Abstract
Asymmetric epoxidation catalyzed with styrene monooxygenase (SMO) is a powerful enzymatic process producing enantiopure styrene epoxide derivatives. To establish a more diversified reservoir of SMOs, a new SMO from Bradyrhizobium sp. ORS 375, named BrSMO, was mined from the database and characterized. BrSMO was constituted of an epoxygenase component of 415 amino acid residues and an NADH-dependent flavin reductase component of 175 residues. BrSMO catalyzed the epoxidation of styrene and 7 more styrene derivatives, yielding the corresponding (S)-epoxides with excellent enantiomeric excesses (95– > 99% ee), with the highest activity achieved for styrene. BrSMO also catalyzed the asymmetric sulfoxidation of 7 sulfides, producing the corresponding (R)-sulfoxides (20–90% ee) with good yields.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enantiopure epoxides are important synthons for the production of pharmaceuticals and fine chemicals [1,2,3,4,5,6,7,8]. The synthesis of chiral epoxides in high yield and with excellent enantiopurity is a challenge in organic synthesis. Until now, many chemical and enzymatic strategies have been developed to synthesize chiral epoxides, among which, the biocatalytic epoxidation of alkenes has been approved as a powerful route to enantiopure epoxides [9,10,11,12].
Styrene monooxygenase (SMO; EC 1.14.14.11), also known as styrene epoxygenase, is an excellent biocatalyst for the epoxidation of many aromatic conjugated and unconjugated alkenes [13,14,15,16,17,18,19,20,21], as well as several aliphatic alkenes with exceptional stereoselectivity, producing the corresponding epoxides with enantiomeric excesses of up to > 99% ee [22,23,24]. They also show good activity in the sulfoxidation of sulfides [25,26,27] and the oxidative coupling of indole to form indigo dyes, most probably through an unstable epoxide intermediate [28].
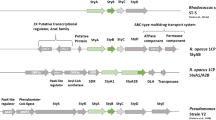
SMO is a two-component flavoprotein composed of a FAD-specific styrene epoxidase (StyA) and NADH-specific flavin reductase (StyB), which catalyzes the formation of reduced FAD [29, 30]. Up until now, 48 SMO genes have been identified from styrene-degrading strains or metagenomes, or through database mining, and functionally characterized, including a very recent work involving the assay of 31 monooxygenases from different microbes with 23 of which showed activity on the epoxidation of styrene with high enantioselectivity [31, 32]. Some of them are E1-type flavin-dependent monooxygenases, which are two-protein systems composed of a monooxygenase and a reductase, and the others belong to group E2-type, where the oxygenase and reductase components are fused to form a self-sufficient protein [26, 33].
The first one was identified from Pseudomonas taiwanensis VLB120 (StyAB) [34], heterologously expressed and developed to catalyze the pilot-scale production of (S)-styrene oxide from styrene, resulting in producing 388 g of styrene oxide in a two-liquid phase 30-L fed-batch system (170 U per liter) [35]. Further efforts have improved the volumetric productivities to 1800 U per liter [36], and the product concentration doubled to 36.3 g Ltot−1, making it one of the most productive biocatalytic oxyfunctionalization processes [37, 38]. Then, more SMOs were identified from Pseudomonas species, such as P. fluorescens ST [21], P. putida SN1 [39], and Pseudomonas sp. LQ26 [22], with the protein sequences of StyA sharing a high similarity of > 90%. SMOs from microbes other than Pseudomonas share lower similarity at protein level, such as those from the genus of Rhodococcus (R. opacus 1CP, Rhodococcus sp. ST-5 and ST-10) [18], a metagenome library [40], and strains of Variovorax paradoxus EPS [26] and Gordonia rubripertincta CWB2 [41]. Nevertheless, all the abovementioned SMOs are of (S)-selectivity, producing (S)-epoxides from styrene and derivatives with good to excellent enantioselectivity.
In our continuous effort to exploit the potential of new SMOs, we have recently identified two new SMOs from marine microbes Paraglaciecola agarilytica NO2 and Marinobacterium litorale DSM 23545 [28], as well as a new epoxidase component from Streptomyces sp. NRRL S-31 [42]. While the former two have displayed the same (S)-selectivity as expected, the latter one has shown excellent (R)-selectivity, complementary to all the known SMOs, yielding the (R)-epoxides with excellent enantioselectivity (91–99% ee) [42].
The present work reports the epoxidation and sulfoxidation activity of a new SMO (BrSMO) from Bradyrhizobium sp. ORS 375 [43], which belongs to the class of α-proteobacteria. Very recently, another hypothetic monooxygenase (GenBank No. WP_012044467) from the same genus (Bradyrhizobium sp. BTAi1) has been analyzed by Tischler et al., but it did not show epoxidation activity toward styrene [32]. Here, BrSMO was found to catalyze the asymmetric (S)-epoxidation of styrene and derivatives with excellent stereoselectivity, and the sulfoxidation of thioanisole with moderate stereoselectivity.
Materials and methods
Chemicals
Substrates of 1a-15a were purchased from Alfa-Aesar (Tianjin, China) or Acros Organics (Geel, Belgium) and used without further purification. Racemic styrene epoxide (1b) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Racemic epoxides were prepared from the corresponding substrates using m-chloroperoxybenzoic acid (2b-8b) [22] or H2O2 (9b-15b) as the oxidant [44].
Plasmids construction and gene manipulation
StyA from Pseudomonas sp. LQ26 (PsStyA) [22] was used as the query template to perform BLASTP [45] search in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The hypothetic monooxygenase (BrSMO) from Bradyrhizobium sp. ORS 375 (GenBank no: NZ_CAFI01000479.1) having a 48% identity with PsStyA was selected. The accession numbers are WP_009031806.1 and WP_009031805.1 for BrStyA and BrStyB, respectively. The DNA sequence encoding BrSMO was synthesized by GenScript Biotech Co. (Nanjing, China). The synthesized DNA fragment and pET-28a (+) (Novagen, Madison, WI) were digested with BamH I and Sac I (New England Biolabs, Beverly, MA, USA), and ligated to achieve the expression plasmid pET-BrSMO. The DNA sequence encoding BrSMO was verified by sequencing the plasmid at Invitrogen Biotechnologies (Shanghai, China).
Protein expression of recombinant BrSMO
The plasmid pET-BrSMO was introduced into E. coli BL21 (DE3). A single colony was picked up and grown in LB medium containing kanamycin (50 mg/L) at 37 °C and 180 rpm. Then 200 mL Terrific Broth containing kanamycin (50 mg/l) was inoculated with the overnight culture and incubated at 37 °C and 220 rpm. After OD600 reached 0.8, IPTG (0.05 mM) was added, and the incubation was continued at 20 °C for 20 h. Cells were harvested by centrifugation (8 × 103g and 4 °C) for 10 min, washed twice. The collected cells were then used for SDS-PAGE analysis (stained with Coomassie blue) and as a catalyst for biotransformation, as described in the next section.
Whole-cell bioconversion and product analysis
Specific epoxidation activity was measured using whole cells following the literature [23, 46, 47]. Briefly, the biotransformation was carried out in 4 mL potassium phosphate buffer (100 mM, pH 7.0) containing 0.5 g CDW/L of the recombinant cells, 0.5% (wt/vol) glucose, and incubated at 30 °C on a rotary shaker at 200 rpm. After 10 min, styrene was added to a final concentration of 1.5 mM (30 mM stock solution in ethanol). The reaction was continued for 5 min. The mixtures were extracted by ethyl acetate and analyzed with gas chromatography (GC). Activities were calculated from the amount of product formed and were expressed in units per g CDW, with one unit (U) being defined as the activity that produces 1 μmol of oxide per min.
For optimal reaction pH and temperature determination, the biotransformation was carried out in 5 mL potassium phosphate buffer or Tris-HCl buffer (100 mM) containing 0.05 g CDW of the cells expressed BrSMO, 0.5% (wt/vol) glucose, 2% (v/v) cyclohexane, and 8 mM styrene. The reaction was carried out at a varied temperature for 15 min with shaking at 200 rpm, terminated by extraction with ethyl acetate, and analyzed with GC.
The relative activities of the whole cells toward various substrates were determined in 5 mL potassium phosphate buffer (100 mM, pH 7.5) containing 0.05 g CDW of the cells expressed BrSMO, 0.5% (wt/vol) glucose, 2% (v/v) cyclohexane, and 8 mM substrate. The reaction was carried out at 30 °C for 30 min with shaking at 200 rpm, terminated by extraction with ethyl acetate, and analyzed with GC.
The yield and optical purity of the whole cells toward various substrates were determined in 5 mL potassium phosphate buffer (100 mM, pH 7.0) containing 0.2 g CDW of the resting cells, 10% (v/v) cyclohexane, and 5 mM substrate. The reactions were performed at 30 °C and 200 rpm for 4 h, and then the mixtures were extracted by ethyl acetate. The organic phases were combined, dried with anhydrous sodium sulfate, and concentrated under vacuum for analysis.
GC analysis was performed on an Agilent Technologies 7890B GC system connected to an FID detector using a Cyclodex-B column (30 m × 0.25 mm × 0.25 μm, USA). The temperatures of the injector and detector were set at 260 °C and 280 °C, respectively. The column temperatures were set at 100 °C for 1b, 140 °C for 2b-4b and 6b-7b, 90 °C for 5b, and 160 °C for 8b-15b.
The optical purities were determined by chiral HPLC using a Daicel Chiralpak IC column (1b, 2b, 6b, and 8b) or Chiralpak OJ-H column (3b) or Chiralpak OB-H column (9b-15b). (See Table S1 and Table S2 of the Supporting information for separation conditions and retention times). The absolute configuration of each product was determined by comparing the elution orders on chiral HPLC or GC (1b, 3b, 6b, 8b [42], 2b [48], 4b [49], 5b [50], 7b [22], and 9b-15b [51]) with published data.
Results and Discussion
Sequence Analysis of BrSMO
StyA from Pseudomonas sp. LQ26 was used as the reference to mine new SMOs from the NCBI nr protein database. Pseudomonas belongs to the class of γ-proteobacteria in the phylum of proteobacteria, which is a major phylum of Gram-negative bacteria [52]. The search returned a hypothesized monooxygenase (BrStyA) from Bradyrhizobium sp. ORS 375, which is a kind of plant symbiotic bacteria and belongs to the class of α-proteobacteria. Previously two SMOs were identified from α-proteobacteria, either from Sphingopyxis sp. Kp5.2 [53] or Gemmobacter nectariphilus DSM 15620 [32]. Both of them could catalyze styrene epoxidation to produce enantiopure (S)-oxide.
BrStyA contains 415 amino acid residues, similar to known SMOs (about 406-427 residues). Its amino acid sequence shares 48% identity to the reference sequence from Pseudomonas sp. LQ26 [22], and 47% identity to StyAST-10 from Rhodococcus sp. ST-10 [18]. Further analysis showed that the DNA coding BrStyA is followed by a 528 bp sequence coding a putative flavin reductase (BrStyB). However, other genes in a typical styrene degradation operon, such as styrene oxide isomerase (styC) gene or phenylacetaldehyde dehydrogenase (styD) gene, were not found in the neighboring region of styAB. BrStyB contains 175 amino acid residues and has 41% and 37% identities to that from Pseudomonas sp. LQ26 and Rhodococcus sp. ST-10, respectively. Based on known mechanisms of SMO, together BrStyA and BrStyB would form a two-component flavoprotein, named BrSMO.
Multiple sequence alignment (Fig. 1) showed that BrSMO contained several common sequence motifs compare to other SMOs of known function. In the reductase component, the signature motif of TANxxxSx10S, important for interacting with isoalloxazine and NADH [54, 55], is conserved in the N-terminus (residues 37–54) (Fig. 1b). In the epoxygenase component, the elements include the FAD-binding fingerprints GxGxxG, GG, and Dx6G (residues 9–14, 117–118, and 138–145, respectively) and the putative dual function motif GDx6P (residues 298–306) related to FAD and/or NAD(P)H interaction (Fig. 1a). The residue Thr47, which is considered functionally important to catalytic activity in StyA was also conserved in BrSMO (Fig. 1a). Notably, a recently identified fingerprint motif is present in BrSMO as N46-V48-H50-Y73-H76-S97 (Fig. 1a), which corresponds to the N46-V48-H50-Y73-H76-S96 motif in PpStyA S12 from P. putida S12 (PDB ID: 3IHM) [56]. This motif is shared by many SMOs and found to lead to (R)-enantioselectivity in sulfoxidation, while the motif S46-Q48-M50-V/I73-I76-A96 in indole monooxygenase often leads to the (S)-enantiomer [32].
Multiple sequence alignment of the two subunits of SMOs, StyA (a) and StyB (b). Several sequence motifs conserved in the flavoprotein hydroxylase family are indicated. A (+) indicates a residue that is part of the new fingerprint motif (N-V-H-Y-H-S) that defines the enantioselectivity of SMOs in sulfoxidation. Shown are BrStyA, BrStyB (Bradyrhizobium sp. ORS 375, WP_009031806, and WP_009031805), SN1StyA, SN1StyB (Pseudomonas putida SN1, ABB03727, and ABB03728), VLB120StyA, VLB120StyB (Pseudomonas taiwanensis VLB120, AAC23718, and AAC23719), LQ26StyA, LQ26StyB (Pseudomonas sp. LQ26, ADE62390, and ADE62391), RhStyA, RhStyB (Rhodococcus sp. ST-10, BAL04129, and BAL04130), STStyA, and STStyB (Pseudomonas fluorescens ST, CAB06823, and CAB06824)
Heterologous expression and functional screening
E. coli BL21 (DE3) cells harboring the pET-BrSMO construct were grown at 20 °C to expressing soluble proteins of BrStyA and BrStyB. SDS-PAGE analysis showed a prominent band at 45 kDa, as expected from the size of BrStyA (Fig. 2), and only vague bands appeared at around 19 kDa, indicating low expression of BrStyB (Fig. 2). In previous works, several SMOs have also been reported of much higher expression of StyA than StyB [18, 28, 29]. The overexpression of BrStyB is often achieved by using the plasmid containing only the coding DNA of StyB but not of StyA [18, 29], which is also the case for BrStyB (Fig. 2).
During the cultivation of E. coli BL21 (DE3)/pET-BrSMO, the formation of blue dye (indigo) was distinctly observed, which was in accordance with the known indigo-forming capacity of SMOs [42, 57], indicating the functional expression of BrSMO. After harvesting, the whole cells expressing BrSMO were used as the catalyst for the epoxidation of styrene, yielding styrene epoxide with > 99% ee, which indicated the BrSMO did have the function as a styrene monooxygenase.
The specific epoxidation activity of BrSMO was determined to be 86 ± 2 U/g CDW, which was comparable to that of the other SMOs measured under similar conditions, such as that from P. taiwanensis VLB120 (79 ± 5 U/g CDW) [23], P. putida SN1 (55 ± 5 U/g CDW) [46], and Pseudomonas sp. LQ26 (66.5 U/g CDW) [47].
Effects of reaction temperature and pH
The effect of reaction temperature on the epoxidation of styrene was measured at temperatures ranging from 20 to 40 °C in potassium phosphate buffer at pH 7.0 using whole cells (Fig. 3). The epoxidation appeared to be sensitive to the reaction temperature. The highest conversion of 47% was achieved at 30 °C after 15-min incubation. The conversion dropped to < 35% when the reaction temperature was maintained at 20 °C or 40 °C.
Effect of reaction temperature on the whole-cell-catalyzed epoxidation. Reaction conditions: a mixture of the resting cells (0.05 g CDW), glucose (0.5%), cyclohexane (2%), and styrene (8 mM) in 5 mL potassium phosphate buffer (0.1 M, pH 7.0) was incubated at varied temperature with shaking at 200 rpm for 15 min
The effect of reaction pH was measured in potassium phosphate buffer (pH 6.0 to 8.0) or Tris-HCl buffer (pH 7.5 to 9.0). BrSMO yielded the highest conversion at pH 7.5 in potassium phosphate buffer and a slightly lower conversion in Tris-HCl buffer of the same pH (Fig. 4). In all cases, the epoxidation yielded (S)-styrene oxide with excellent enantioselectivity (> 99% ee).
Effect of reaction pH on the whole-cell-catalyzed epoxidation. Reaction conditions: a mixture of the resting cells (0.05 g CDW), glucose (0.5%), cyclohexane (2%), and styrene (8 mM) in 5 mL (0.1 M) potassium phosphate buffer (black circle) or Tris-HCl buffer (black square) was incubated at 30 °C with shaking at 200 rpm for 15 min
BrSMO-catalyzed asymmetric epoxidation and sulfoxidation
Fourteen more substrates besides styrene were tested using the whole-cell biotransformation system to probe the substrate spectrum and stereoselectivity of BrSMO. The results showed that BrSMO could catalyze the asymmetric epoxidation of 8 aromatic alkenes (Table 1), as well as the sulfoxidation of 7 sulfides (Table 2).
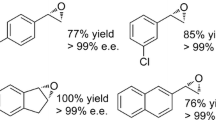
The epoxidation of styrene derivatives afforded the corresponding (S)-epoxides with excellent enantiomeric excesses of > 99% ee (for 1a, 3a-4a, 6a-8a), 97% ee (for 2a), and 95% ee (for 5a) (Table 1). The enantioselectivity of BrSMO is similar to that of SMOs from P. taiwanensis VLB120 [29], Pseudomonas sp. LQ26[22], Rhodococcus sp. ST-5 and ST-10 [18], metagenome [33, 40], and Marinobacterium litorale DSM 23545 and Paraglaciecola agarilytica NO2 [28], which produce the (S)-epoxides with enantiomeric excesses ranging from 95to > 99% ee.
BrSMO showed the highest relative activity toward styrene (1a), and the activity decreased when a substituent was present on the benzene ring (Table 1). Compared to the meta-substituted styrene (3a and 6a), the relative activities of ortho- and para-substituents (2a, 4a, 5a, and 7a) were low. Consequently, their product yields after 4-h reaction were also lower than meta-substituted styrenes. In addition, each of the chloro-substituted substrates showed higher relative activity than their bromo-substituted counterparts (2a vs 5a, 3a vs 6a, 4a vs 7a) (Table 1). The results indicated that the position and type of substituents affected the enantioselectivity and conversion rate. Besides, aliphatic alkenes, such as 1-heptene, were also tested, but no product was detected.
BrSMO also catalyzed the sulfoxidation of sulfides, affording (R)-sulfoxide with medium stereoselectivity (59–90% ee), except 2-chlorothioanisole (10a) (20% ee). Among them, 4-chlorothioanisole (12a) and 4-bromothioanisole (15a) yielded products with the highest enantiomeric excess of 89% ee and 90% ee, respectively (Table 2). The relative activity of BrSMO was the highest toward thioanisole (9a), followed by 3-chlorothioanisole (11a) and 4-bromothioanisole (15a). The product yields of 9a, 11a, 12a, and 15a reached 100% after 4-h reaction, while others ranged from 67 to 87% (Table 2).
The (R)-selectivity of BrSMO is complementary to some (S)-selective P450s [51, 58], and is in accordance with the fingerprint motif of N46-V48-H50-Y73-H76-S97 that defines the enantioselectivity of SMOs in sulfoxidation [32]. Other SMOs with varied stereoselectivity have also been reported. For example, SMO from P. taiwanensis VLB120 catalyzed sulfoxidation reactions of thioanisole, but with poor enantioselectivity (26% ee) [25]. SMOs from Gordonia polyisoprenivorans catalyzed sulfoxidation reactions and produced the (R)-enantiomer with excellent enantioselectivity (98.6% ee) [59]. At the same time, several StyAs were able to catalyze the sulfoxidation and produced the (S)-enantiomer with excellent enantioselectivities (98–99% ee) [26, 32, 55, 59, 60].
Conclusion
In summary, we have identified a new two-component styrene monooxygenase, BrSMO, from the genome of Bradyrhizobium sp. ORS 375 and functionally expressed it in E. coli. The resulting whole-cell biocatalyst can catalyze the asymmetric epoxidation of styrene derivatives, yielding (S)-epoxides with excellent enantiomeric excesses; it can also catalyze the oxidation of sulfides, producing (R)-sulfoxide. The results presented here would enrich the diversity of styrene monooxygenase and implicate great potential for bio-epoxidation reactions.
References
Davis, R. L., Stiller, J., Naicker, T., Jiang, H., & Jørgensen, K. A. (2014). Asymmetric Organocatalytic Epoxidations: Reactions, Scope, Mechanisms, and Applications. Angewandte Chemie International Edition., 53(29), 7406–7426.
Farina, V., Reeves, J. T., Senanayake, C. H., & Song, J. J. (2006). Asymmetric Synthesis of Active Pharmaceutical Ingredients. Chemical Reviews., 106(7), 2734–2793.
Schmid, A., Dordick, J. S., Hauer, B., Kiener, A., Wubbolts, M., & Witholt, B. (2001). Industrial biocatalysis today and tomorrow. Nature., 409(6817), 258–268.
Yuan, J., Lukito, B. R., & Li, Z. (2019). De NovoBiosynthesis of (S)- and (R)-Phenylethanediol in YeastviaArtificial Enzyme Cascades. ACS Synthetic Biology., 8(8), 1801–1808.
Liu, Y. C., Guo, C., Liu, Y., Wang, H. B., & Wu, Z. L. (2017). Enzymatic cascades for the stereo-complementary epimerisation of in situ generated epoxy alcohols. Organic & Biomolecular Chemistry., 15(12), 2562–2568.
Wu, S., Liu, J., & Li, Z. (2017). Biocatalytic Formal Anti-Markovnikov Hydroamination and Hydration of Aryl Alkenes. Acs Catalysis., 7(8), 5225–5233.
Wu, S., Zhou, Y., Seet, D., & Li, Z. (2017). Regio- and Stereoselective Oxidation of Styrene Derivatives to Arylalkanoic AcidsviaOne-Pot Cascade Biotransformations. Advanced Synthesis & Catalysis., 359(12), 2132–2141.
Liu, Y. C., & Wu, Z. L. (2016). Switchable asymmetric bio-epoxidation of α,β-unsaturated ketones. Chem Commun (Camb)., 52(6), 1158–1161.
Hussain, M. M., & Walsh, P. J. (2008). Tandem Reactions for Streamlining Synthesis: Enantio- and Diastereoselective One-Pot Generation of Functionalized Epoxy Alcohols. Accounts of Chemical Research., 41(8), 883–893.
Lin, H., Liu, J.-Y., Wang, H.-B., Ahmed, A. A. Q., & Wu, Z.-L. (2011). Biocatalysis as an alternative for the production of chiral epoxides: A comparative review. Journal of Molecular Catalysis B Enzymatic., 72(3-4), 77–89.
Chang, D., Zhang, J., Witholt, B., & Li, Z. (2004). Chemical and Enzymatic Synthetic Methods for Asymmetric Oxidation of the C–C Double Bond. Biocatalysis., 22(2), 113–131.
Sello, G., Tiziano, F., & Orsini, F. (2006). Recent Developments in Epoxide Preparation. Current Organic Synthesis., 3(4), 457–476.
Lin, H., Liu, Y., & Wu, Z. L. (2011). Highly diastereo- and enantio-selective epoxidation of secondary allylic alcohols catalyzed by styrene monooxygenase. Chem Commun (Camb)., 47(9), 2610–2612.
Toda, H., Ohuchi, T., Imae, R., & Itoh, N. (2015). Microbial Production of Aliphatic (S)-Epoxyalkanes by Using Rhodococcus sp. Strain ST-10 Styrene Monooxygenase Expressed in Organic-Solvent-Tolerant Kocuria rhizophila DC2201. Applied and Environmental Microbiology., 81(6), 1919–1925.
Andreas, S., Karin, H., Hans-Juergen, F., Frank, H., & Bernard, W. (2001). Advanced Synthesis & Catalysis, 343, 732–737.
Hollmann, F., Hofstetter, K., & Schmid, A. (2006). Non-enzymatic regeneration of nicotinamide and flavin cofactors for monooxygenase catalysis. Trends in Biotechnology., 24(4), 163–171.
Tischler, D., Kermer, R., Groning, J. A. D., Kaschabek, S. R., van Berkel, W. J. H., & Schlomann, M. (2010). StyA1 and StyA2B from Rhodococcus opacus 1CP: a Multifunctional Styrene Monooxygenase System. Journal of Bacteriology., 192(19), 5220–5227.
Toda, H., Imae, R., Komio, T., & Itoh, N. (2012). Expression and characterization of styrene monooxygenases of Rhodococcus sp. ST-5 and ST-10 for synthesizing enantiopure (S)-epoxides. Appl Microbiol Biotechnol., 96(2), 407–418.
Gursky, L. J., Nikodinovic-Runic, J., Feenstra, K. A., & O’Connor, K. E. (2010). In vitro evolution of styrene monooxygenase from Pseudomonas putida CA-3 for improved epoxide synthesis. Applied Microbiology & Biotechnology., 85(4), 995–1004.
Nikodinovic-Runic, J., Coulombel, L., Francuski, D., Sharma, N. D., Boyd, D. R., Ferrall, R. M., & Ke, O. C. (2013). Applied Microbiology & Biotechnology, 97, 4849–4858.
Gennaro, P. D., Colmegna, A., Galli, E., Sello, G., Pelizzoni, F., & Bestetti, G. (1999). A New Biocatalyst for Production of Optically Pure Aryl Epoxides by Styrene Monooxygenase from Pseudomonas fluorescensST. Applied & Environmental Microbiology., 65(6), 2794–2797.
Lin, H., Qiao, J., Liu, Y., & Wu, Z. L. (2010). Styrene monooxygenase from Pseudomonas sp. LQ26 catalyzes the asymmetric epoxidation of both conjugated and unconjugated alkenes. Journal of Molecular Catalysis B: Enzymatic., 67(3-4), 236–241.
Panke, S., Witholt, B., Schmid, A., & Wubbolts, M. G. (1998). Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120. Appl Environ Microbiol., 64(6), 2032–2043.
Dunn, H. D., Curtin, T., O’Riordan, M. A., Coen, P., Kieran, P. M., Malone, D. M., & O’Connor, K. E. (2005). FEMS Microbiol Lett, 249, 267–273.
Hollmann, F., Lin, P. C., Witholt, B., & Schmid, A. (2003). Stereospecific Biocatalytic Epoxidation: The First Example of Direct Regeneration of a FAD-Dependent Monooxygenase for Catalysis. J Am Chem Soc., 125(27), 8209–8217.
Tischler, D., Schwabe, R., Siegel, L., Joffroy, K., Kaschabek, S. R., Scholtissek, A., & Heine, T. (2018). VpStyA1/VpStyA2B of Variovorax paradoxus EPS: An Aryl Alkyl Sulfoxidase Rather than a Styrene Epoxidizing Monooxygenase. Molecules., 23(4), 809.
Boyd, D. R., Sharma, N. D., Mcmurray, B., Haughey, S. A., & O’Connor, K. E. (2011). Organic & Biomolecular Chemistry., 10, 782–790.
Pu, W., Cui, C., Guo, C., & Wu, Z. L. (2018). Characterization of two styrene monooxygenases from marine microbes. Enzyme Microb Technol., 112, 29–34.
Otto, K., Hofstetter, K., Rothlisberger, M., Witholt, B., & Schmid, A. (2004). Biochemical Characterization of StyAB from Pseudomonas sp. Strain VLB120 as a Two-Component Flavin-Diffusible Monooxygenase. Journal of Bacteriology., 186(16), 5292–5302.
Montersino, S., Tischler, D., Gassner, G. T., & Berkel, W. J. H. V. (2011). Catalytic and Structural Features of Flavoprotein Hydroxylases and Epoxidases. Advanced Synthesis & Catalysis., 353(13), 2301–2319.
Huijbers, M. M., Montersino, S., Westphal, A. H., Tischler, D., & van Berkel, W. J. (2014). Flavin dependent monooxygenases. Arch Biochem Biophys., 544, 2–17.
Heine, T., Scholtissek, A., Hofmann, S., Koch, R., & Tischler, D. (2020). Accessing Enantiopure Epoxides and Sulfoxides: Related Flavin‐Dependent Monooxygenases Provide Reversed Enantioselectivity. ChemCatChem., 12(1), 199–209.
Tischler, D., Eulberg, D., Lakner, S., Kaschabek, S. R., van Berkel, W. J., & Schlomann, M. (2009). Identification of a Novel Self-Sufficient Styrene Monooxygenase from Rhodococcus opacus 1CP. Journal of Bacteriology., 191(15), 4996–5009.
Velasco, A., Alonso, S., García, J. L., Perera, J., & Díaz, E. (1998). Genetic and Functional Analysis of the Styrene Catabolic Cluster of Pseudomonas sp. Strain Y2. Journal of Bacteriology., 180(5), 1063–1071.
Panke, S., Held, M., Wubbolts, M. G., Witholt, B., & Schmid, A. (2002). Pilot-scale production of (S)-styrene oxide from styrene by recombinantEscherichia coli synthesizing styrene monooxygenase. Biotechnol Bioeng., 80(1), 33–41.
Park, J. B., Buhler, B., Habicher, T., Hauer, B., Panke, S., Witholt, B., & Schmid, A. (2006). The efficiency of recombinantEscherichia coli as biocatalyst for stereospecific epoxidation. Biotechnol Bioeng., 95(3), 501–512.
Kuhn, D., Kholiq, M. A., Heinzle, E., Buhler, B., & Schmid, A. (2010). Intensification and economic and ecological assessment of a biocatalytic oxyfunctionalization process. Green Chemistry., 12(5), 815–827.
Kuhn, D., Buhler, B., & Schmid, A. (2012). Production host selection for asymmetric styrene epoxidation: Escherichia coli vs. solvent-tolerant Pseudomonas. Journal of Industrial Microbiology & Biotechnology., 39(8), 1125–1133.
Bae, J. W., Han, J. H., Park, M. S., Lee, S. G., Lee, E. Y., Jeong, Y. J., & Park, S. (2006). Development of recombinantPseudomonas putida containing homologous styrene monooxygenase genes for the production of (S)-styrene oxide. Biotechnology and Bioprocess Engineering., 11(6), 530–537.
Van Hellemond, E. W., Janssen, D. B., & Fraaije, M. W. (2007). Discovery of a Novel Styrene Monooxygenase Originating from the Metagenome. Appl Environ Microbiol., 73(18), 5832–5839.
Heine, T., Zimmerling, J., Ballmann, A., Kleeberg, S. B., Rückert, C., Busche, T., Winkler, A., Kalinowski, J., Poetsch, A., Scholtissek, A., Oelschlägel, M., Schmidt, G., & Tischler, D. (2018). Applied and Environmental Microbiology., 84, e00154–e00118.
Cui, C., Guo, C., Lin, H., Ding, Z.-Y., Liu, Y., & Wu, Z.-L. (2020). Functional characterization of an (R)-selective styrene monooxygenase from streptomyces sp. NRRL S-31. Enzyme and Microbial Technology., 132, 109391.
Damien, M., Lucie, M., Gilles, B., Nico, N., André, V., David, V., Smith, A. A. T., Eric, G., Claudine, M., & Lionel, M. (2011). Genes, 3, 35–61.
Hartman, T., Šturala, J., & Cibulka, R. (2015). Two-Phase Oxidations with Aqueous Hydrogen Peroxide Catalyzed by Amphiphilic Pyridinium and Diazinium Salts. Advanced Synthesis & Catalysis., 357(16-17), 3573–3586.
Jiang, J., Chen, X., Zhang, D., Wu, Q., & Zhu, D. (2015). Characterization of (R)-selective amine transaminases identified by in silico motif sequence blast. Applied Microbiology and Biotechnology., 99(6), 2613–2621.
Park, M. S., Bae, J. W., Han, J. H., Lee, E. Y., Lee, S. G., & Park, S. (2006). Journal of Microbiology & Biotechnology., 16, 1032–1040.
Lin, H., Tang, D. F., Ahmed, A. A., Liu, Y., & Wu, Z. L. (2012). Mutations at the putative active cavity of styrene monooxygenase: Enhanced activity and reversed enantioselectivity. J Biotechnol., 161(3), 235–241.
Liao, S., Leutzsch, M., Monaco, M. R., & List, B. (2016). Catalytic Enantioselective Conversion of Epoxides to Thiiranes. J Am Chem Soc., 138(16), 5230–5233.
Li, A., Liu, J., Pham, S. Q., & Li, Z. (2013). Engineered P450pyr monooxygenase for asymmetric epoxidation of alkenes with unique and high enantioselectivity. Chem Commun (Camb)., 49(98), 11572–11574.
Toda, H., Imae, R., & Itoh, N. (2012). Efficient biocatalysis for the production of enantiopure (S)-epoxides using a styrene monooxygenase (SMO) and Leifsonia alcohol dehydrogenase (LSADH) system. Tetrahedron: Asymmetry., 23(22-23), 1542–1549.
Guo, C., & Wu, Z. L. (2017). Construction and functional analysis of a whole-cell biocatalyst based on CYP108N7. Enzyme & Microbial Technology., 106, 28–34.
Gupta, R. S. (2000). The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiology Reviews., 24(4), 367–402.
Oelschlagel, M., Zimmerling, J., Schlomann, M., & Tischler, D. (2014). Styrene oxide isomerase of Sphingopyxis sp. Kp5.2. Microbiology., 160(11), 2481–2491.
Thotsaporn, K., Sucharitakul, J., Wongratana, J., Suadee, C., & Chaiyen, P. (2004). Cloning and expression of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii: evidence of the divergence of enzymes in the class of two-protein component aromatic hydroxylases. Biochim Biophys Acta., 1680(1), 60–66.
Groning, J. A., Kaschabek, S. R., Schlomann, M., & Tischler, D. (2014). A mechanistic study on SMOB-ADP1: an NADH:flavin oxidoreductase of the two-component styrene monooxygenase of Acinetobacter baylyi ADP1. Arch Microbiol., 196(12), 829–845.
Ukaegbu, U. E., Kantz, A., Beaton, M., Gassner, G. T., & Rosenzweig, A. C. (2010). Structure and Ligand Binding Properties of the Epoxidase Component of Styrene Monooxygenase. Biochemistry., 49(8), 1678–1688.
Heine, T., Tucker, K., Okonkwo, N., Assefa, B., Conrad, C., Scholtissek, A., Schlömann, M., Gassner, G., & Tischler, D. (2017). Engineering Styrene Monooxygenase for Biocatalysis: Reductase-Epoxidase Fusion Proteins. Applied Biochemistry and Biotechnology., 181(4), 1590–1610.
Fruetel, J. A., Mackman, R. L., Peterson, J. A., & Montellano, P. R. O. D. (1994). Relationship of active site topology to substrate specificity for cytochrome P450terp (CYP108). Journal of Biological Chemistry., 269(46), 28815–28821.
Heine, T., Grossmann, C., Hofmann, S., & Tischler, D. (2019). Indigoid dyes by group E monooxygenases: mechanism and biocatalysis. Biol Chem., 400(7), 939–950.
Willrodt, C., Gröning, J. A. D., Nerke, P., Koch, R., Scholtissek, A., Heine, T., Schmid, A., Bühler, B., & Tischler, D. (2020). Highly efficient access to (S)-Sulfoxides utilizing a promiscuous flavoprotein monooxygenase in a whole-cell biocatalyst format. ChemCatChem, 12, 1–9.
Funding
This work was financially supported by the National Natural Science Foundation of China (21572220 and 21708038) and CAS Key Laboratory of Environmental and Applied Microbiology & Environmental Microbiology Key Laboratory of Sichuan Province, Chengdu Institute of Biology, CAS (No. KLCAS-2016-08 and KLCAS-2018-1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 666 kb)
Rights and permissions
About this article
Cite this article
Cui, C., Lin, H., Pu, W. et al. Asymmetric Epoxidation and Sulfoxidation Catalyzed by a New Styrene Monooxygenase from Bradyrhizobium. Appl Biochem Biotechnol 193, 65–78 (2021). https://doi.org/10.1007/s12010-020-03413-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03413-8