C and 1 mM β-D-1-thiogalactopyranoside (IPTG) was added into the medium, the induction process lasted for 12 h, and the cells were harvested at 8000 × g for 10 min and stored at 4 °C.
Enzymatic Activity Assay
The enzymatic reaction system was composed of 10 mM pyruvate, 20 mM (R)-α-MBA, 0.5 mM PLP, an appropriate amount of biocatalyst and 100 mM Na2HPO4-NaH2PO4 buffer (pH 7.5) in a total volume of 10 mL. The reaction was performed at 30 °C for 30 min and stopped by addition of 800 μL of 50% (v/v) acetonitrile in water. Acetophenone formed in the reaction mixture was subjected to high-performance liquid chromatography (HPLC) analysis. One unit (U) of ω-TA activity was defined as the amount of biocatalyst required to produce 1 μmol acetophenone per min under the assay condition described above.
Analytical Methods
The amount of acetophenone was performed with an Agilent HPLC system (Agilent, US) and an Agilent 2414 ultraviolet detector (Agilent, US). The analytical column was Eclipse XDB-C18 column (250 × 4.6 mm, 5 μm) (Agilent, US). The mobile phase was 50% (v/v) acetonitrile in water. The column temperature was set at 30 °C, the flow rate was maintained at 1.0 mL/min, and UV detection was carried out at 205 nm. The retention time of acetophenone was 5.9 min.
For quantitative chiral analysis of (L)- and (D)-Ala using HPLC, the derivatization reagent was firstly prepared as follow: 10 mL absolute ethanol solution containing 15 mM OPA and NAC, was diluted to 50 mL using 0.1 M borate buffer (pH 9.8). The samples and derivatization reagent were blended according to volume ratio of 1:2 and reacted at 30 °C for 15 min. Then, the derivatization products were detected by an Agilent HPLC system (Agilent, US) equipped with an Agilent 2414 fluorescence detector (Agilent, US). The analytical column was a C18 column (250 × 4.6 mm, 5 μm) (Agilent, US). The mobile phase was comprised of 50 mM sodium acetate and methanol in a volume ratio of 9:1. The column temperature was set at 35 °C, and the flow rate was 1 mL/min. The excitation and emission wavelengths were 360 and 405 nm, respectively. The retention times of derivatization products of (L)- and (D)-Ala were 14.6 and 13.1 min, respectively.
Enantiomeric excess (ee) was calculated using equation: \(ee \left(\%\right)=\frac{{\mathrm{P}}_{D}-{\mathrm{P}}_{L}}{{\mathrm{P}}_{D}+{\mathrm{P}}_{L}}\times 100\), where PL and PD were the mole fractions of generated (L)- and (D)-alanine during the enzymatic reaction.
Homology Modeling and Docking Simulation
The homology model of Gibberella zeae TA (GzTA) was built using SWISS-MODEL based on the crystal structure of Nectria haematococca TA (PDB ID: 4CMD) [23], which possessed higher identity (76.6%), Global Model Quality Estimation (GMQE, 0.94) and Quaternary Structure Quality Estimation (QSQE, 0.96). Model geometry and quality were checked with web servers molprobity (http://molprobity.biochem.duke.edu/). The molecule pyruvate-quinonoid intermediate was optimized by applying the force field CHARMm. Molecular docking was performed using AutoDock 4.2. The molecular docking poses were ranked according to score functions which were used to predict their binding affinities and conformations of the molecules at the active site of enzymes [24]. Putative substrate access tunnels and binding sites were analyzed using CAVER 3.0 [25]. Structure analysis of the generated model was done by PyMOL software.
Site-Directed Mutagenesis
The pET28b/GzTA plasmid was isolated using the AxyPrep Plasmid Miniprep Kit. Each substituted mutant was constructed using QuickChange site-directed mutagenesis kit (Jiangsu, China) with pET28b/GzTA as template. For the construction of combined mutants, the product of the first mutagenesis PCR was applied as the template for the next round of PCR. The primers in this experiment were listed in Table 1. The PCR products were treated with Dpn I (Thermo Fisher Scientific, US) followed by transformed to E. coli BL21 (DE3). The mutations were confirmed by DNA sequencing.
Purification of Enzymes
According to Jia et al. [26], the aforementioned cells were resuspended in 10 mL of 100 mM phosphate buffer (pH 7.5) and sonicated on ice for 10 min (70 W, 1 s working, 1 s resting), the soluble supernatant via centrifugation, the obtained supernatants were loaded onto a 10 mL Nickel-NTA superflow column pre-equilibrated with binding buffer (20 mM NaH2PO4, 300 mM NaCl, pH 8.0). After removing non-specifically bound protein, the column was linearly eluted with a 10–500 mM imidazole gradient by combining binding buffer and elution buffer (20 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0). All the operations were conducted at 4 °C. After dialysis against 20 mM Na2HPO4/NaH2PO4 buffer (pH 7.5), purified enzymes were applied for biological characteristics analysis.
Enzymatic Kinetics Analysis
The kinetic constants of purified enzymes toward pyruvate and α-MBA were determined using the enzymatic activity assay but a varied substrate or donor concentration. Specifically, the measurement of the constants toward pyruvate was performed with its concentration ranging from 1 to 20 mM and α-MBA at a fixed concentration of 20 mM. On the contrary, the constants measurement for α-MBA was conducted with its concentration ranging from 1 to 20 mM and pyruvate at a fixed concentration of 20 mM. The Km and Vmax values were calculated through a nonlinear curve fit by fitting with Michaelis–Menten plot using OriginPro 9.0.
Enzymatic Biological Characteristics
The effect of reaction temperature on enzymatic activity was studied using the standard activity assay expect for using temperatures ranging from 33 to 57 °C. The effect of pH on enzymatic activity was investigated using the standard activity assay in 100 mM HAc-NaAc buffer (pH 5.0–6.0), 100 mM Na2HPO4-NaH2PO4 buffer (pH 6.0–8.0) or 100 mM Tris–HCl buffer (pH 8.0–9.0). The thermostability of the purified enzyme was investigated by incubation at 35 °C and pH 8.0 for 160 min. Samples were withdrew at a certain time intervals and their activities were measured using the standard activity assay method.
Substrate Specificity
To investigate the substrate spectrum, the activities of GzTA and its mutants against selected substrates 1a-1f were determined using the standard activity assay method. The substrate concentration was controlled at 10 mM. The activity of substrate 1a was taken as 100%.
Converting Utility of Recombinant Enzymes
The catalysis efficiency of wide-type GzTA and mutant F113L/V60A/S214A was performed by the measurement of conversion rate. The reaction system was composed of 50 mM 1d, 200 mM α-MBA, 2 mM PLP, 25 g/L wet cells containing recombinant enzyme, and appropriate amount of 100 mM sodium phosphate buffer (pH 7.0) in a total volume of 10 mL. The reaction was conducted at 35 °C for 3 h, and the conversion of substrate 1d was determined by HPLC.
Statistical Analysis
Unless specifically noted, all experiments in this study were performed in triplicate. Analysis of variance was performed using the SAS program 8.1. Least significant difference was computed at p < 0.05. All of the figures in this study were drawn using the OriginPro 9.0 software.
Results
Screening the Optimum (R)-ω-TA for Research
α-Ketonic acids are the important sources for synthesis of amino acids and derivatives. To obtain the optimum (R)-ω-TA acting on α-ketonic acids, the performance of eight previously synthesized ω-TAs was investigated in terms of amination of pyruvate using (R)-α-MBA as amine donor. As shown in Table 2, six recombinant strains respectively containing HhTA, AoTA, FfTA, MvTA, GzTA, and HnTA showed certain degrees of (R)-selective activities. Among them, GzTA possessed the highest activity of 894.8 U/g and e.e. value of > 99%. Therefore, GzTA was selected for the subsequent study.
(L)-Ala Scanning Mutagenesis
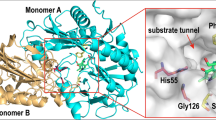
The homology model of GzTA was simulated using Swiss-Model server based on the crystal structure of Nectria haematococca TA (PDB ID: 4CMD) [27], due to its relatively high identity (76.6%), GMQE (0.94) and QSQE (0.96) to GzTA. The built model was deemed reliable providing an accurate evaluation of the target structures. In addition, the model structure of GzTA docking with pyruvate-quinonoid intermediate was exhibited in Fig. 1. The active site residues of all ω-TAs derive from two adjacent subunits, and the minimal actively catalysis unit is a homodimer [28]. The docking model identified 18 active site residues that participated in the formation of the active-site surface and entry tunnel. Furthermore, sequences alignment analysis between GzTA and aforementioned (R)-ATA117 (PDB ID 5FR9, 39.6% amino acid homology with GzTA) revealed that active-site residues including H74, R77, W147, N180, Q182, G213, F216, N217, L234, G236, I237, C272, and G276 were conserved (Fig. S1) and excluded from analysis. The remainder five residues including V60, F113, F186, S214, and T273 were selected, with three residues (F186, S214, T273) laying in the active pocket and two neighboring residues (F113, V60) locating at the entrance of access tunnel. To get a better understanding about their function, these five residues were individually substituted with (L)-Ala which has no functional side chain using site-directed mutagenesis. The activities of the resultant mutants toward pyruvate were shown in Fig. 2. Probably ascribed to contribution of relieved steric hindrance in the active pocket and tunnel, mutants V60A and S214A showed 186% and 231% of activity relative to that of wild-type GzTA. Besides, mutants F113A, F186A and T273A exhibited decreased activities, in particular, mutant F113A showed merely 24% that of the wild-type GzTA.
The modeled structure of GzTA docking with pyruvate-quinonoid intermediate. (A) enlarged view, (B) close view. Monomer A and B were shown in orange and cyan, respectively. Pyruvate-quinonoid intermediate was exhibited in violet, K179 and R126 were presented in yelloworange, and the selected amino acids were highlighted in greencyan color
Site-Directed Mutagenesis and Combinatorial Mutagenesis
Compared to V60, F113 locating at the border of the access tunnel might have more important function than V60. In this point of view, F113 was site-directed mutated to eleven amino acids carrying diverse side chains according to Midelfort et al. [29], their activities were measured following the standard activity assay. As shown in Fig. 2, when Phe113 was mutated to Cys, Asn, Asp, Leu, Ile, and His, the activities of mutants were obviously increased. In particular, the activity of F113L exhibited 259% that of wild-type GzTA. It indicated that substitution of the phenyl group of F113 with correspondingly smaller side chains was suitable for improving activity, but the selected residue was not verified to be the smaller the better [19].
Combinatorial mutagenesis was further conducted based on stacking the positive single-mutants V60A, F113L and S214A. As shown in Fig. 2, double-mutants F113L/V60A, F113L/S214A and V60A/S214A showed about 214%, 151%, and 30% of activity of wild-type GzTA, respectively. The triple-mutant F113L/V60A/S214A showed an activity of 116% that of wild-type GzTA. It was observed that on one hand the stacking effect was negatively influenced once deletion of F113L in the combinatorial mutagenesis, and on the other hand the activities of all combinatorial mutants were lower than that of mutant F113L. This result was not analogous to the viewpoint that overlaying the positive single-mutant sites could synergistically improve activity [30]. One reasonable explanation was that introduction of redundant neutral (L)-Ala residues into F113L mutant seemed to have severely impact on enzymatic hydrophobicity and further affected the enzymatic activity, since extra space might be occupied by water molecules and thus the diffusion of hydrophobic substrates was disfavored [31].
Kinetics Analysis of Wide-Type GzTA and Mutants
For the reaction catalyzed by ω-TA, both the effect of substrate and donor on the catalysis performance should be considered together resulting from structural changes induced by mutation. Wild-type GzTA and mutants were purified to electrophoretic homogeneity (Fig. S2), their kinetic parameters were determined at 30 °C (Table 3). For pyruvate substrate and α-MBA donor, the Km values of F113L, F113L/V60A and F113L/V60A/S214A mutants were increased respectively as compared to those of wide-type GzTA, indicating that the affinities of all the mutants toward both pyruvate and α-MBA were decreased. Crucially, all the mutants displayed obviously improvement of turnover numbers (kcat) toward pyruvate and α-MBA as compared to those of wide-type GzTA. Finally, due to kcat values of double- and triple-mutant exhibited larger increase than their corresponding Km values, thereby higher catalytic efficiency (kcat/Km) values toward pyruvate and α-MBA were observed for all the mutants, as compared to those of wide-type GzTA.
Enzymatic Properties of Wide-Type Enzyme and Mutants
The effect of temperature on the activity of these enzymes was analyzed over a temperature range between 33 and 57 °C (Fig. 3A). Wild-type enzyme exhibited a maximum activity at 37 °C. No changes in optimum temperature of mutants F113L and F113L/V60A were observed. However, mutant F113L/V60A/S214A showed a downward shift in temperature optimal to be around 35℃. Interestingly, modeling analysis of the intramolecular interactions revealed that mutant F113L/V60A/S214A lacked a hydrogen bond between residue 214 and 186 (Fig. S3). It was generally accepted that the formed hydrogen bonds in the active pocket are highly associated with enzyme stability [32], and fewer hydrogen bonds might cause the reduction in optimum reaction temperature of triple-mutant. The effect of pH on the activity of these enzymes was explored in the pH range between 5.0 and 9.0 (Fig. 3B). The optimum pH of GzTA was confirmed at around pH 8.0, and no substantial changes in optimum pH were observed in the mutants. This optimum pH was similar to most of reported TAs, such as optimum pH 8.5 for ATA-117 [21], pH 8.0 for Ruegeria sp. TA [20] and Aspergillus terreus TA [32]. To evaluate the effect in thermostability, the remaining activities of GzTA and mutants were measured after incubation at 35 °C and pH 8.0 for different times (Fig. 3C), wild-type enzyme and mutants showed similar remaining activities after temperature exposure, retained more than 53.8% of the initial activity after incubation for 20 min, and reserved 19.2% of the initial activity with the incubation time extending to 160 min.
Enzymatic characteristics of purified enzymes. (A) Effect of temperature on activity, (B) effect of pH on activity, (C) effect of temperature on thermostability. In the first two experiments, the maximum activities of each enzyme were defined as 100%, respectively. In the thermostability experiment, original activity of each enzyme was taken as 100%, respectively
Substrate Specificity
To determine whether wild-type GzTA and mutants could act on bulky α-keto acids substrates, the activities of these enzymes toward six different α-keto acids (1a-f) were analyzed using α-MBA as amine donor. As shown in Table 4, wild-type GzTA showed the highest activity using pyruvate 1a as substrate, and the activity was obviously decreased toward α-keto acid substrates (1b-f) carrying larger side chain. Since the steric hindrance in the tunnel has been released, mutants F113L and F113L/V60A exhibited approximate 1.2- to 5.8-fold activity improvement toward substrates 1b-f in comparison with usage of wild-type GzTA. Further expansion of the pocket, triple-mutant F113L/V60A/S214A afforded about 1.1- to 1.9-fold activity increase toward substrate 1b-f, in comparison with that using double-mutant F113L/V60A.
Amination Utility Toward Bulkier α-Keto Acids
To exploit the effectiveness of relaxed steric constraint of the mutant, the triple-mutant F113L/V60A/S214A was applied for asymmetric amination of substrate 1d, with wide-type GzTA as control. After converting 50 mM 1d for 2 h, as shown in Fig. 4, mutant F113L/V60A/S214A with higher activity for bulkier substrate afforded a high conversion of 25%, in contrast, low activity of wide-type GzTA toward substrate 1d led to only 6.5% conversion after the same reaction time. The obtained effect provided an outset for in-depth research for enzymatic application for amination of bulky α-keto acids substrates.
Discussion
ω-TAs have been increasingly considered as efficient biocatalysts ascribing to their ability to synthesize a wide range of enantiomerically pure amino acids and derivatives. As compared to (S)-TAs, scarce (R)-ω-TAs have attracted more attention [33]. In this work, six of in-house ω-TAs exhibited certain degrees of (R)-selective activities, and GzTA possessed the highest activity.
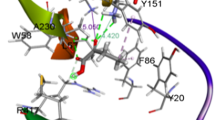
For natural ω-TAs, severe steric constraint in the tunnel or pockets restricts their application [15]. Herein, protein engineering policy was applied to improve enzymatic catalysis performance. When V60 at the entrance tunnel was replaced by A60, the activity toward pyruvate increased by 1.9-fold as compared to that of wild-type GzTA. Figure 5A and B depicted that although residue 60 (either V or A) would generate two hydrogen bonds with D59 and T274 respectively, the mutant V60A could provide a more accessible tunnel to diffuse reactants, since the diameter of the tunnel was increased from 8.7 Å of the original enzyme to 9.4 Å of the mutant V60A. For residue 113, there is literature reporting that the residues at the rim of tunnel are usually hydrophobic [34, 35], such that mutation of F113 to A113 decreased the activity to a very low level. Substitution F113 with L113 not only maintained hydrophobicity at the entrance of tunnel but also created an increasement of 1.4 Å in the tunnel diameter (Fig. 5C, D); thus, the mutant with a 2.6-fold improvement in activity was obtained. For residue 214, the reduction in the size of S214 to A214 increased the activity by 2.3-fold. Molecular modeling suggested that the side chain of S214 located at the dorsal part of substrate, substitution with A214 resulted in the distance between K179 and residue 214 increasing from 8.4 Å of wide-type GzTA to 8.9 Å of the mutant (Fig. 5E, F).
Molecular docking models of GzTA and mutants. (A, C, E) wild-type GzTA. (B) V60A, (D) F113L, (F) S214A. Pyruvate-quinonoid intermediate presented in violet, the selected amino acid residues were all highlighted in chromatic color. The distance between two residues denoted red dotted line, and the hydrogen bond between two residues were presented as black dotted line. The residue with asterisk belonged to the other monomer of dimer
The ω-TA reaction mechanism mainly includes two steps, oxidative deamination from amine donors to PLP generating PMP, and reductive amination of amine acceptors to form products and thereby regenerate PLP [36]. Previous researchers described that the eliminating enzymatic steric hindrance in the active pocket contributed to better activity and turnover numbers (kcat) [37]. Herein, GzTA as well as its mutants were allowed to determine the kinetic parameters in order to confirm which reaction step was benefited from the alleviation of enzymatic steric hindrance. As shown in Table 3, both the kcat and kcat/Km values of mutant F113L were increased as compared to those of wide-type GzTA, suggesting that both the efficiencies of oxidative deamination and reductive amination processes were enhanced. However, for specific substrate and donor, excessive expanding the space might result in partial water molecules entering into the pocket, which might negatively affect kcat values of single and double mutants toward pyruvate and α-MBA, in comparison with that of mutant F113L [38].
The application capability of GzTA and its mutants for the acceptance of bulkier α-ketonic acids was further investigated. As compared to the reported (R)-ω-TAs, such as Aspergillus terreus [32] and Aspergillus fumigatus [16], GzTA showed broader activities toward different α-ketonic acids. Although the double- and triple-mutants ameliorate activities toward substrates 1b to 1f, the activities were still less than 12.3% of that using pyruvate as substrate. During the following converting experiment, the triple-mutant F113L/V60A/S214 showed a 3.9-fold improvement in amination ability toward substrate 1d, compared with usage of wide-type GzTA. Dourado et al. [39] employed a rational design strategy to obtain a mutant W57F/R88H/V153S/K163F/I259M/R415A/V422A, increased the reaction rate by 1,716-fold toward bulky ketone for producing (S)-amine. Oue et al. [40] redesigned a mutant enzyme with seventeen amino acid substitutions and showed over 2,100,000-fold increase in the catalytic efficiency for non-native substrate (L)-Val. Therefore, this research paved an upstanding way to further improve the practical application of GzTA for asymmetric amination of specific bulky α-ketonic acids.
Conclusion
The present research described identification and characterization of a high activity (R)-ω-TA from G. zeae, and succeeded in creating single-, double- and triple-mutant that showed better specificity and higher efficiency toward bulkier α-keto acids. The obtained original enzymes and mutants were a valuable supplement to the scarce (R)-TAs library, and the gained knowledge regarding the effect of mutations on the catalytic efficiency and substrate specificity were useful for further improvement of more industrially important (R)-ω-TAs for synthesis of more valuable amine products.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Koszelewski, D., Lavandera, I., Clay, D., Rozzell, D., & Kroutil, W. (2008). Asymmetric synthesis of optically pure pharmacologically relevant amines employing ω-transaminases. Advanced Synthesis & Catalysis, 350, 2761–2766. https://doi.org/10.1002/adsc.200800496
Guo, F., & Berglund, P. (2017). Transaminase biocatalysis: Optimization and application. Green Chemistry, 19, 333–360. https://doi.org/10.1039/C6GC02328B
Mathew, S., & Yun, H. (2012). ω-Transaminases for the production of optically pure amines and unnatural amino acids. ACS Catalysis, 2, 993–1001. https://doi.org/10.1021/cs300116n
Shin, J. S., & Kim, B. G. (1997). Kinetic resolution of α-methylbenzylamine with ω-transaminase screened from soil microorganisms: Application of a biphasic system to overcome product inhibition. Biotechnology and Bioengineering, 55, 348–358. https://doi.org/10.1002/(SICI)1097-0290(19970720)55:2%3c348::AID-BIT12%3e3.0.CO;2-D
Shin, J. S., Yun, H., Jang, J. W., Park, I., & Kim, B. G. (2003). Purification, characterization, and molecular cloning of a novel amine:Pyruvate transaminase from Vibrio fluvialis JS17. Applied Microbiology and Biotechnology, 61, 463–471. https://doi.org/10.1007/s00253-003-1250-6
Yun, H., Lim, S., Cho, B. K., & Kim, B. G. (2004). ω-Amino acid:Pyruvate transaminase from Alcaligenes denitrificans Y2k–2: A new catalyst for kinetic resolution of β-Amino acids and amines. Applied and Environment Microbiology, 70, 2529–2534. https://doi.org/10.1128/AEM.70.4.2529-2534.2004
Hanson, R. L., Davis, B. L., Chen, Y. J., Goldberg, S. L., Parker, W. L., Tully, T. P., Montana, M. A., & Patel, R. N. (2008). Preparation of (R)-amines from racemic amines with an (S)-amine transaminase from Bacillus megaterium. Advanced Synthesis & Catalysis, 350, 1367–1375. https://doi.org/10.1002/adsc.200800084
Chen, D. Z., Wang, Z., Zhang, Y. J., Sun, Z. Y., & Zhu, Q. (2008). An amine: Hydroxyacetone aminotransferase from Moraxella lacunata WZ34 for alaninol synthesis. Bioprocess and Biosystems Engineering, 31, 283–289. https://doi.org/10.1007/s00449-007-0158-4
Schätzle, S., Höhne, M., Redestad, E., Robins, K., & Bornscheuer, U. T. (2009). Rapid and sensitive kinetic assay for characterization of ω-transaminases. Analytical Chemistry, 81, 8244–8248. https://doi.org/10.1021/ac901640q
Kim, J., Kyung, D. H., Yun, H. D., Cho, B. K., & Kim, B. G. (2006). Screening and purification of a novel transaminase catalyzing the transamination of aryl beta-amino acid from Mesorhizobium sp. LUK. Journal of Microbiology and Biotechnology, 16, 1832–1836. https://kmbase.medric.or.kr/KMID/0545120060160111832.
Iwasaki, A., Yamada, Y., Kizaki, N., Ikenaka, Y., & Hasegawa, J. (2006). Microbial synthesis of chiral amines by (R)-specific transamination with Arthrobacter sp. KNK168. Applied Microbiology and Biotechnology, 69, 499–505. https://doi.org/10.1007/s00253-005-0002-1
Höhne, M., Schätzle, S., Jochens, H., Robins, K., & Bornscheuer, U. T. (2010). Rational assignment of key motifs for function guides in silico enzyme identification. Nature Chemical Biology, 6, 807–813. https://doi.org/10.1038/nchembio.447
Jiang, J. J., Chen, X., Zhang, D. L., Wu, Q. Q., & Zhu, D. M. (2015). Characterization of (R)-selective amine transaminases identified by in silico motif sequence blast. Applied Microbiology and Biotechnology, 99, 2613–2621. https://doi.org/10.1007/s00253-014-6056-1
Iglesias, C., Panizza, P., & Rodriguez Giordano, S. R. (2017). Identification, expression and characterization of an R-ω-transaminase from Capronia semiimmersa. Applied Microbiology and Biotechnology, 101, 5677–5687. https://doi.org/10.1007/s00253-017-8309-2
Genz, M., Vickers, C., Bergh, T. V. D., Joosten, H. J., Dörr, M., Höhne, M., & Bornscheuer, U. T. (2015). Alteration of the donor/acceptor spectrum of the (S)-amine transaminase from Vibrio fluvialis. International Journal of Molecular Sciences, 16, 26953–26963. https://doi.org/10.3390/ijms161126007
Park, E. S., Dong, J. Y., & Shin, J. S. (2014). Active site model of (R)-selective ω-transaminase and its application to the production of D-amino acids. Applied Microbiology and Biotechnology, 98, 651–660. https://doi.org/10.1007/s00253-013-4846-5
Iwasaki, A., Matsumoto, K., Hasegawa, J., & Yasohara, Y. (2012). A novel transaminase, (R)-amine: Pyruvate aminotransferase, from Arthrobacter sp. KNK168 (FERM BP-5228): Purification, characterization, and gene cloning. Applied Microbiology and Biotechnology, 93, 1563–1573. https://doi.org/10.1007/s00253-011-3580-0
Pavlidis, I. V., Weiß, M. S., Genz, M., Spurr, P., Hanlon, S. P., Wirz, B., Iding, H., & Bornscheuer, U. T. (2016). Identification of (S)-selective transaminases for the asymmetric synthesis of bulky chiral amines. Nature Chemistry, 8, 1076–1082. https://doi.org/10.1038/nchem.2578
Nobili, A., Steffen-Munsberg, F., Kohls, H., Trentin, I., Schulzke, C., Höhne, M., & Bornscheuer, U. T. (2015). Engineering the active site of the amine transaminase from Vibrio fluvialis for the asymmetric synthesis of aryl-alkyl amines and amino alcohols. ChemCatChem, 7, 757–760. https://doi.org/10.1002/cctc.201403010
Weiß, M. S., Pavlidis, I. V., Spurr, P., Hanlon, S. P., Wirz, B., Iding, H., & Bornscheuer, U. T. (2016). Protein-engineering of an amine transaminase for the stereoselective synthesis of a pharmaceutically relevant bicyclic amine. Organic & Biomolecular Chemistry, 14, 10249–10254. https://doi.org/10.1039/C6OB02139E
Savile, C. K., Janey, J. M., Mundorff, E. C., Moore, J. C., Tam, S., Jarvis, W. R., Colbeck, J. C., Krebber, A., Fleitz, F. J., Brands, J., Devine, P. N., Huisman, G. W., & Hughes, G. J. (2010). Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science, 329, 305–309. https://doi.org/10.1126/science.1188934
Jia, D. X., Zhou, L., & Zheng, Y. G. (2017). Properties of a novel thermostable glucose isomerase mined from Thermus oshimai and its application to preparation of high fructose corn syrup. Enyzme and Microbial Technology, 99, 1–8. https://doi.org/10.1016/j.enzmictec.2017.01.001
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., Kiefer, F., Cassarino, T. G., Bertoni, M., Bordoli, L., & Schwede, T. (2014). SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42, 252–258. https://doi.org/10.1093/nar/gku340
Zhang, D. L., Chen, X., Chi, J., Feng, J. H., Wu, Q. Q., & Zhu, D. M. (2015). Semi-rational engineering a carbonyl reductase for the enantioselective reduction of β-amino ketones. ACS Catalysis, 5, 2452–2457. https://doi.org/10.1021/acscatal.5b00226
Chovancova, E., Pavelka, A., Benes, P., Strnad, O., Brezovsky, J., Kozlikova, B., Gora, A., Sustr, V., Klvana, M., Medek, P., Biedermannova, L., Sochor, J., & Damborsky, J. (2012). CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Computational Biology, 8, e1002708–e1002708. https://doi.org/10.1371/journal.pcbi.1002708
Jia, D. X., Liu, Z. J., Xu, H. P., Li, J. L., Li, J. J., Jin, L. Q., Cheng, F., Liu, Z. Q., Xue, Y. P., & Zheng, Y. G. (2019). Asymmetric synthesis of L-phosphinothricin using thermostable alpha-transaminase mined from Citrobacter koseri. Journal of Biotechnology, 302, 10–17. https://doi.org/10.1016/j.jbiotec.2019.06.008
Tian, Y., Gao, Y., Chen, Y., Liu, G., & Ju, X. (2019). Identification of the fipronil resistance associated mutations in Nilaparvata lugens GABA receptors by molecular modeling. Molecules, 24, 4116. https://doi.org/10.3390/molecules24224116
Bezsudnova, E. Y., Popov, V. O., & Boyko, K. M. (2020). Structural insight into the substrate specificity of PLP fold type IV transaminases. Applied Microbiology and Biotechnology, 104, 2343–2357. https://doi.org/10.1007/s00253-020-10369-6
Midelfort, K. S., Kumar, R., Han, S., Karmilowicz, M. J., McConnell, K., Gehlhaar, D. K., Mistry, A., Chang, J. S., Anderson, M., Villalobos, A., Minshull, J., Govindarajan, S., & Wong, J. W. (2013). Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin. Protein Engineering Design and Selection, 26(1), 25–33. https://doi.org/10.1093/protein/gzs065
Choi, Y. H., Kim, J. H., Park, B. S., & Kim, B. G. (2016). Solubilization and iterative saturation mutagenesis of α1,3-fucosyltransferase from Helicobacter pylori to enhance its catalytic efficiency. Biotechnology and Bioengineering, 113, 1666–1675. https://doi.org/10.1002/bit.25944
Xu, L., Liu, X. H., Yin, Z. H., Liu, Q., Lu, L. L., & Xiao, M. (2016). Site-directed mutagenesis of α-L-rhamnosidase from Alternaria sp. L1 to enhance synthesis yield of reverse hydrolysis based on rational design. Applied Microbiology and Biotechnology, 100, 10385–10394. https://doi.org/10.1007/s00253-016-7676-4
Huang, J., Xie, D. F., & Feng, Y. (2017). Engineering thermostable (R)-selective amine transaminase from Aspergillus terreus through in silico design employing B-factor and folding free energy calculations. Biochemical and Biophysical Research Communications, 483, 397–402. https://doi.org/10.1016/j.bbrc.2016.12.131
Steffen-Munsberg, F., Vickers, C., Kohls, H., Land, H., Mallin, H., Nobili, A., Skalden, L., Bergh, T. V. D., Joosten, H. J., Berglund, P., Höhne, M., & Bornscheuer, U. T. (2015). Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnology Advances, 33, 566–604. https://doi.org/10.1016/j.biotechadv.2014.12.012
Dawood, A. W. H., Weiß, M. S., Schulz, C., Pavlidis, I. V., Iding, H., De Souza, R. O. M. A., & Bornscheuer, U. T. (2018). Isopropylamine as amine donor in transaminase-catalyzed reactions: Better acceptance through reaction and enzyme engineering. ChemCatChem, 10, 3943–3949. https://doi.org/10.1002/cctc.201800936
Chen, C. K. M., Lee, G. C., Ko, T. P., Guo, R. T., Huang, L. M., Liu, H. J., Ho, Y. F., Shaw, J. F., & Wang, A. H. J. (2009). Structure of the alkalohyperthermophilic archaeoglobus fulgidus Lipase Contains a unique C-terminal domain essential for long-chain substrate binding. Journal of Molecular Biology, 390, 672–685. https://doi.org/10.1016/j.jmb.2009.05.017
Cassimjee, K. E., Humble, M. S., Miceli, V., Colomina, C. G., & Berglund, P. (2011). Active site quantification of an ω-transaminase by performing a half transamination reaction. ACS Catalysis, 1, 1051–1055. https://doi.org/10.1021/cs200315h
Wu, K., Yang, Z., Meng, X., Chen, R., Huang, J., & Shao, L. (2020). Engineering an alcohol dehydrogenase with enhanced activity and stereoselectivity toward diaryl ketones: Reduction of steric hindrance and change of the stereocontrol element. Catalysis Science & Technology, 10, 1650. https://doi.org/10.1039/c9cy02444a
Cho, B. K., Park, H. Y., Seo, J. H., Kim, J., Kang, T. J., Lee, B. S., & Kim, B. G. (2008). Redesigning the substrate specificity of ω-aminotransferase for the kinetic resolution of aliphatic chiral amines. Biotechnology and Bioengineering, 99, 275–284. https://doi.org/10.1002/bit.21591
Dourado, D. F. A. R., Pohle, S., Carvalho, A. T. P., Dheeman, D. S., Caswell, J. M., Skvortsov, T., Miskelly, I., Brown, R. T., Quinn, D. J., Allen, C. C. R., Kulakov, L., Huang, M., & Moody, T. S. (2016). Rational design of a (S)-selective-transaminase for asymmetric synthesis of (1S)-1-(1,1’-biphenyl-2-yl)ethanamine. ACS Catalysis, 6, 7749–7759. https://doi.org/10.1021/acscatal.6b02380
Oue, S., Okamoto, A., Yano, T., & Kagamiyama, H. (1999). Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. Journal of Biological Chemistry, 274, 2344–2349. https://doi.org/10.1074/jbc.274.4.2344
Funding
This work was supported by the Program of the National Key Research and Development Project (2020YFA0908400), the National Defense Science and Technology Innovation Zone Foundation of China, and the National Natural Science Foundation of China (31700693).
Author information
Authors and Affiliations
Contributions
Conceptualization: Dong-Xu Jia. Methodology: Chen Peng, Jun-Liang Li. Data analysis: Chen Peng, Jun-Liang Li. Software: Fan Wang. Writing original manuscript: Dong-Xu Jia, Jun-Liang Li, Chen Peng. Review and revising manuscript: Dong-Xu Jia, Chen Peng, Fan Wang. Funding acquisition: Zhi-Qiang Liu, Yu-Guo Zheng. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jia, DX., Peng, C., Li, JL. et al. Redesign of (R)-Omega-Transaminase and Its Application for Synthesizing Amino Acids with Bulky Side Chain. Appl Biochem Biotechnol 193, 3624–3640 (2021). https://doi.org/10.1007/s12010-021-03616-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03616-7