Abstract
A bacterium Pseudomonas sp. TRMK1 capable of utilizing various phenylpropanoids was isolated from agro-industrial waste by enrichment culture technique. It is gram-negative, motile, aerobic, and able to utilize three different phenolic acids such as p-coumaric, ferulic, and caffeic acids at concentrations of 5, 10, and 15 mM in 18 h of incubation. The residual concentration of phenolic acids was analyzed by HPLC. The catabolic pathway of p-coumaric, ferulic, and caffeic acids is suggested based on the characterization of metabolic intermediates by GC, GC-HRMS, and different enzymatic assays. Further, Pseudomonas sp. TRMK1 utilizes a wide range of mixture of phenolic acids present in the synthetic effluent.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Agro-industrial operations cause major environmental pollution owing to the discharge of natural phenolic compounds. These effluents are discharged by wine distilleries, during the extraction of olive oil, and in cork processing [1–3]. Chemically, agro-industrial wastes comprise sugars, tannins, polyphenols, polyalcohols, pectins, lipids, and a variety of simple aromatic compounds [4]. The major phenolic compounds in agro-industrial waste include p-coumaric, caffeic, ferulic, cinnamic, protocatechuic, syringic, vanillic, veratric, and p-hydroxyphenylacetic acids, tyrosol, and hydroxytyrosol [5].
Cinnamic acid derivatives, namely, ferulic and p-coumaric acids, are plant secondary products and are widely distributed in the cell walls of gramineous plants extensively ester-linked to lignin or polysaccharides [6]; these are found in soil as breakdown products of lignin [7]. The wastewater released by olive mills contains phenolic compounds and causes a major environmental problem [2]. High accumulation of phenolic acids in soil inhibits the growth of plants in their vicinity [8–10] and causes partition and loss of cell membrane of the fermenting organisms by reducing cell growth and sugar assimilation [11]. Above 1 mM accumulation, phenolic acids in soil inhibit the growth of many species of ruminal bacteria [12]. They also delay the metabolism of sugars and citric acid by wine lactic acid bacteria (LAB) [13, 14]. Phenolic compounds have considerable inhibitory effect and are more toxic than furfural and hydroxymethylfurfural [11].
The biodegradation of phenolic acids is important in global carbon cycle [15], where carbon in the atmosphere photosynthesizes to produce new plant materials. Several reports have described the biodegradation of p-coumaric, ferulic, and caffeic acids by fungi [16, 17] and yeast [18], but limited work is reported on bacteria [9, 19–21].
Biodegradation is an eco-friendly and cost-effective process to reduce the toxic level of phenolic acids in contaminated soil; biotransformation also yields valuable products. The phenolic acids are converted to hydroxybenzoic acids, and their derivatives find important application as dietary antioxidants [22], natural flavors [23], preservatives, and medicines and as monomers for liquid crystal polymers currently used in various electronic devices [24]. Ferulic acid is used as a starting material to produce vanillin (4-hydroxy-3-methoxybenzaldehyde), an important aroma compound used in beverages and other food industries, such as bread, cake, ice cream, chocolate, and confectionery products, as well as fragrances [25].
In the present communication, a bacterium proficient in utilizing various phenolic compounds was isolated and identified as Pseudomonas sp. TRMK1. The capability of this strain to utilize most of the phenylpropanoids, mainly p-coumaric, ferulic, and caffeic acids, has been demonstrated. Its ability was demonstrated in the utilization of a wide range of mixtures of phenolic acids present in the synthetic effluent. Further, catabolic pathways for these compounds have been proposed based on metabolite characterization and enzymatic assays.
Materials and Methods
Isolation, Media, and Growth Conditions
A bacterium capable of utilizing phenolic acids as the sole source of carbon and energy was isolated by a selective enrichment culture method [26]. One gram of agro-industrial waste was added to a 250 ml Erlenmeyer flask containing 50 ml of an autoclaved mineral salt (MS1) medium of pH 7 comprising the following (g L−1): K2HPO4 6.8; KH2PO4 1.2; NH4NO3 1.0; MgSO4.7H2O 0.1; MnSO4.4H2O 0.1; CaCl2.2H2O 0.1; FeSO4.7H2O 0.1; and Na2MoO7.2H2O 0.006 supplemented with 5 mM p-coumaric acid. Flasks were incubated in an orbital shaker (Labtech, India) (180 rpm) at 30 °C for 15 days. Further enrichment was carried out by transferring 5 % inoculum to fresh MS1 medium supplemented with 5 mM p-coumaric acid. After several such transfers, the culture was streaked on MS1 agar plates spread with p-coumaric acid. The purity of the culture was checked by plating on Luria–Bertani (LB) agar.
16S rDNA Sequencing
The 16S ribosomal DNA (rDNA) of the potent isolate TRMK1 was amplified by PCR using the universal primer pair F27, 5′-AGAGTTTGATCMTGGCTCAG-3′ and R1492 5′-TACGGYTACCTTGTTACGACTT-3′ [27]. The amplified products were examined by 0.7 % (w/v) agarose gel electrophoresis and purified by using a DNA extraction kit (Qiagen). The 16S ribosomal DNA sequencing was done by ABI PRISM 310 DNA sequencer and ABI PRISM BigDye Terminator Cycle Sequencing Kit. DNA sequence analysis was then executed by BLAST network services at the National Center for Biotechnology Information (NCBI). The arrangement of the sequences was done using CLUSTALQ pro V1.82 at the European bioinformatics site (www.ebi.ac.uk/clustalw). The sequence was polished manually after double-checking with the raw data to evacuate ambiguities and was submitted to GenBank. Using MEGA version 4, molecular evolutionary analysis was made and a phylogenetic tree was constructed by a Neighbor-Joining method (data not shown) [28]. The 16S rDNA sequence of strain TRMK1 was deposited and is accessible in the NCBI GenBank database with accession numbers KT717679.
Utilization of Phenolic Acids
Utilization of phenolic acids was carried out by inoculating Pseudomonas sp. TRMK1 into 250 ml flasks containing 50 ml of MS1 medium supplemented with p-coumaric, ferulic, and caffeic acids individually. Three different concentrations, 5, 10, and 15 mM phenolic acids were used to study the influence of their concentrations on the growth of this strain. All these flasks were incubated in an orbital shaker at 180 rpm and 30 °C. The utilization of phenolic acids was determined by checking their residual concentration at different time intervals. At regular intervals of 3 h up to the final incubation period of 18 h, the culture was withdrawn and centrifuged (8000 rpm, 10 min). The resulting supernatant was filtered using a 0.2 μm filter paper, and the residual concentrations of phenolic acids were estimated by HPLC.
Phenolic acid % utilization, ή, at different periods of incubation is defined by the following expression:
where S0 represents the initial concentration of phenolic acid (%) prior to utilization by Pseudomonas sp. TRMK1 and S is the residual concentration (%) after the utilization.
Extraction and Analysis of Metabolites
The plausible metabolites in the p-coumaric acid catabolic pathway are 4-vinyl phenol, p-hydroxybenzaldehyde, and p-hydroxybenzoic acid; 4-vinyl guaiacol, vanillin, and vanillic acid in ferulic acid; and 4-vinyl catechol and 3,4-dihydroxy benzaldehyde in caffeic acid. Protocatechuic acid may be the terminal aromatic metabolite in all the three pathways.
In order to demonstrate the catabolic pathways of these phenolic acids in this strain, the culture was grown on respective phenolic acids. The culture broth was withdrawn and centrifuged at 8000 rpm for 10 min. The spent medium was acidified to pH 2 with 6 M HCl and extracted twice with an equal volume of ethyl acetate. The ethyl acetate fraction was dried over anhydrous Na2SO4 and concentrated under vacuum in a rotary evaporator (Heidolph, Laborota 4000). Aliquots of extract were resuspended in 1 ml of methanol.
The metabolic extract was derivatized to strengthen the volatility or thermal stability of the compounds and for more accessibility to gas chromatography (GC). The samples were treated with N-methyl-N-trimethylsilyl-trifluoroacetamide (MSTFA). The method involved the addition of 50 μl of MSTFA to 200 μl of metabolite extract in a small reaction vial constantly agitating for 12 h at 30 °C to form TMS esters and ethers [29]. The trimethylsilyl (TMS)-derivatized samples were analyzed with GC (Agilent technology 7820) and GC-HRMS (AccuTOF GCv from Jeol Asia Ltd.).
Preparation of Cell Free Extract
Pseudomonas sp. TRMK1 cells, grown individually on p-coumaric, ferulic, and caffeic acids, were harvested at the mid-logarithmic growth phase by centrifugation at 8000 rpm (10 min at 4 °C). The cell pellet was washed twice with 25 mM potassium phosphate buffer of pH 7.0 and resuspended in the same buffer. The cell suspension was disrupted four times using a Vibra-Cell ultrasonicator (model 375, USA) at nominal power of 70 W for 30 s periods, each of 30 s duration, followed by a 1 min off-cycle during which the disrupted cells and the oscillator probe are cooled in ice. Unbroken cells and cell debris were removed by centrifugation at 12,000 rpm (30 min at 4 °C). This cell free supernatant was used for different enzymatic assays [30].
Enzyme Assay
Phenolic Acid Decarboxylase
One milliliter of reaction mixture consisting of 1 mM p-coumaric, ferulic, and caffeic acids (for respective PAD assays), 25 mM sodium phosphate buffer (pH 6.0), and an appropriate amount of crude enzyme was incubated at 37 °C for 40 min. The reaction was terminated by adding 20 folds of 25 mM stop buffer (Tris, 0.3 % SDS, pH 6.0) [32, 33]. The decarboxylase activity of p-coumaric, ferulic, and caffeic acids was assayed by the disappearance of respective absorbance peaks at 287, 314, and 313 nm and the appearance of new product peaks of 4-vinyl phenol, 4-vinyl guaicol, and 4-vinyl catechol at 258, 262, and 265 nm, respectively [31, 32].
p-Hydroxybenzaldehyde Dehydrogenase
One milliliter of reaction mixture comprises 100 mM phosphate buffer (pH 7.0), 60 μl of 600 nmol of NADP+, 100 μl of 40 nmol of p-hydoxybenzaldehyde, and a suitable amount of enzyme. The reaction was initiated by adding p-hydoxybenzaldehyde. This enzyme assay was monitored spectrophotometrically by the decrease in the absorbance at 330 nm, which is due to the conversion of p-hydoxybenzaldehyde to p-hydroxybenzoic acid [33].
p-Hydroxybenzoic Acid Hydroxylase
One milliliter of reaction mixture comprises 100 mM phosphate buffer (pH 7.0), 25 μl of 25 μM NADPH, and an appropriate amount of enzyme. The reaction was started by adding 60 μl of 400 μM p-hydroxybenzoic acid. The activity was examined by measuring the rate of decrease in absorbance at 340 nm due to the substrate-dependent oxidation of NADPH [34].
Protocatechuate 4,5-Dioxygenase
The reaction mixture for the assay of protocatechuate 4,5-dioxygenase comprised 25 mM phosphate buffer (pH 7.0), 50 μl of 15 mM l-cysteine, and an appropriate amount of enzyme. The reaction was initiated by adding 40 μl of 1 mM protocatechuic acid. The formation of γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde was observed by the increase in absorbance at 340 nm [35].
Vanillin Dehydrogenase
The assay mixture comprises 100 mM phosphate buffer (pH 7.0), 40 μl of 1 mM NAD(P)+, and an appropriate amount of enzyme. The reaction was initiated by adding 80 μl of 1 mM vanillin. The activity was determined spectrophotometrically by measuring the decrease in absorbance at 340 nm due to the substrate-dependent oxidation of NAD(P)H [36].
Specific activity of enzymes was expressed as micromoles of substrate-oxidized/product formed per milligram of protein under assay conditions. The protein concentration was determined by Lowry’s method using BSA as a standard [37].
Utilization of Various Phenolic Compounds
Pseudomonas sp. TRMK1 was inoculated into 50 ml of MS1 medium in 250 ml Erlenmeyer flasks supplemented with syringic acid, vanillin, and p-hydroxybenzaldehyde (5 mM), cinnamic acid (10 mM), vanillic acid (10 mM), and p-hydroxybenzoic and protocatechuic acid (25 mM), as a source of carbon and energy. The flasks were kept in a rotary incubator at 180 rpm for 30 °C. The culture was withdrawn at regular intervals of 6 h up to the final incubation period of 36 h. The cells were separated by centrifugation at 8000 rpm for 10 min. The resulting supernatant was filtered through a 0.2 μm filter paper, and the residual concentration of respective phenolic acids was examined by HPLC.
Utilization of Mixture of Phenolic Acids from the Synthetic Effluent
The typical composition of the effluent discharged from the agro-industrial waste is high content of organic substances, including sugars, tannins, polyphenols, polyalcohols, pectins, and lipids [38, 39]. A synthetic effluent was prepared by mixing 1.5 mM each of p-coumaric, ferulic, caffeic, cinnamic, protocatechuic, syringic, and vanillic acids in 50 ml MS1 medium in a 250 ml conical flask. The flask was incubated in a rotatory orbital shaker (180 rpm) at 30 °C. The growth was monitored at 600 nm, and the residual concentration of phenolic acids was analyzed by HPLC as mentioned above.
Analytical Methods
Growth of the bacterium and the presence of various enzymatic activities were checked spectrophotometrically (SPECORD 50). HPLC analysis was carried out by Waters 2489 equipped with a dual-beam wavelength UV-Vis detector and a C18 column, and the solvent system used was acetonitrile/water/acetic acid (30:69.5:0.5, v/v) at a flow rate of 1 ml min−1 [4]. Detection was achieved at 310 nm for p-coumaric acid, 280 nm for ferulic acid, 315 nm for caffeic acid, 280 nm for cinnamic acid, 256 nm for p-hydroxybenzoic acid, 275 nm for syringic acid, 320 nm for p-hydroxybenzaldehyde, 345 nm for vanillin, and 260 nm for vanillic and protocatechuic acids.
The derivatized metabolic extracts of p-coumaric, ferulic, and caffeic acids were analyzed by GC system consisting of a DB-5 column (length 30 m, inner diameter 0.25 mm, 0.25 μm film) with a flame ionization detector. The analysis was performed by splitless mode. Nitrogen was used as a carrier gas with a flow rate of 1 ml min−1. The GC conditions for the analysis were as follows: injector temperature 250 °C, detector temperature 300 °C, and column temperature 60 °C for the initial 5 min, followed by 15 °C rise per minute for 5 min, and then 10 °C min−1 for the final hold time of 4 min [40]. The trimethylsilyl (TMS)-derivatized samples were also analyzed with a GC-HRMS equipped with HP5 column (length 30 m, diameter 0.25 mm ID, and thickness 0.25 μm) with the ionization voltage of 70 eV, and the carrier gas was helium (Sophisticated Analytical Instrument Facility, Indian Institute of Technology, Bombay, India).
Results
Isolation and Identification of a Bacterium
The enrichment culture technique was used to isolate the p-coumaric acid-utilizing bacterium. The strain Pseudomonas sp. TRMK1 has the capability to grow on p-coumaric acid in the MS1 medium supplemented with 5 mM p-coumaric acid as a source of carbon and energy. This strain is also potential to degrade ferulic and caffeic acids. Morphological, physiological, and biochemical features of the strain are given in Table 1. The strain is gram-negative, motile, small rods, and appears as short chains. On a p-coumaric acid-MS1 agar plate, the strain TRMK1 formed sticky circular colonies. The strain shows positive tests for catalase, lipase, indole production, urease, carbohydrate fermentation, casein and starch hydrolysis, but exhibits negative results for gelatin liquefaction, Voges–Proskauer, methyl red, endospore and H2S production.
Initial investigation of 16S rDNA sequence (1261 bp) of the strain was done at NCBI, where significant groupings from these databases were downloaded for further examination. The sequence showed 97 % likeness with Pseudomonas taiwanensis. In the phylogenetic examination, it is clustered with P. taiwanensis strain BCRC and Pseudomonas plecoglossicida strain NBRC confirming its identity as Pseudomonas sp. and is named as Pseudomonas sp. TRMK1.
Utilization of Phenolic Acids
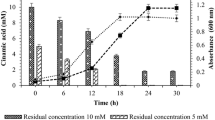
Strain Pseudomonas sp. TRMK1 utilizes all the three phenolic acids within 18 h of incubation. The bacterium exhibits lag, log, and stationary phase. The growth of bacterium increases with increase in the incubation period. When the strain was grown in all the three phenolic acids, it exhibits 3 h of lag phase and then enters into log phase that lasts 12–15 h. At an initial concentration of 5 mM p-coumaric acid, the strain utilizes 0.49 mM per hour with 99.94 % degradation efficiency during its log phase. When the concentration of p-coumaric acid is increased to 10 and 15 mM, the strain utilized 1 and 1.18 mM per hour with the degradation efficiency of 97 and 94.5 %, respectively (Fig. 1). At 5 mM initial concentration of ferulic acid, the strain utilized 1.3 mM per hour with 98.4 % degradation efficacy. When its concentration was increased to 10 mM, the strain utilized 0.58 mM per hour with 92 % degradation efficiency (Fig. 2). Its degradation ability with caffeic acid was also checked. At 5 mM initial concentration, the strain utilized 0.62 mM per hour of incubation with degradation efficiency of 98.4 %. With 10 mM caffeic acid, the strain utilized 0.59 mM per hour at 96 % degradation efficiency (Fig. 3). However, the growth of the strain is inhibited beyond 10 mM ferulic and caffeic acids because of their toxic effect.
Utilization of different concentrations of p-coumaric acid by Pseudomonas sp. TRMK1. [Residual residual concentration of 5, 10, and 15 mM p-coumaric acid at different time intervals; Absorbance growth absorbance of the strain at 5, 10, and 15 mM concentrations of p-coumaric acid at different time intervals]
Analysis of Metabolites
GC analysis of metabolic extract confirmed the degradation of p-coumaric, ferulic, and caffeic acids by this strain. The metabolic intermediates were identified based on the comparison of retention times with those of authentic compounds. The p-coumaric acid pathway metabolic intermediates such as p-hydroxybenzaldehyde, p-hydroxybenzoic acid, protocatechuic acid; vanillin, and vanillic and protocatechuic acids were observed as metabolic intermediates in the ferulic acid pathway. In the caffeic acid catabolic pathway, protocatechuate is the only metabolic intermediate detected.
N-Methyl-N-trimethylsilyl trifluoroacetamide (MSTFA)-derivatized metabolic intermediates were tentatively identified by matching their retention times with those of respective authentic compounds by GC. For further confirmation, they were characterized by GC-HRMS analysis (Table 2). The GC-HRMS mass spectrum of metabolite I (4-vinyl phenol) showed a molecular ion at m/z 177 and with the fragmentation pattern with the loss of CH3 (M+– 15) followed by m/z 162 (M+– CH3), 147 (M+– 2CH3), 133 (M+– 2CH3– CH2), 115, 102, 89, and 59. GC-HRMS of metabolite II (p-hydroxybenzaldehyde) gave a molecular ion (M+– 2) at m/z 196 and subsequently at 166 (M+– CH2O), 152 (M+– CH2O– CH3), 138 (M+– CH2O– 2CH3), 124 (M+– CH2O– 3CH3), 111, 98, 85, 68, and 55. The mass spectrum of metabolite III (p-hydroxybenzoic acid) showed a molecular ion (M+– 2) at m/z 282 and subsequently ion fragments at 252 (M+– 2CH3), 209 (M+– TMS), 196 (M+– TMS –CH3), 181 (M+– TMS –2CH3), 168, 153, 139, 125, 111, 96, 83, 69, and 57. The GC-HRMS spectrum of metabolite IV (4-vinyl guaiacol) showed an ion fragment at m/z 224 (M+– 2) and subsequent fragments at 196 (M+– C2H4), 181 (M+– C2H4– CH3), 168 (M+– C2H4– 2CH3), 154 (M+– C2H4– 3CH3), 139 (M+– C2H4– 4CH3), 125, 110 (M+– C2H4– 4CH3– TMS), 84, 68, and 56. The fragmentation of metabolite V (vanillin) gave the molecular ion peak at m/z 224 (M+) followed by 196 (M+– CH2O), 181 (M+– CH2O– CH3), 168 (M+– CH2O– 2CH3), 154 (M+– CH2O– 3CH3), 139 (M+– CH2O– 4CH3), 125, 111, 97, 70, and 55. Mass spectrum of metabolite VI (vanillic acid) exhibited molecular ion fragment at m/z 314 (M+– 2) and successive fragments at 298 (M+– CH3), 283 (M+– 2CH3), 269 (M+– 3CH3), 239 (M+– TMS), 226 (M+– TMS– CH3), 208, 182, 149, 134, and 75. The GC-HRMS mass spectrum of metabolite VII (4-vinyl catechol) revealed the molecular ion peak at m/z 280 (M+– 2) and consecutive fragments at 263 (M+– CH3), 230, 207 (M+– TMS), 189 (M+– 6CH3), 175, 151, 125, 101, 88, and 76. The GC-HRMS spectrum of metabolite VIII (3,4-dihydroxybenzaldehyde) showed a molecular ion peak at m/z 284 (M+– 2) and sequential fragmentation at 267 (M+– CH3), 253 (M+– CHO’), 241, 226, 213, 198, 185, 170, 157, 129, 114, 98, and 83.
Enzyme Assay
The activities of various enzymes involved in the catabolism of phenolic acids were detected in the cell free extract of TRMK1 cells grown on different phenolic acids. They are summarized in Table 3. Protocatechuate 4,5-dioxygenase exhibited good activity in the cell free extract and is responsible for ring fission leading to the formation of TCA cycle intermediates. Further, p-hydroxybenzaldehyde dehydrogenase and p-hydroxybenzoic acid hydroxylase showed less activity and the rest of the enzymes’ moderate activity.
Proposed Catabolic Pathway of p-Coumaric, Ferulic, and Caffeic Acids
The catabolic pathway of p-coumaric, ferulic, and caffeic acids is proposed based on GC-HRMS analysis of metabolites and enzyme assay studies. The decarboxylation of p-coumaric acid leads to the formation of 4-vinyl phenol, which is confirmed by the presence of p-coumaric acid decarboxylase. 4-Vinyl phenol is further oxidized to p-hydroxybenzaldehyde. p-Hydroxybenzaldehyde dehydrogenase catalyzed the conversion of p-hydroxybenzaldehyde to p-hydroxybenzoic acid. p-Hydroxybenzoic acid upon hydroxylation gives protocatechuic acid catalyzed by p-hydroxybenzoic acid hydroxylase. The formed protocatechuic acid further undergoes ring cleavage to produce γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde (Fig. 4A).
Ferulic acid on decarboxylation leads to the formation of 4-vinyl guaiacol, which is catalyzed by ferulic acid decarboxylase. 4-Vinyl guaiacol is converted to vanillin. Vanillin is further oxidized to vanillic acid by vanillin dehydrogenase. Vanillic acid is further demethoxylated to the formation of protocatechuic acid. Finally, protocatechuic acid undergoes meta ring cleavage to produce γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde, which is confirmed by protocatechuate 4,5-dioxygenase activity (Fig. 4B). Caffeic acid decarboxylase catalyzes the decarboxylation of caffeic acid to 4-vinyl catechol, which is oxidized to 3,4-dihydroxy benzaldehyde and then converted to protocatechuic acid (Fig. 4C).
Utilization of Various Phenolic Compounds
The ability of Pseudomonas sp. TRMK1 is further checked for its growth and utilization of other phenolic compounds at different concentrations. The strain showed maximum utilization of p-hydroxybenzoic and protocatechuic acids. When the concentrations of both these phenolics were provided at 25 mM, the strain showed utilization of 99.68 % at 30 h and 99.64 % at 36 h, respectively. Further, when 10 mM concentrations of cinnamic, vanillin, and vanillic acids were supplied, the strain utilized 96.8 % at 18 h, 34 % at 24 h, and 90.8 % at 24 h, respectively. When p-hydroxybenzaldehyde and syringic acid were provided at a concentration of 5 mM, the strain utilized 58 % at 24 h and 86.6 % at 18 h of incubation, respectively (Fig. 5). At 5 mM concentration of cinnamic, p-hydroxybenzoic, and protocatechuic acids and vanillin, the strain utilized 99.37 % at 8 h, 98.24 % at 18 h, 93.2 % at 18 h, and 100 % at 24 h, respectively. When 10, 15, and 20 mM concentrations of p-hydroxybenzoic and protocatechuic acids were supplied, the strain utilized 99.30 and 97.87 % at 24 h, 99.62 and 98.7 % at 24 h, and 99.69 and 99.39 % at 30 h, respectively (data not shown).
Utilization of Mixture of Phenolic Acids from the Synthetic Effluent
In order to check the ability of this strain for the utilization of a mixture of phenolic acids, the synthetic effluent containing the mixture of phenolic acids was prepared and inoculated with the strain TRMK1. The strain utilizes 99.4, 94.8, 99.8, 100, 99.3, 99.3, and 100 % of 1.5 mM each of p-coumaric, ferulic, caffeic, cinnamic, protocatechuic, syringic, and vanillic acids, respectively, after 12 h of incubation showing the maximum growth of 0.958 at 600 nm (Table 4).
Discussion
In this communication, we present a novel bacterial strain isolated from agro-industrial waste capable of degrading high concentrations of p-coumaric, ferulic, and caffeic acids individually. This strain has been identified as Pseudomonas sp. based on various physiological, morphological, and biochemical tests and 16S rDNA sequencing. Many reports are available on the utilization and catabolic pathway of these phenolic compounds by fungi [16, 17] and yeast [18], but little information is available on bacteria.
Pseudomonas sp. TRMK1 is more potent in utilizing various phenolic acids. It is reported that Halomonas sp. IMPC utilizes 100 % of 10 mM p-coumaric acid [21], and Lactobacillus plantarum LPNC8 utilizes 1.2 mM p-coumaric acid completely [32]. Whereas, TRMK1 utilizes 95.4 % of 15 mM p-coumaric acid within 18 h of incubation and 92 % of 10 mM ferulic acid in 18 h of incubation. The Cupriavidus sp. B-8 isolate showed the degradation efficiency of 96.84 % of 1 mM ferulic acid at 12 h [9]. Bacillus coagulans BK07 utilized 95 % of ferulic acid completely at a concentration of 2.13 mM in 7 h [20]. Strain TRMK1 utilized 96 % of 10 mM caffeic acid in 18 h. Table 5 shows the superiority of strain TRMK1 in the degradation of these phenolic acids over previously reported bacterial strains.
Bacterial catabolism of these phenolic acids begins with the cleavage of the side chain. Generally, two types of side chain cleavages occur: C1 cleavage of aliphatic side chain is involved in the formation of vinyl derivatives by non-oxidative decarboxylation [41, 42], whereas C2 cleavage leads to the formation of aldehydes [43, 44]. In strain TRMK1, the catabolism of these phenolic acids followed C1 cleavage, which leads to the formation of vinyl derivatives by phenolic acid decarboxylase [33]. The presence of this enzyme is noticed in the cell free extracts of TRMK1 grown on these compounds.
In strain TRMK1, the catabolism of p-coumaric acid begins with decarboxylation leading to the formation of 4-vinyl phenol catalyzed by p-coumarate decarboxylase. Similar conversion is noticed in Bacillus pimilus PS213 for the catabolism of p-coumaric acid. Further metabolism of 4-vinyl phenol was not traced in this bacterium [32]. However, in strain TRMK1, the formed 4-vinyl phenol undergoes oxidation to form p-hydroxybenzaldehyde. p-Hydroxybenzaldehyde dehydrogenase catalyzes the conversion of p-hydroxybenzaldehyde to p-hydroxybenzoic acid. This is evidenced by the presence of this enzyme in the cell free extract and detection of p-hydroxybenzoic acid in the culture supernatant. p-Hydroxybenzoic acid is hydroxylated at the aromatic nucleus by p-hydroxybenzoate hydroxylase to form protocatechuate, a terminal aromatic metabolite. Further, protocatechuate undergoes ring cleavage by protocatechuate 4,5-dioxygenase to form γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde. Acinetobacter calcoaceticus DSM 586 utilizes p-coumaric acid with the sequential formation of p-hydoxybenzoic and protocatechuic acids. However, the formed protocatechuate undergoes ring cleavage by protocatechuate 3,4-dioxygenase [45].
Ferulic acid is decarboxylated by ferulic acid decarboxylase to 4-vinyl guaiacol, which is further oxidized to vanillin. Vanillin dehydrogenase catalyzes the conversion of vanillin to vanillic acid, which undergoes demethoxylation to form protocatecuate in strain TRMK1. The formed protocatecuate further undergoes ring cleavage to form γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde by protocatechuate 4,5-dioxygenase. Such pathways were reported for catabolism of ferulic acid in Cupriavidus sp. B-8 [9], Pseudomonas fluorescens BF13 [46], P. fluorescens [47], and Bacillus coagulans BK07 [20]. The catabolism of caffeic acid undergoes non-oxidative decarboxylation to 4-vinyl catechol by caffeic acid decarboxylase. The formed 4-vinyl catechol is oxidized to 3,4-dihydroxy benzaldehyde and then to protocatechuic acid in this strain. Protocatechuate is the terminal aromatic metabolite formed during the catabolism of all the three phenolic acids. Protocatechuate undergoes ring cleavage by protocatechuate 4,5-dioxygenase to form γ-carboxy-α-hydroxy cis,cis-muconic semialdehyde that enters into TCA cycle intermediates.
Conclusion
Pseudomonas sp. TRMK1, a potent strain for the degradation of different phenolic compounds, isolated by enrichment culture technique, has been identified based on 16S rDNA sequencing. The strain has the capability to utilize a wide range of phenolic compounds including cinnamic, vanillic, p-hydroxybenzoic, syringic, and protocatechuic acids, p-hydroxy benzaldehyde, and vanillin as a sole source of carbon and energy. This strain has the ability to utilize maximum concentration of p-coumaric acid (15 mM), ferulic acid (10 mM), and caffeic acid (10 mM) in 18 h of incubation with degradation efficiency of 95.4, 92, and 96 %, respectively. Strain TRMK1 also showed potential degradation of the mixture of phenolic acids present in the synthetic effluent. Further, the catabolic pathways of p-coumaric, ferulic, and caffeic acids in Pseudomonas sp. TRMK1 have been proposed based on the identification of metabolic intermediates and the activities of various enzymes.
References
Borja, R., Martin, A., Maestro, R., Luque, M., & Duran, M. M. (1993). Enhancement of the anaerobic digestion of wine distillery wastewater by the removal of phenolic inhibitors. Bioresource Technology, 45, 99–104.
Lesage-Meessen, L., Navarro, D., Maunier, S., Sigoillot, J. C., Lorquin, J., Delattre, M., Simon, J. L., Asther, M., & Labat, M. (2001). Simple phenolic content in olive oil residues as a function of extraction systems. Food Chemistry, 75, 501–507.
Minhalma, M., Dias, C. R., & De Pinho, M. N. (2000). Adsorptive fouling in ultrafiltration of cork processing wastewaters. Advances in Environmental Research, 3, 539.
Chamkha, M., Labat, M., Bharat, K. C. P., & Garcia, J. L. (2001). Isolation of a cinnamic acid-metabolizing Clostridium glycolicum strain from oil mill wastewaters and emendation of the species description. International Journal of Systematic and Evolutionary Microbiology, 51, 2049–2054.
Labat, M., Augur, C., Perraud-Gaime, I., Roussos, S., & Sayadi, S. (2000). In T. Sera, C. R. Soccol, A. Pandey, & S. Roussos (Eds.), Biotechnological potentialities of polyphenolic compounds of coffee and comparison with olive, in coffee biotechnology and quality (pp. 517–531). Dordrecht: Kluwer.
Pan, G. X., Bolton, J. L., & Leary, G. J. (1998). Determination of ferulic and p-coumaric acids in wheat straw and the amounts released by mild acid and alkaline peroxide treatment. Journal of Agricultural and Food Chemistry, 46, 5283–5288.
Boudet, A. M., Kajita, S., Grima-Pettenati, J., & Goffner, D. (2003). Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends in Plant Science, 8, 576–581.
Kuiters, A. T., & Sarink, H. M. (1986). Leaching of phenolic compounds from leaf and needle litter of several deciduous and coniferous trees. Soil Biology and Biochemistry, 18, 475–480.
Yuan, C. L., Wei-chun, Y., Yong-hua, Z., Zhi-hui, Y., Yu, Z., & Yue-hui, C. (2013). Biodegradation of ferulic acid by a newly isolated strain of Cupriavidus sp. B-8. Journal of Central South University, 20, 1964–1970.
Zeng, R. S., & Mallik, A. U. (2006). Selected ectomycorrhizal fungi of black spruce (Picea mariana) can detoxify phenolic compounds of Kalmia angustifolia. Journal of Chemical Ecology, 32, 1473–1489.
Pienkos, P. T., & Zhang, M. (2009). Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose, 16, 743–762.
Borneman, W. S., Akin, D. E., & Vaneseltine, W. P. (1986). Effect of phenolic monomers on ruminal bacteria. Applied and Environmental Microbiology, 52, 1331–1339.
Rozes, N., Arola, L., & Bordons, A. (2003). Effect of phenolic compounds on the co-metabolism of citric acid and sugars by Oenococcusoeni from wine. Letters in Applied Microbiology, 36, 337–341.
Campos, F. M., Figueiredo, A. R., Hogg, T. A., & Couto, J. A. (2009). Effect of phenolic acids on glucose and organic acid metabolism by lactic acid bacteria from wine. Food Microbiology, 26, 409–414.
Nonaka, K., Ohata, H., Santo, Y., & Hosokawa, K. (2008). Utilization of phenylpropanoids by Pseudomonas putida soil isolates and its probable taxonomic significance. Microbes and Environments, 23, 360–364.
Alvarado, I. E., Lomascolo, A., Navarro, D., Delattre, M., Asther, M., & Lesage-Meessen, L. (2001). Evidence of a new biotransformation pathway of p-coumaric acid into p-hydroxybenzaldehyde in Pycnoporus cinnabarinus. Applied Microbiology and Biotechnology, 57, 725–730.
Sachan, A., Ghosh, S., & Mitra, A. (2006b). Biotransformation of p-coumaric acid by Paecilomyces variotii. Letters in Applied Microbiology, 42, 35–41.
Shashwati, G., Sachan, A., & Mitra, A. (2006). Formation of vanillic acid from ferulic acid by Paecilomyces variotii MTCC 6581. Current Science, 90, 825–829.
Sutherland, J. B., Crawford, D. J., & Pometto, A. L. (1983). Metabolism of cinnamic, p-coumaric and ferulic acids by Streptomyces setonii. Canadian Journal of Microbiology, 29, 1253–1257.
Karmakar, B., Vohar, R. M., Nandanwar, H., Sharma, P., Gupta, K. G., & Sobti, R. C. (2000). Rapid degradation of ferulic acid via 4-vinylguaiacol and vanillin by a newly isolated strain of Bacillus coagulans. Journal of Biotechnology, 80, 195–202.
Abdelkafi, S., Labat, M., Casalot, L., Chamkha, M., & Sayadi, S. (2006). Isolation and characterization of Halomonas sp. strain IMPC, a p-coumaric acid-metabolizing bacterium that decarboxylates other cinnamic acids under hypersaline conditions. FEMS Microbiol. Lett., 255, 108–114.
Toma’s-Barbera’n, F. A., & Clifford, M. N. (2000). Flavanones, chalcones and dihydrochalcones—nature, occurrence and dietary burden. Journal of the Science of Food and Agriculture, 80, 1073–1080.
Walton, N. J., Mayer, M. J., & Narbad, A. (2003). Vanillin. Phytochem., 63, 505–515.
Mcqualter, R. B., Chong, B. F., Meyer, K., Van, D. D., O'shea, M. G., Walton, N. J., Vitanen, P. V., & Brumbley, S. M. (2005). Initial evaluation of sugarcane as a production platform for p-hydroxybenzoic acid. Plant Biotechnol., 3, 29–41.
Tanruean, K., Chandet, N., & Rakariyatham, N. (2013). Bioconversion of ferulic acid into high value metabolites by white rot fungi isolated from fruiting-body of the polypore mushroom. J. Med. Biol. Eng., 2, 168–172.
Mukram, I., Anand, S. N., Kirankumar, B., Monisha, T. R., Pooja, V. R., & Karegoudar, T. B. (2015). Isolation and identification of a nitrile hydrolyzing bacterium and simultaneous utilization of aromatic and aliphatic nitriles. International Biodeterioration & Biodegradation, 100, 165–171.
Lane, D. J. (1991). Small subunit ribosomal RNA sequences and primers. Large subunit ribosomal RNA sequences and primers. In E. Stackebrandt & M. Goodfellow (Eds.), Nucleie Aeid Teehniques in bacterial systematics (pp. 148–175). Chichester: Wiley.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Sepic, E., Bricelj, M., & Leskovsek, H. (1998). Degradation of fluoranthene by Pasteurella sp. IFA and Mycobacterium sp. PYR-1: isolation and identification of metabolites. Journal of Applied Microbiology, 85, 746–754.
Santoshkumar, M., Veeranagouda, Y., Lee, K., & Karegoudar, T. B. (2011). Utilization of aliphatic nitrile by Paracoccus sp. SKG isolated from chemical waste samples. Int. biodeter. Biodegr., 65, 153–159.
Degrassi, G., Polverino De Laureto, P., & Bruschi, C. V. (1995). Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Applied and Environmental Microbiology, 61, 326–332.
Barthelmebs, L., Divies, C., & Cavin, J. F. (2000). Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Applied and Environmental Microbiology, 66, 3368–3375.
Whited, G. M., & Gibson, D. T. (1991). Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. Journal of Bacteriology, 173, 3017–3020.
Suarez, M., Martín, M., Ferrer, E., & Garrido-Pertierra, A. (1995). Purification and characterization of 4-hydroxybenzoate 3-hydroxylase from a Klebsiella pneumoniae mutant strain. Archives of Microbiology, 164, 70–77.
MacDonald, D. L., Stanier, R. Y., & Ingraham, J. L. (1954). The enzymatic formation of β-carboxymuconic acid. The Journal of Biological Chemistry, 210, 809–820.
Ding, W., Si, M., Zhang, W., Zhang, Y., Chen, C., Zhang, L., Lu, Z., Chen, S., & Shen, X. (2015). Functional characterization of a vanillin dehydrogenase in Corynebacterium glutamicum. Scientific Reports, 5, 8044.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the folin phenol reagent. The Journal of Biological Chemistry, 193, 265–275.
Balice, V., & Cera, O. (1984). Acidic phenolic fraction of the juice of olives determined by gas chromatographic method. Grasas y Aceites, 25, 178–180.
D’Annibale, A., Crestini, C., Vinciguerra, V., & Sermanni, G. G. (1998). The biodegradation of recalcitrant effluents from an olive mill by a white-rot fungus. Journal of Biotechnology, 61, 209–218.
Mercy, O. A., Simeon, O. O., Saheed, A., Ayokunle, O., & Temitope, A. E. (2014). Analysis of phenolic compounds, phytosterols, lignans and stilbenoids in garlic and ginger oil by gas chromatography. J. Food Chem. Nutr., 02, 53–60.
Mathew, S., Abraham, T. E., & Sudheesh, S. (2007). Rapid conversion of ferulic acid to 4-vinyl guaiacol and vanillin metabolites by Debaryomyces hansenii. Journal of Molecular Catalysis B: Enzymatic, 44, 48–52.
Baqueir-Peña, I., Rodriguez-Serrano, G., Gonalez-Zamor, E., Augur, C., Loera, O., & Saucedo-Castañeda, G. (2010). Biotransformation of ferulic acid to 4-vinylguaiacol by a wild and a diploid strain of Aspergillus niger. Bioresource Technology, 101, 4721–4724.
Toms, A., & Wood, J. M. (1970). The degradation of trans-ferulic acid by Pseudomonas acidovorans. Biochemistry, 9, 337–343.
Karnath, N. G. K., & Reber, H. (1979). Regulation of the utilization of 4-hydroxybenzoate and 4-hydroxycimmate in batch and continuous cultures of Pseudomonas testosterone. Archives of Microbiology, 120, 97–103.
Delneri, D., Degrassi, G., Rizzo, R., & Bruschi, C. V. (1995). Degradation of trans-ferulic and p-coumaric acid by Acinetobacter calcoaceticus DSM 586. Biochim. Biophysica Acta., 1244, 363–367.
Civolani, C., Barghini, P., Roncetti, R. A., Ruzzi, M., & Alma, S. (2000). Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Applied and Environmental Microbiology, 66, 2311–2317.
Narbad, A., & Gasson, M. J. (1998). Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly- isolated strain of Pseudornonas fluorescens. Microbiology, 144, 1397–1405.
Barthelmebs, L., Divies, C., & Cavin, J. F. (2001). Molecular characterization of the phenolic acid metabolism in the lactic acid bacteria Lactobacillus plantarum. Le Lait, 81, 161–171.
Acknowledgements
One of the authors Mr. Monisha T R would like to thank University Grants Commission (UGC), New Delhi, India, for the financial assistance in the mode of BSR Fellowship in Sciences for meritorious students as well as the Indian Institute of Technology Bombay, India, for GC-HRMS analysis. The financial support from UGC in the form of UGC-SAP program [No. F.4-27/2015/DRS -II (SAP-II)] sanctioned to the department is highly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
T R, M., I, M., B, K. et al. Utilization of Phenylpropanoids by Newly Isolated Bacterium Pseudomonas sp. TRMK1. Appl Biochem Biotechnol 182, 1240–1255 (2017). https://doi.org/10.1007/s12010-017-2396-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2396-5