Abstract
Two new endo-1,4-beta-xylanases encoding genes EpXyn1 and EpXyn3 were isolated from mesophilic fungus Eupenicillium parvum 4–14. Based on analysis of catalytic domain and phylogenetic trees, the xylanases EpXYN1 (404 aa) and EpXYN3 (220 aa) belong to glycoside hydrolase (GH) family 10 and 11, respectively. Both EpXYN1 and EpXYN3 were successfully expressed in Pichia pastoris and the recombinant enzymes were characterized using beechwood xylan, birchwood xylan, or oat spelt xylan as substrates, respectively. The optimum temperatures and pH values were 75 °C and 5.5 for EpXYN1, and 55 °C and 5.0 for EpXYN3. EpXYN1 exhibited a high stability at high temperature (65 °C) or at pH values from 8 to 10. EpXYN3 kept over 80% enzymatic activity after treatment at pH values from 3 to 10. The specific activities of EpXYN1 and EpXYN3 were 384.42 and 214.20 U/mg using beechwood xylan as substrate, respectively. EpXYN1 showed lower Km values and higher specific activities toward different xylans compared to EpXYN3. Thin-layer chromatography analysis indicated that the hydrolysis profiles of xylans or xylo-oligosacharides were different by EpXYN1and EpXYN3. EpXYN3 had a higher efficiency than EpXYN1 in production of feruloylated oligosaccharides (FOs) from de-starched wheat bran. The maximum levels of FOs released by EpXYN1 and EpXYN3 were 11.1 and 14.4 μmol/g, respectively. In conclusion, the two xylanases are potential candidates for various industrial applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hemicellulose predominantly consists of heteroxylans and is a renewable resource for production of liquid fuels, value-added chemicals, or biomaterials in energy, functional foods, or pharmaceutical industries [1, 2]. Heteroxylans have a complex structure with backbones of β-1,4-linked xylose residues and side chains of various substituent units such as arabinose, glucuronic acid, or 4-O-methylglucuronic acid [3, 4]. Enzymatic hydrolysis is a key step in industrial preparation of fermentable monosaccharides, xylo-oligosaccharides (XOS), or other chemicals from heteroxylans [5, 6]. Due to its structural complexity, the complete degradation of heteroxylans requires a diverse range of cooperatively acting enzymes, primarily endo-1,4-β-xylanase and β-xylosidase, and additional enzymes such as α-l-arabinofuranosidase, α-d-glucuronidase, and acetyl xylan esterase [5, 7]. Endo-β-1,4-xylanase (EC 3.2.1.8) catalyzes the hydrolysis of internal β-1,4-d-xylosidic bonds in xylans and generates various unsubstituted or branched XOS, and plays a crucial role in effective degradation of hemicellulose [3, 5]. As an important enzyme, xylanase has been widely used in biofuel production, animal feed production, food and health industry, or pulping and laundry industry [8,9,10].
Xylanases can be produced by different organisms including fungi, bacteria, marine algae, protozoa, snails, crustaceans, insect, and seeds [6, 10]. Among these organisms, filamentous fungi are more suitable for large-scale production of xylanases because of their strong ability to produce extracellular enzymes. Many species of Aspergillus, Trichoderma, or Penicillium have been used widely in industrial production of xylanase for various applications [10,11,12]. T. reesei endo-xylanase II and endo-xylanase from A. niger var. awamori have been applied in paper and pulp industry or bread baking, respectively [10, 11]. To date, a large number of endo-β-1,4-xylanase genes have been reported and showed diverse sequences. According to sequences comparisons of the catalytic domains, they were classified to glycoside hydrolases (GH) families 5, 8, 10, 11, 30, or 43, in the Carbohydrate-Active enZYmes (CAZy) database (http://www.cazy.org/) [5]. The majority of endo-β-1,4-xylanases belong to GH10 and GH11 families, and were extensively studied for the important value in industrial applications [11,12,13]. GH10 family xylanases have an (α/ß)8 barrel structure which forms a bowl shape, and show higher catalytic activity on highly branched xylan backbone. GH11 family xylanases are densely packed and consist mainly of ß-sheets that form a ß-jelly roll fold structure and have higher substrate specificity compared to GH10 family xylanases [3, 5, 13]. A carbohydrate-binding module (CBM), which aids binding to cellulose, are usually found at the N-terminus of some GH10 xylanases but not GH11 xylanases [14]. The functions of CBM domain in xylanases involved in the binding ability of enzymes to natural substrates and enzymatic thermostability [14, 15].
Xylanases from mesophilic microorganism generally show a low optimum temperature (50–55 °C) and poor thermal stability (below 65 °C), and cannot withstand the high temperature in many industrial processes [15, 16]. Thermotolerant xylanases have many advantages in industrial applications including higher specific activity (lower cost of enzyme), increment of substrate and product solubility, and decrement of microbial contamination [17]. Some thermophilic fungi (e.g., Humicola insolens) and bacteria (e.g., Caldicellulosiruptor sp.) or hyperthermophilic archaea (e.g., Thermococcus zilligii) have been reported to produce thermotolerant xylanases, which exhibit increased specific activity and higher thermostability [5, 15, 18]. Recently, some researchers enhanced the thermostable of GH11 xylanases through protein engineering [16, 19]. Development of novel and thermotolerant xylanases is significant for biodegradation of hemicellulose.
Wheat bran, a major agricultural by-product, is rich in arabinoxylans and ferulic acid (4 to 6 mg/g) [20]. Ferulic acid (4-hydroxy-3-methoxycinnamic acid, FA) is a very attractive phenolic compound, which displays antioxidative ability [21]. In wheat bran, FA cross-links arabinoxylan molecules via arabinose residues and strengthens cell wall by providing connection between lignin and heteroxylan [22, 23]. Feruloylated oligosaccharides (FOs) owing their nutritional functions to both FA and oligosaccharides possess potential applications in functional food and pharmaceutical industries [24]. FOs from wheat bran displayed strong antioxidant activity against free radical-induced oxidative damage in rat erythrocytes and human lymphocytes [23, 25]. FOs can be efficiently released from wheat bran by enzymatic hydrolysis with single endo-β-1,4-xylanase [26]. The aim of this study was to find new xylanases for preparation of FOs from wheat bran. In previous study, thermotolerant (hemi)cellulolytic enzymes producer Eupenicillium parvum 4–14 showed a high efficiency in extracting FA from wheat bran, suggesting efficient endo-β-1,4-xylanase produced by the fungus [27]. In this study, two genes encoding GH10 or GH11 family xylanases were isolated from E. parvum 4–14, and the recombinant enzymes were characterized and compared in the releasing of FOs from wheat bran.
Materials and Methods
Strain, Media, and Growth Conditions
The filamentous fungus Eupenicillium parvum 4–14 (CCTCC M2015404), a producer of thermotolerant (hemi)cellulolytic enzymes, was preserved on potato dextrose agar (PDA) slant by our lab, and deposited in the China Center for Type Culture Collection (CCTCC, Wuhan, China) [27]. Pichia pastoris KM71H strain was employed to express the target proteins. Escherichia coli DH5α was used for plasmid propagation and grown at 37 °C in Luria-Bertani (LB) medium with or without antibiotic.
Cloning of Xylanase Genes from E. parvum
Two integrated genes encoding endo-xylanase (named EpXyn1 and EpXyn3, respectively) were found in the transcriptome data of E. parvum 4–14 (unpublished data). To clone the genes, specific primers were designed by software Primer Premier 6.0 (Table 1). E. parvum was cultured in solid state fermentation (SSF) medium for 72 h at 37 °C [27]. The collected mycelia were subjected to total RNA extraction with a TransZol Plant kit (TransGen, Beijing, China) according to the commercial manual. First-strand cDNA was synthesized by EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (oligo (dT) was used) (TransGen, Beijing, China). The cDNA of genes were amplified by primer sets enXyn1_f1/enXyn1_r1 for EpXyn1 or enXyn3_f1/enXyn3_r1 for EpXyn3, and inserted into the vector pEASY-Blunt (Transgene, Beijing, China). DNA sequences were determined by the BioSune Biotechnologies Co. Lid (Shanghai, China) using Sanger sequencing.
Expression of Xylanases in Yeast and Protein Purification
The genes EpXyn1 and EpXyn3 (removed native signal sequences) were amplified by specific primer sets enXyn1_f2/enXyn1_r2 or enXyn3_f2/enXyn3_r2, respectively. After gel purifications, the corresponding DNA fragments were ligated into plasmid pPICZαB (terminated with EcoRI-XbaI) by Hieff Clone™ One Step Cloning Kit (Yeasen, Shanghai, China), and yielding plasmid pPIC-EpXyn1 or pPIC-EpXyn3, respectively. After linearization with SacI, the recombinant plasmids were introduced into P. pastoris KM71H competent cells by electroporation with a Gene Pulser® II Electroporation System (Bio-Rad, Hercules, CA, USA). Yeast transformants were selected on YPD plates containing 100 μg/mL of Zeocin (Invitrogen, USA). Randomly selected yeast transformants were cultured in 5 mL BMMY medium containing 0.8% methanol according to the Pichia Expression Kit Instruction Manual (Invitrogen, USA). Yeast transformants with high enzymatic activity were extendedly cultured in 25 mL BMMY medium containing 1% methanol at 28 °C and 200 rpm for 8 days. After centrifugation at 5000×g for 10 min, the fermentation supernatant was applied to the Ni-NTA affinity column (Qiagen, Valencia, CA, USA) for protein purification. After dialysis treatment at 4 °C for 24 h, the pure proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12%, SDS-PAGE) and enzymatic activity determination. Proteins were quantified with a BCA assay kit (Thermo Tech, USA).
Enzyme Assay
Xylanase activity was determined in 50 mM sodium citrate buffer (pH 4.8) using 0.5% (w/v) of beechwood xylan, birchwood xylan, or oat spelt xylan (Sigma, USA) as substrates. The enzymatic reaction system includes 1.0 mL of buffer, 100 μL of pure enzyme (0.2–0.5 μg), and 400 μL of substrate solution. Reaction mixture was incubated at the experimentally determined optimal temperature (55 °C for EpXYN3 or 75 °C for EpXYN1) for 10 min, and the released reducing sugar was quantified by Somogyi-Nelson method [28]. Briefly, the 1.5 mL reaction mixture was added with 0.5 mL of Somogyi reagent and treated at 99 °C for 5 min. After cooling to room temperature, 0.5 mL of Nelson reagent was added and incubated at 25 °C for 20 min. Per 200 μL of reaction solution was transfer to 96-well microplate, measured the absorbance at 520 nm with a SpectraMAX190 Microplate Reader (Melecular Devices, USA). Reducing sugar was quantified according to xylose standard curve. One unit of xylanase activity was defined as the amount of enzyme required to liberate 1 μmol of xylose from the substrate per minute under the assay conditions.
Effect of pH or Temperature on Enzymatic Activity and Stability
The effects of pH or temperature on recombinant xylanases were detected in universal pH buffers (pHs values range from 3.0 to 9.5 with 0.5 interval) [29], or at temperatures range from 30 to 90 °C with 5 °C interval. Relative activity was calculated using the highest activity as 100%. For test of pH stability, pure enzyme (1 μg/μL) was mixed with universal pH buffer (v/v, 1:2) and stored at 4 °C for 12 h. After 100- or 200-fold dilution with ddH2O, the residual enzymatic activity was assayed under standard conditions. Thermal stability of enzyme was estimated by determination of the residual activities under the conditions as described above after pre-incubation of enzyme sample (1 μg/μL) at different temperatures for 5, 10, 20, 30, 60, or 90 min. Relative activities (%) were calculated according to the radio of the activities of treated enzyme and untreated enzyme, using the activity of untreated enzyme as 100%.
Effect of Metal Ions or Chemicals on Enzymatic Activity
Effects of metal ions or chemical reagents on different xylanase were estimated by addition of these substances in reaction system individually. The additive concentrations were 1 or 5 mM for MgCl2, FeCl3, MnCl2, CoCl2, ZnCl2, CuCl2, CaCl2, and NiSO4, or 0.1 to 1 mM for EDTA, DTT, and SDS, respectively. Enzyme activities were assayed under standard conditions. Relative activities (%) were calculated using the activity of untreated enzyme as 100%.
Determination of Kinetic Parameters
The kinetic parameters (Vmax and Km) of different xylanases were determined using beechwood xylan, oat spelt xylan (insoluble), or birchwood xylan (Sigma, Louis, Missouri, USA) as substrates, respectively. Per 0.4 μg of pure enzyme was mixed with 0.125, 0.25, 0.5, 1.0, 1.5, 2.0, or 2.5 mg/mL substrate, respectively. The enzymatic activity was determined under standard conditions except that the reaction time was 5 min. The Vmax and Km values were calculated by the software GraphPad Prism 5.0 using nonlinear regression (http://www.graphpad.com/ prism/). Turnover number (kcat) was calculated as the ration of Vmax to the concentration of enzyme used in the reaction [30].
Thin-Layer Chromatography Analysis of the Hydrolysis Products
The hydrolyzed products of different xylans or xylo-oligosaccharides (XOS, BZ Oligo, Qingdao, China) by the recombinant xylanases were analyzed by thin-layer chromatography (TLC). In 2 mL sodium citrate buffer (pH 5.0, 50 mM), beechwood xylan (10 mg), birchwood xylan (20 mg), or oat spelt xylan (20 mg) were mixed with 25 μg of pure enzyme, and incubated at 55 °C (for EpXYN3) or 75 °C (for EpXYN1) for an appropriate time. In the same buffer, 1 mg/mL of XOS (from xylobiose to xylohexaose, X2 to X6) was hydrolyzed by different xylanases (10 μg/mL) at the same temperatures for 24 h. The enzymatic reactions were stopped by boiling for 10 min. After centrifugation at 5000×g for 10 min, per 3 μL of supernatants were spotted onto TLC Silica gel 60 F254 glass plates (Merck, Whitehouse Station, NJ, USA). The plates were developed in a solvent system consisting of acetonitrile–ethyl acetate–2-propanol–water (v/v/v/v, 17:5:11:10) [31]. After air drying for 10 min, the plates were visualized by spraying a methanol-sulfuric acid mixture (v/v, 9:1) with 0.2% orcinol and heating at 85 °C for 5 min [32]. Xylose and X2–X6 were used as standards.
Releasing of FOs from Wheat Bran by Enzymatic Hydrolysis
De-starched wheat bran (DSWB) was prepared by the previous method [27]. To release FOs from DSWB, single or double xylanases was put into a 2-mL centrifuged tube containing 1 mL of sodium citrate buffer (pH 5, 50 mM) and 50 mg of substrate. For single enzyme hydrolysis, 0–160 U/g (dry substrate) of enzyme was added and incubated at 60 °C (for EpXYN1) or 50 °C (for EpXYN3), or 80 U/g of enzyme was added and incubated at different temperatures. For double enzymes hydrolysis, 40 or 80 U/g of EpXYN1 with the same amount of EpXYN3 were added and incubated at 50 °C. The reaction mixtures were incubated at 1,500 rpm up to 48 h on a thermo shaker, and stopped at 100 °C for 15 min. After centrifugation at 10,000g for 10 min, 100 μL of suspension was diluted with 100 mM MOPS (3-(N-morpholino)-propanesulfonic acid, pH 6.0) buffer. The same enzyme was deactivated by incubation at 100 °C for 20 min and used as a control. FO content in the suspension was determined spectrophotometrically by measuring absorbance at 286 and 323 nm, and calculated according to the following formula [26].
The letter b indicates the path length of sample (cm). The molar absorption coefficients, which determined at pH 6.0 in 100 mM MOPS, were Ɛ286 = 14,176 mol/L/cm and Ɛ323 = 10,350 mol/L/cm for free FA, and Ɛ′286 = 12,465 mol/L/cm and Ɛ′323 = 19,345 mol/L/cm for esterified FA [26]. To verify the efficiency of spectrophotometry measure, free FA in hydrolytic mixture with or without alkaline treatment was determined by an Agilent 1260 Infinity high-performance liquid chromatography (HPLC) system (Agilent, USA) equipped with a ZORBAX Eclipse Plus C18 reversed-phase column [27]. A mobile phase of methanol to 0.1% (v/v) acetic acid (v/v, 35:65) was used and monitored at 320 nm.
Bioinformatics Analysis
BioEdit 7.0 software was used to assemble and translate the nucleic acid sequences. Protein sequences were analyzed by BLAST on line (http://www.NCBI.nim.nih.gov/). Putative signal peptides were predicted with SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Theoretical pI and molecular weight (Mw) of target proteins were calculated with ExPASy Server (http://web.expasy.org/compute_pi/). ClustalX 1.83 software and Mega 6 program were used for multiple sequences alignment and construction of phylogenetic tree, respectively [33]. Structure-based sequence alignment was generated using ESPript 3.0 [34].
Statistics Analysis
For statistical analysis, one-way analysis of variance (ANOVA) was performed with the SPSS 20 software using LSD test. A P value of less than 0.05 was considered statistically significant.
Results
Molecular Characterization of EpXYN1 and EpXYN3
The integrated encoding region of xylanases genes EpXyn1 (1212 bp) and EpXyn3 (660 bp) were cloned from E. parvum 4–14, and the cDNA sequences were deposited in GenBank under the accession numbers MF682063 and MF682064, respectively. Signal peptides were predicted at the N-terminus of deduced proteins EpXYN1 or EpXYN3 by the SignalP 4.1 Server, indicating that the two proteins are extracellular enzymes. Theoretical pI and Mw are 4.97 and 40.75 kDa for mature EpXYN1, or 5.21 and 21.58 kDa for mature EpXYN3. By analysis of protein sequences with BLAST tool, EpXYN1 has a typical catalytic domain of GH10 xylanases, a Ser/Thr-rich linker region, and a carbohydrate-binding module (CBM), and EpXYN3 contains a typical catalytic domain of GH11 xylanase (Fig. 1a, b). Structure-based sequence alignments revealed that EpXYN1 had typical (β/α)8 elements of GH10 xylanases, EpXYN3 had a typical β-Jelly roll fold of GH11 xylanases, and the conserved catalytic residues Glu existed in these protein sequences (Fig. 1a, b).
Structure-based sequence alignment of enzymes EpXYN1 (a) or EpXYN3 (b) with related xylanases. a The secondary structural elements of EpXYN1 using the xylanase FoXyn10a from Fusarium oxysporum as a template (PDB NO. 3U7B). XYN-Ps, xylanase D (OKO90312.1) from Penicillium subrubescens; XYN-Pb, a xylanase (OOQ87260.1) from P. brasilianum; XYN-An, a xylanase from Aspergillus niger (AIC36735), CBM1, carbohydrate-binding module family 1. b The secondary structural elements of EpXYN3 using the xylanase TcXylC from Talaromyces cellulolyticus as a template (PDB NO. 3WP3). XynG1, a xylanase from A. fischeri (KF233756); XYNB-An, xylanase B from Aspergillus niger (ACA24724.1); XYN-Ao, a xylanase from A. oryzae (AFA51067); GH11-Po, a member of glycoside hydrolase family 11 from P. expansum (XP_016599740). The secondary-structure elements α-helices, β-strands, and strict β-turns are denoted as squiggles, arrows, and letters TT, respectively. The conserved catalytic residues Glu are rendered as arrows. The figure was constructed with ESPript 3.0
BLAST analysis indicated that EpXYN1 was mostly related to endo-1,4-beta-xylanase (GenBank: OOQ87260) from P. brasilianum with a 82% identity, and EpXYN3 was mostly related to putative endo-1,4-beta-xylanase (XynG1, GenBank: OOQ87260.1) from A. fischeri with a 68% identity. The constructed phylogenetic trees showed that EpXYN1 and EpXYN3 were affiliated with GH10 or GH11 xylanases, respectively (supplementary Fig. S1).
Expression and Purification of EpXYN1 and EpXYN3
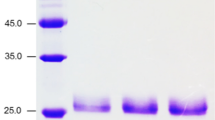
Xylanase activities were detected in the fermentation liquid of selected yeast transformants. The highest activities were produced after 5 or 7 days induction with methanol for xylanases EpXYN1 and EpXYN3, respectively (supplementary Fig. S2). The recombinant proteins were purified to near-homogeneity and analyzed on SDS-PAGE gel (Fig. 2). The result of electrophoresis showed that the actual Mw (approximately 53 kDa) of recombinant EpXYN1 was obviously larger than the theoretical value of 41.58 kDa (including His-tag). At the same time, the Mw of EpXYN1 did not change after treatment with endoglycosidases Endo Hf. The actual Mw of recombinant EpXYN3 was similar to the theoretical value (Fig. 2).
Optimal pHs and Temperatures
Recombinant EpXYN1 and EpXYN3 displayed the maximum activities at pH values 5.5 and 5.0 toward three xylans, respectively (Fig. 3a, b). The relative enzyme activities decreased to less than 50% when adjusted the pH to 7.5 for EpXYN1, or 7.0 for EpXYN3. Reducing the pH to 3.5 led to 47–63% decrements of these activities. The two enzymes maintained a high activity (above 87%) at the pH values from 4.5 to 5.5 using beechwood xylan or birchwood xylan as substrate. EpXYN1 showed the maximum activity at 75 °C and kept about 90% activity at 80 °C toward different xylans (Fig. 3c). At a lower temperature, the enzyme activity was declined. At the same time, the optimal temperature of EpXYN3 was 55 °C, and quickly lost the activity with the increment of temperature (Fig. 3d).
Effect of pH and temperature on the activities of recombinant xylanases EpXYN1 or EpXYN3. a, b Optimal pHs of recombinant EpXYN1 and EpXYN3, respectively. The enzyme activities were assayed at 55 °C for EpXYN1 or 75 °C for EpXYN3 in universal pH buffers (pH values 3.0–9.5). c, d Optimal temperatures of recombinant EpXYN1 and EpXYN3, respectively. The enzymatic activities were determined in 50 mM sodium citrate buffer (pH 4.8) at different temperatures. Beechwood xylan (BeXy), birchwood xylan (BiXy), or oat spelt xylan (OaXy) were using as substrates, respectively. The relative activities were calculated using the maximum enzyme activity as 100%. Error bars represent standard deviations from three repeated detections
pH Stability and Thermostability of Recombinant Xylanases
The results indicated that EpXYN1 was stable at low pH values (3.0 to 4.0) and high pH values (9.0 to 10), and lost 24–35% activity after treated at the pH values from 5.0 to 8.0. EpXYN3 kept over 80% activity at different pH values, and the lost activity less than 3% at the pH values from 7.0 to 9.0 (Fig. 4). The thermostability test showed EpXYN1 was stable at 65 °C, and sharply lost the activity when treated at higher temperatures (Fig. 5a). At the same time, EpXYN3 quickly lost the activity at 55 °C, and displayed high stability at 45 °C (Fig. 5b).
pH stability of recombinant xylanases EpXYN1 or EpXYN3. Purified enzyme (1 μg/μL) were stored in universal buffers with different pH values for 12 h at 4 °C, and the residual activities were determined under standard conditions using beechwood xylan as substrate. The relative activities were calculated using the enzyme activity of untreated samples as 100%. Error bars represent standard deviations from three repeated detections
Thermostability of recombinant xylanases EpXYN1 (a) or EpXYN3 (b). Purified enzymes (1 μg/μL) were incubated at different temperatures up to 90 min. The residual activities were determined under standard conditions using beechwood xylan as substrate. The relative activities were calculated using the enzyme activity of untreated samples as 100%. Error bars represent standard deviations from three repeated detections
Effect of Metal Ions and Chemicals on Enzymatic Activity
The effects of metal ions or chemicals on the activities of recombinant xylanases were evaluated using beechwood xylan as substrate. As described in Table 2, the activities of EpXYN1 and EpXYN3 were slightly decreased when addition of Ca2+, Zn2+, Mg2+, Cu2+, Co2+, or Ni2+ at a high concentration (5 mM). High concentration Fe3+ showed strong repression effects on these activities. The addition of 0.1 or 1 mM EDTA led to 16.4 and 17.2% increments on the activity of EpXYN1. SDS with a final concentration of 3.5 mM displayed sharp inhibition on the activities of EpXYN1 and EpXYN3.
Specific Activity and Kinetic Parameters
The specific activities of EpXYN1 with three xylans as substrates were higher than these of EpXYN3. For the two enzymes, the specific activities toward birchwood xylan were higher than the other two xylans (Table 3). The kinetic constants of two enzymes toward different substrates were also listed in Table 3. EpXYN1 and EpXYN3 had the lowest Michaelis constant (Km) values of 0.85 ± 0.12 or 0.95 ± 0.18 mg/mL toward beechwood xylan, respectively. Using birchwood xylan or oat spelt xylan as substrate, the Km values were 1.13 ± 0.08 or 2.07 ± 0.35 mg/mL for EpXYN1, and 3.84 ± 0.61 or 3.58 ± 0.46 mg/mL for EpXYN1, respectively. The maximum initial velocities (Vmax) of EpXYN1 on beechwood or oat spelt xylan were higher than those of EpXYN3. The two enzymes had similar Vmax values on birchwood xylan. The kcat values of EpXYN1 were higher than these of EpXYN3 toward three xylans.
TLC Analysis of Hydrolytic Products
TLC analysis showed that different xylans and XOS (except xylobiose) were efficiently hydrolyzed by the two xylanases (Fig. 6). For EpXYN1, the main hydrolytic products of three xylans was xylobiose, followed by xylotriose and unidentified longer XOS, and appeared a small amount of xylose with the extended hydrolysis time (Fig. 6a–c). All of the XOS except xylobiose were hydrolyzed by EpXYN1, and xylobiose with a small amount of xylose were the main hydrolytic product (Fig. 6d). For EpXYN3, xylotriose, xylobiose, and some longer XOS were the main hydrolytic products from the three xylans (Fig 6e–g). Xylobiose could not be hydrolyzed by EpXYN3. Xylotriose was partially degraded to xylobiose and xylose by the enzyme. The longer XOS (X4 to X6) were efficiently hydrolyzed by EpXYN3 (Fig. 6h).
TLC analysis of the hydrolytic products of various xylans or xylo-oligosaccharides by recombinant xylanases. a–d Hydrolysis profiles of beechwood xylan, birchwood xylan, oat spelt xylan, or xylo-oligosaccharides by EpXYN1, respectively. e–h Hydrolysis profiles of beechwood xylan, birchwood xylan, oat spelt xylan, or xylo-oligosaccharides by EpXYN3, respectively. In a 2 mL sodium citrate buffer (pH 4.8, 50 mM), 20 mg xylans (10 mg of beechwood xylan) were mixed with 25 μg of pure enzymes and incubated at standard temperatures for different time. Different xylo-oligosaccharides (X2 to X6, 1 mg/mL) were hydrolyzed by the enzymes (10 μg/mL) under the same conditions for 24 h. S, xylose (X1) and xylo-oligosaccharides (X2 to X6) were used as standards
Production of Feruloylated Oligosaccharides from Wheat Bran
Different amounts of FOs were extracted from DSWB by different enzymatic treatments. The addition of 8 U/g of EpXYN1 or EpXYN3 led to release 4.4 or 9.2 μmol/g FOs from DSWB, respectively. Increasing of the enzyme dosage to 40 U/g led to the extract levels of FOs up to 7.5 μmol/g for EpXYN1 or 13.0 μmol/g for EpXYN3. Further increment of enzyme dosage significantly enhanced the FOs release by EpXYN1 but EpXYN3 (Fig. 7a). The optimum enzyme dosages were 80 U/g for EpXYN1 and 40 U/g for EpXYN1 in releasing FOs from DSWB.
Release of feruloylated oligosaccharides (FOs) from de-starched wheat bran (DSWB) by enzymatic hydrolysis. a Comparison of the FOs release from DSWB by different enzyme dosage. Total 0–160 U/g (dry substrate) of xylanase was added into the reaction mixture, and incubated at 60 °C (EpXYN1) or 50 °C (EpXYN3) for 24 h. b, c Time course of FOs release from DSWB by EpXYN1 (b) or EpXYN3 (c) under different temperatures. The enzyme dosage was 80 U/g for each treatment. d Comparison of the FOs release from DSWB by single xylanase and double xylanases. Single enzyme group, 80 U/g of enzyme was added and incubated at 60 °C (EpXYN1) or 50 °C (EpXYN3) for 48 h. Double enzymes I, the amount of EpXYN1 and EpXYN3 was 40 U/g. Double enzymes II, the amount of EpXYN1 and EpXYN3 was 80 U/g. The hydrolysis of double enzymes was conducted at 50 °C for 48 h. The concentration of FOs was determined by spectrophotometry method. Each data is the average of three replications. Different letters above the error bars indicate significant differences by LSD test (P ≤ 0.05)
The time curves of FOs release from DSWB by xylanases were analyzed at different temperatures. For EpXYN1, the highest level of FOs was released when enzymatic hydrolysis at 60 °C. The FOs levels were decreased at higher or lower incubation temperatures (Fig. 7b). Enzymatic hydrolysis with EpXYN3, the releasing level of FOs at 50 °C was higher than that at 45 °C. Raising the temperature of incubation led to a lower extract level of FOs compared to that at 50 or 45 °C (Fig. 7c). For the two enzymes, the optimum time was 36 h of incubation in releasing FOs from DSWB (Fig. 7b, c). At the same time, the releasing levels of FOs were compared among different treatments with single enzyme or double enzymes. For the single hydrolysis, EpXYN3 released more FOs from the substrate than that by EpXYN1 under the optimal temperatures. EpXYN3 combined with EpXYN1 did not significantly change the extract level compared to single EpXYN3 (Fig. 7d). Under the experimental conditions, the maximum levels of released FOs were 11.1 μmol/g for EpXYN1, and 14.4 μmol/g for EpXYN3. For same samples, the FOs contents determined by spectrophotometry measure (10.66 ± 0.56 and 12.93 ± 0.90 μmol/g) were close to those of HPLC analysis (9.97 ± 0.41 and 11.80 ± 1.10 μmol/g) (supplementary Table S1).
Discussion
Xylanases are attractive enzymes in many industrial fields including biofuel production, paper and pulp industry, and food and health industry [6, 10]. In this study, two new xylanases encoding genes EpXyn1 and EpXyn3 were isolated from E. parvum 4–14. EpXyn1 gene encodes a high MW xylanase EpXYN1 (404 aa) belonging to GH10 family. As some other GH10 xylanases [35, 36], a CBM family 1 domain was predicted in the C terminal of EpXYN1, suggesting its potential ability of binding to cellulose. EpXyn3 gene encodes a low MW of xylanase EpXYN3 (220 aa) which belongs to GH11 family. EpXYN1 and EpXYN3 have low identities (82 and 68%) with known xylanases.
Pure xylanases were obtained by inducible expression of EpXyn1 or EpXyn3 genes in P. pastoris cells. The actual Mw of recombinant EpXYN1 was larger than the theoretical value of the protein. Deglycosylation treatment with EndoHf (action on N-linked glycosylation) could not decrease the size of EpXYN1. Shibata et al. reported a same phenomenon on the GH10 xylanase PspXyn10 from Penicillium sp. [35]. They speculated that O-linked glycosylation led to the enlargement of the molecular size, because the linker region of fungal cellulases with a CBM1 domain was highly modified by O-glycosylation [35]. This explanation may be the case of EpXYN1. EpXYN1 and EpXYN3 displayed best catalytic activities at low pH values (4.5 to 5.5) that is in conformity with the action condition of many industrially important cellulolytic enzymes (such as from T. reesei). Acid pretreatments have gained much interest as one of important methods for promoting enzymatic saccharification of biomass materials [37]. These acid enzymes are more suitable for bioconversion of biomass such as preparation of monosaccharides and oligosaccharides from brans, especially with an acid pretreatment procedure [38]. EpXYN1 showed the best catalytic ability at 75 °C and a high thermal stability, which is in conformity with previous study [27]. Thermostable enzymes have excellent performances in bioprocessing applications [17]. The new thermophilic xylanase EpXYN1 could be used in many fields with high-temperature process such as textile, pulp, and paper industries. Enhanced thermostability of xylanases involved in many unique strategies such as an increment of salt bridges and hydrogen, introduction of disulfide bridges at the N- or C- termini or in the helix regions, and improved internal packing [5]. It was reported that the CBM1 domain and linker region was important for the thermostability of GH10 xylanase Xyn10A from A. fumigatus Z5 [36]. Structural analysis of EpXYN1 protein should be done for understanding the thermotolerant mechanism. At the same time, the optimum temperature of EpXYN3 indicates that it is a mesophilic xylanase.
The specific activities and kinetic parameters of EpXYN1 or EpXYN3 toward different xylans were similar to those of reported fungal xylanases [7, 37, 38]. The two xylanases displayed higher substrate affinities (lower Km values) toward beechwood xylan than birchwood xylan and oat spelt xylan, that is inconsistent with other xylanases from P. oxalicum, T. reesei, and A. nidulans [7, 39, 40]. Beechwood xylans have good solubility and more enzyme access due to its few modifications and containing approximately 94% xylose [7]. It is suggested that beechwood xylan is more suitable substrate for these fungal xylanases. TLC analysis indicated that the test xylans were mainly hydrolyzed into xylobiose or longer XOS by EpXYN1 and EpXYN3. It was suggested that the enzymes randomly cleaved the xylan backbone. XOS possess prebiotic effects, which can be used in functional foods, animal feed, and pharmaceuticals [13, 41, 42]. The two new xylanases are valuable for the production of XOS from biomass. In addition, xylotriose could be hydrolyzed into xylobiose and xylose by EpXYN1 but EpXYN3. This is in agreement with the conclusion that GH11 family xylanases have higher substrate specificity than the xylanases in GH10 family [5].
As excellent functional ingredients, FOs can be used in health foods and pharmaceutical industries [24, 25]. FOs widely exist in agricultural by-products including corn bran and wheat bran [25]. A GH10 family xylanase from Thermoascus aurantiacus released 12 μmol of FeXOSs (feruloylated xylo-oligosaccharides) from 1 g of pretreated corn cobs [26]. In this study, the xylanases EpXYN1 and EpXYN3 had good performance at releasing of FOs from DSWB. Under the test conditions, the maximum levels of 11.1 and 14.4 μmol/g of FOs were released by EpXYN1 and EpXYN3, respectively. Previous study indicated that 26.3 μmol/g (0.51%, w/w) of FA was released from DSWB by alkaline extraction [27]. Calculation based on this data, the releasing rates of FOs were 42.2 and 54.8% by EpXYN1 and EpXYN3, respectively. The enzymatic hydrolysis with double enzymes did not enhance the extract level compared to single EpXYN3. GH11 xylanase EpXYN3 showed a higher efficiency than GH10 xylanase EpXYN1 at releasing of FOs from DSWB under the test conditions. This result is in agreement with the findings of Beaugrand et al., which compared the liberation of XOS from wheat bran by GH11 and GH10 xylanases from Thermobacillus xylanilyticus [43]. Wheat cell walls are composed of a complex interconnected network with pore radius ranging from 1.5 to 4 nm [44]. Proteins with molecular weights greater than 20 kDa (Stokes radius 2.5 nm) would be excluded from most numerous of the pores in the wheat samples [44]. It is suggested that the high MW of xylanase EpXYN1 (approximately 53 kDa) would have more difficulty to penetrate the bran tissues compared to the low MW of EpXYN3 (21.58 kDa).
In conclusion, we have reported the genes, characterizations, and application of two new endo-1,4-beta-xylanases from E. parvum 4–14. The most important feature of EpXYN1 lies in its high-temperature catalysis (75 °C) and high thermostability. Main hydrolytic products of three commercial xylans by the two xylanases are xylobiose and xylotriose. EpXYN3 showed a higher efficiency in releasing FOs from wheat bran. The characteristics of two xylanases provide a potential way for industrial preparations of FOs or XOS from biomass. In future studies, understanding the synergistic reactions of the enzymes with other hemicelluloses is important for industrial applications.
References
Peng, F., Peng, P., Xu, F., & Sun, R. C. (2012). Fractional purification and bioconversion of hemicelluloses. Biotechnology Advances, 30(4), 879–903.
Liu, J., Chinga-Carrasco, G., Cheng, F., Xu, W., Willför, S., Syverud, K., & Xu, C. (2016). Hemicellulose-reinforced nanocellulose hydrogels for wound healing application. Cellulose, 23(5), 3129–3143.
Li, H., Wu, H., Jiang, F., Wu, J., Xue, Y., Gan, L., Liu, J., & Long, M. (2018). Heterologous expression and characterization of an acidic GH11 family xylanase from Hypocrea orientalis. Applied Biochemistry and Biotechnology, 184, 228–238.
Zhou, X., Li, W., Mabon, R., & Broadbelt, L. J. (2017). A critical review on hemicellulose pyrolysis. Energy Technology, 5(1), 52–79.
Collins, T., Gerday, C., & Feller, G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiology Reviews, 29(1), 3–23.
Chakdar, H., Kumar, M., Pandiyan, K., Singh, A., Nanjappan, K., Kashyap, P. L., & Srivastava, A. K. (2016). Bacterial xylanases: Biology to biotechnology. 3 Biotech, 6(2), 150.
Liao, H., Zheng, H., Li, S., Wei, Z., Mei, X., Ma, H., Shen, Q., & Xu, Y. (2015). Functional diversity and properties of multiple xylanases from Penicillium oxalicum GZ-2. Scientific Reports, 5,12631.
Juodeikiene, G., Basinskiene, L., Vidmantiene, D., Makaravicius, T., & Bartkiene, E. (2012). Benefits of β-xylanase for wheat biomass conversion to bioethanol. Journal of the Science of Food and Agriculture, 92(1), 84–91.
Murugesan, G. R., & Persia, M. E. (2015). Influence of a direct-fed microbial and xylanase enzyme on the dietary energy uptake efficiency and performance of broiler chickens. Journal of the Science of Food and Agriculture, 95(12), 2521–2527.
Juturu, V., & Wu, J. C. (2012). Microbial xylanases: Engineering, production and industrial applications. Biotechnology Advances, 30(6), 1219–1227.
Chutani, P., & Sharma, K. K. (2015). Biochemical evaluation of xylanases from various filamentous fungi and their application for the deinking of ozone treated newspaper pulp. Carbohydrate Polymers, 127, 54–63.
Zhang, H.-M., Wang, J.-Q., Wu, M.-C., Gao, S.-J., Li, J.-F., & Yang, Y.-J. (2014). Optimized expression, purification and characterization of a family 11 xylanase (AuXyn11A) from Aspergillus usamii E001 in Pichia pastoris. Journal of the Science of Food and Agriculture, 94(4), 699–706.
Morgan, N. K., Wallace, A., Bedford, M. R., & Choct, M. A. (2017). Efficiency of xylanases from families 10 and 11 in production of xylo-oligosaccharides from wheat arabinoxylans. Carbohydrate Polymers, 167, 290–296.
Miao, Y., Li, P., Li, G., Liu, D., Druzhinina, I. S., Kubicek, C. P., Shen, Q., & Zhang, R. (2017). Two degradation strategies for overcoming the recalcitrance of natural lignocellulosic xylan by polysaccharides-binding GH10 and GH11 xylanases of filamentous fungi. Environmental Microbiology, 19(3), 1054–1064.
Meng, D.-D., Ying, Y., Chen, X.-H., Lu, M., Ning, K., Wang, L.-S., & Li, F.-L. (2015). Distinct roles for carbohydrate-binding modules of glycoside hydrolase 10 (GH10) and GH11 xylanases from Caldicellulosiruptor sp. strain F32 in thermostability and catalytic efficiency. Applied and Environmental Microbiology, 81(6), 2006–2014.
Tang, F., Chen, D., Yu, B., Luo, Y., Zheng, P., Mao, X., Yu, J., & He, J. (2017). Improving the thermostability of Trichoderma reesei xylanase 2 by introducing disulfide bonds. Electronic Journal of Biotechnology, 26, 52–59.
Kumar, V., Marín-Navarro, J., & Shukla, P. (2016). Thermostable microbial xylanases for pulp and paper industries: Trends, applications and further perspectives. World Journal of Microbiology and Biotechnology, 32(2), 34.
Du, Y., Shi, P., Huang, H., Zhang, X., Luo, H., Wang, Y. R., & Yao, B. (2013). Characterization of three novel thermophilic xylanases from Humicola insolens Y1 with application potentials in the brewing industry. Bioresource Technology, 130, 161–167.
Silva, S. B., Pinheiro, M. P., Fuzo, C. A., Silva, S. R., Ferreira, T. L., Lourenzoni, M. R., Nonato, M. C., Vieira, D. S., & Ward, R. J. (2017). The role of local residue environmental changes in thermostable mutants of the GH11 xylanase from Bacillus subtilis. International Journal of Biological Macromolecules, 97, 574–584.
Gopalan, N., Rodriguez-Duran, L. V., Saucedo-Castaneda, G., & Nampoothiri, K. M. (2015). Review on technological and scientific aspects of feruloyl esterases: A versatile enzyme for biorefining of biomass. Bioresource Technology, 193, 534–544.
Kumar, N., & Pruthi, V. (2014). Potential applications of ferulic acid from natural sources. Biotechnology Reports, 4, 86–93.
Uraji, M., Arima, J., Inoue, Y., Harazono, K., & Hatanaka, T. (2014). Application of two newly identified and characterized feruloyl esterases from Streptomyces sp. in the enzymatic production of ferulic acid from agricultural biomass. PLoS One, 9(8), e104584.
Lapierre, C., Pollet, B., Ralet, M.-C., & Saulnier, L. (2001). The phenolic fraction of maize bran: Evidence for lignin-heteroxylan association. Phytochemistry, 57(5), 765–772.
Ou, J., & Sun, Z. (2014). Feruloylated oligosaccharides: Structure, metabolism and function. Journal of Functional Foods, 7, 90–100.
Xie, C., Wu, Z., Guo, H., & Gu, Z. (2014). Release of feruloylated oligosaccharides from wheat bran through submerged fermentation by edible mushrooms. Journal of Basic Microbiology, 54(S1), S14–S20.
Katapodis, P., & Christakopoulos, P. (2008). Enzymic production of feruloyl xylo-oligosaccharides from corn cobs by a family 10 xylanase from Thermoascus aurantiacus. Lwt-Food Science and Technology, 41(7), 1239–1243.
Long, L., Ding, D., Han, Z., Zhao, H., Lin, Q., & Ding, S. (2016). Thermotolerant hemicellulolytic and cellulolytic enzymes from Eupenicillium parvum 4-14 display high efficiency upon release of ferulic acid from wheat bran. Journal of Applied Microbiology, 121(2), 422–434.
Zheng, F., Huang, J., Liu, X., Hu, H., Long, L., Chen, K., & Ding, S. (2016). N- and C-terminal truncations of a GH10 xylanase significantly increase its activity and thermostability but decrease its SDS resistance. Applied Microbiology and Biotechnology, 10, 3555–3565.
Turner, B. L. (2010). Variation in pH optima of hydrolytic enzyme activities in tropical rain forest soils. Applied and Environmental Microbiology, 76(19), 6485–6493.
Mi, S., Jia, X., Wang, J., Qiao, W., Peng, X., & Han, Y. (2014). Biochemical characterization of two thermostable xylanolytic enzymes encoded by a gene cluster of Caldicellulosiruptor owensensis. PLoS One, 9(8), e105264.
Amel, B.-D., Nawel, B., Khelifa, B., Mohammed, G., Manon, J., Salima, K.-G., Farida, N., Hocine, H., Bernard, O., Jean-Luc, C., & Marie-Laure, F. (2016). Characterization of a purified thermostable xylanase from Caldicoprobacter algeriensis sp. nov. strain TH7C1T. Carbohydrate Research, 419, 60–68.
Liao, H., Sun, S., Wang, P., Bi, W., Tan, S., Wei, Z., Mei, X., Liu, D., Raza, W., Shen, Q., & Xu, Y. (2014). A new acidophilic endo-β-1,4-xylanase from Penicillium oxalicum: Cloning, purification, and insights into the influence of metal ions on xylanase activity. Journal of Industrial Microbiology and Biotechnology, 41(7), 1071–1083.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729.
Robert, X., & Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(W1), W320–W324.
Shibata, N., Suetsugu, M., Kakeshita, H., Igarashi, K., Hagihara, H., & Takimura, Y. (2017). A novel GH10 xylanase from Penicillium sp. accelerates saccharification of alkaline-pretreated bagasse by an enzyme from recombinant Trichoderma reesei expressing Aspergillus β-glucosidase. Biotechnology for Biofuels, 10(1), 278.
Miao, Y., Kong, Y., Li, P., Li, G., Liu, D., Shen, Q., & Zhang, R. (2018). Effect of CBM1 and linker region on enzymatic properties of a novel thermostable dimeric GH10 xylanase (Xyn10A) from filamentous fungus Aspergillus fumigatus Z5. AMB Express, 8(1), 44.
Rattanaporn, K., Tantayotai, P., Phusantisampan, T., Pornwongthong, P., & Sriariyanun, M. (2018). Organic acid pretreatment of oil palm trunk: Effect on enzymatic saccharification and ethanol production. Bioprocess and Biosystems Engineering, 41(4), 467–477.
Yuan, L., Scanlon, M. G., Eskin, N. A., Thiyam-Hollander, U., & Aachary, A. A. (2015). Effect of pretreatments and endo-1,4-β-xylanase hydrolysis of canola meal and mustard bran for production of oligosaccharides. Applied Biochemistry and Biotechnology, 175(1), 194–208.
He, J., Chen, D., Yu, B., & Zhang, K. (2010). Optimization of the Trichoderma reesei endo-1,4-beta-xylanase production by recombinant Pichia pastoris. Biochemical Engineering Journal, 52(1), 1–6.
Maitan-Alfenas, G. P., Oliveira, M. B., Nagemc, R. A. P., de Vries, R. P., & Guimarães, V. M. (2016). Characterization and biotechnological application of recombinant xylanases from Aspergillus nidulans. International Journal of Biological Macromolecules, 91, 60–67.
Kumar, V., Dangi, A. K., & Shukla, P. (2018). Engineering thermostable microbial xylanases toward its industrial applications. Molecular Biotechnology, 60(3), 226–235.
Ma, R., Bai, Y., Huang, H., Luo, H., Chen, S., Fan, Y., Cai, L., & Yao, B. (2017). Utility of thermostable xylanases of Mycothermus thermophilus in generating prebiotic xylooligosaccharides. Journal of Agricultural and Food Chemistry, 65(6), 1139–1145.
Beaugrand, J., Chambat, G., Wong, V. W. K., Goubet, F., Rémond, C., Paës, G., Benamrouche, S., Debeire, P., O'Donohue, M., & Chabbert, B. (2004). Impact and efficiency of GH10 and GH11 thermostable endoxylanases on wheat bran and alkali-extractable arabinoxylans. Carbohydrate Research, 339(15), 2529–2540.
Chesson, A., Gardner, P. T., & Wood, T. J. (1997). Cell wall porosity and available surface area of wheat straw and wheat grain fractions. Journal of the Science of Food and Agriculture, 75(3), 289–295.
Funding
This work was supported by grants from a 948 Research Project (No. 2013-4-16) from State Forestry Administration of China, the Natural Science Fund for Provincial Colleges and University of Jiangsu Province, China (15KJB220003), the Research Fund for the Advanced Talents, Nanjing Forestry University (GXL201311), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 275 kb)
Rights and permissions
About this article
Cite this article
Long, L., Xu, M., Shi, Y. et al. Characterization of Two New Endo-β-1,4-xylanases from Eupenicillium parvum 4–14 and Their Applications for Production of Feruloylated Oligosaccharides. Appl Biochem Biotechnol 186, 816–833 (2018). https://doi.org/10.1007/s12010-018-2775-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2775-6