Abstract
Plantaricins are a group of ribosomally synthesized antimicrobial peptides in Lactobacillus plantarum that exert antimicrobial activities against some foodborne pathogens. In this study, we observed that plantaricin E in L. plantarum 163 was missing 19 amino acids (plnE mutant amino acid sequence: FNRGGYNFGKSVRH, plnE amino acid sequence: FNRGGYNFGKSVRHVVDAIGSVAGIRGILKSIR). In order to study the effects of mutant plnE, plnE mutant genes with and without the signal peptide were cloned from the L. plantarum 163 genome, linked to the pET32a vector, and expressed via a fusion protein (thioredoxin) in Escherichia coli BL21 (DE3). All target proteins were purified using Ni-NTA, SP FF columns, and RP-HPLC. The purified proteins were stable in an acidic environment and at temperatures below 80 °C, but they were easily degraded under alkaline conditions and by protease treatment. They showed antimicrobial activity against gram-positive bacteria such as Micrococcus luteus, Staphylococcus epidermidis, Lactococcus lactis, Lactobacillus paracasei, and Listeria innocua. In addition, SP-plnE and PlnE exerted stronger activity than nisin. The signal peptide had a positive effect on the activities of PlnE and PlnEm. Thus, these purified proteins may have potential applications in the food industry to control foodborne pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antibiotic resistance is becoming a major public health concern worldwide. “Superbugs” may cause many serious infections that currently available antibiotics cannot treat [3, 17, 35]. Plantaricins, which are considered nontoxic and are relatively safe for use as food preservatives and drugs, are novel antimicrobial peptides with the potential to control “superbugs” [23, 29]. Based on the statistics presented in the antimicrobial peptide database (http://bactibase.pfba-lab-tun.org/main.php), approximately 40% of known bacteriocins are produced by Lactobacillus.
In addition, as many of the species of the genus Lactobacillus have a “generally regarded as safe” status, including Lactobacillus plantarum, they may have applications as probiotics [23]. Many plantaricins are produced by various strains of L. plantarum. For example, plantaricin SIK83 [2] was purified and identified from L. plantarum SIK-83; this peptide inhibits some lactic acid bacteria belonging to Lactobacillus, Leuconostoc, Pediococcus, and Streptococcus. Additionally, plantaricins A, C, S, E, F, J, K, SA6, and UG1 [7, 9, 11, 16, 24, 26, 27, 31] have been purified and detected from the culture supernatants of L. plantarum. To date, more than 36 plantaricins [4, 12, 13, 18, 19, 28, 34, 36, 39, 45] have been identified in L. plantarum, and 16 of these plantaricins are included in the bacteriocin database (http://aps.unmc.edu/AP/).
Plantaricin E is a two-peptide bacteriocin that depends on the complementarity of two different antimicrobial peptides (plantaricin E and plantaricin F) to function [21]. The PlnE gene was cloned from L. plantarum WCFS1, C11, and J51 and was found in the soil metagenome [1, 22, 24, 33]. This gene was then heterologously expressed in Escherichia coli to determine its antimicrobial activity The results indicated that this molecule had activity against Listeria innocua, Micrococcus luteus, and some lactic acid bacteria [24]. The yield of plantaricin E can be as high as 1.2–1.5%, which is much higher than that in the native strain (0.3–0.7%) [25]. The three-dimensional structures of plantaricin E have been analyzed by nuclear magnetic resonance spectroscopy (NMR) in the presence of dodecylphosphocholine (DPC) micelles. Plantaricin E has an α-helix in the N-terminal (residues 10–21) and an α-helix-like structure (residues 25–30) and a small random coil (residues 31–33) in the C-terminal [10]. The mechanism of plantaricin E has been extensively studied, and many hypotheses have been proposed. These studies have indicated that plantaricins E and F have complementary effects and form pores in the cytoplasmic membranes of target cells. Plantaricins E and F dissipate the pH gradient (ΔpH) and electrical potential (ΔΨ) [21]. Recent research results have shown that plantaricins E and F interact in an antiparallel manner and that the C-terminal of plantaricin E and N-terminal of plantaricin F are localized to the outer part of the target cell membrane, whereas the N-terminal of plantaricin E and C-terminal of plantaricin F are localized to the inner part of the membrane [8].
In the present work, we isolated an L. plantarum strain having antimicrobial activity against some gram-positive bacteria [14]. A genome framework map was analyzed to identify the bacteria-associated gene. A plantaricin-associated locus was found; this locus contained five operons: pln123, plnRL, plnEFI, plnABD, and plnGHSTUVW. The function of pln123 was tested and verified in our previous work [20]. In this study, we found that the plnE expressed in L. plantarum 163 lost five bases, resulting in a stop codon (TAG) and shortening of PlnE to 14 amino acids, compared with the 33-amino acid sequence of PlnE in L. plantarum WCSF1 [33] (gene ID of plnE 1064171). To evaluate the influence of the mutant PlnE, the protein was expressed as a fusion protein in E. coli. In addition, the effects of the signal peptide on plantaricin activity were also studied.
Material and Methods
Microbial Strains and Culture Medium
The indicator strains were Lactococcus lactis, L. innocua CICC 10417, Lactobacillus paracasei CICC 20241, methicillin-resistant S. aureus (MRSA), methicillin-resistant Staphylococcus epidermidis (MRSE), M. luteus CMCC63202, and S. epidermidis. E. coli DH5α and E. coli BL21 (DE3) strains used for cloning and protein expression were grown in LB medium at 37 °C. Lactic acid bacteria were grown in MRS medium at 30 °C, Listeria were grown in brain–heart infusion medium at 30 °C, and all other indicator strains were grown in LB medium at 37 °C.
The protein marker, recombinase exnase II, high-fidelity enzyme DNA polymerase, and E. coli strains DH5α and BL21 (DE3) were purchased from Vazyme Biotech (Nanjing, China). A low-molecular-weight protein ladder and restriction endonuclease were purchased from Thermo Scientific (MA, USA). Cation exchanger (HiTrap SP FF) columns and nickel-nitrilotriacetic acid (Ni-NTA) were purchased from GE Healthcare (Pittsburgh, PA, USA). Dialysis bags were purchased from Viskase (IL, USA). Primers were designed as indicated in Table 1.
Construction of Recombinant Plasmids
The genomes of L. plantarum 163 and STIII were extracted as previously described [30]. PlnE and sp-plnE genes were cloned from L. plantarum STIII, and PlnEm and sp-plnEm were cloned from L. plantarum 163 by high-fidelity enzyme DNA polymerase. After a hotstart at 95 °C for 3 min, 30 cycles of the following program were run: denaturation at 95 °C for 20 s, primer annealing at 45 °C for 20 s, and elongation at 72 °C for 30 s, with a final elongation at 72 °C for 10 min. The sequence of the enterokinase site was removed with KpnI and BamHI and then reconstructed by PCR (using the primer enterokinase-F with the reaction conditions described above). Four genes were linked to linearized pET32a (digested with KpnI and BamHI) directly using the infusion method (recombinase exnase II) and transformed into E. coli DH5α as described previously [30]. The plasmid of the positive clone was extracted and transformed into E. coli BL21 (DE3) for protein expression after induction with 0.5 mM IPTG.
Expression and Purification of Recombinant Proteins
One liter of culture was induced with 0.5 mM IPTG (OD600 of 0.6), and the cells were cultured for an additional 8 h at 25 °C. After washing with phosphate-buffered saline (PBS, pH 7.4), cells were suspended in binding buffer (20 mM Na3PO4, 8 M urea, pH 7.8) and then crushed by sonication for 15 min. The supernatant was loaded onto the Ni-NTA resin and eluted with binding buffer (containing 200 mM imidazole). The imidazole, phosphate, and urea were removed by dialysis in enterokinase buffer (10 mM Tris-HCl, pH 8.0, with 10 mM CaCl2) at 4 °C overnight. The fusion protein was removed by enterokinase at 37 °C overnight and analyzed by Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The protein was then purified using an Ni-NTA column again to remove the fusion protein and histidine tag. The flow-through, which contained recombinant plantaricins, was purified using a cation exchanger (SP FF) column in an AKTA purifier UPC10 (GE Healthcare). Proteins were collected, and the antimicrobial activity was tested after desalting by dialysis in citrate buffer (0.1 M, pH 5.0). The concentrations of target proteins were determined using Bradford assays. For further purification, active samples were applied to an AICHROMBond C18 (250 × 4.6 mm) column using an Ultimate 2000 HPLC system (Dionex, CA, USA).
Inhibitory Activities of Purified Proteins
Target protein activities were quantified with a 96-well microtiter plate assay. The density of the indicator bacteria was adjusted to 2 × 106 CFU/mL. Fifty microliters each of the indicator bacteria and gradient dilutions (1, 10, 20, 30, 40, and 50 μM) of target proteins was added to the 96-well plates. Alternatively, 50 μL nisin (0.1, 0.5, 1, 5, 10, or 15 μM) was added as a positive control, and 50 μL citrate buffer (0.1 M, pH 5.0) was added as a negative control. More accurate results of minimum inhibitory concentrations (MICs) were tested by further dilutions by using more refined concentrations. Three replicates were carried out for each experiment. The MICs were defined as the minimum concentrations for which the OD600 did not change significantly (P < 0.05) compared with the initial OD600 value.
Protein Stability in the Presence of Proteases and Variations in pH and Temperature
All target proteins were adjusted to a concentration of 10 μM (pH 5.0). The indicator bacterium was L. innocua. Proteins were then treated with proteinase E, proteinase k, pepsin, or papain in appropriate buffers and temperatures for 1 h. The relative activities, which depended on the diameter of the inhibition zone, were then tested and calculated after the pH was adjusted to 5.0. Additionally, the buffer for the protein was changed to acidic (pH 1, 2, 3, or 4) or alkaline (pH 11, 12, 13, or 14) for 2 h, followed by adjustment to pH 5.0 to calculate the relative stability of the protein under different pH conditions as described above. Moreover, plantaricins were treated by heating (60, 80, 100, or 120 °C) for 30 min, and relative activities of the proteins were calculated as described above. All analyses were carried out in triplicate, and results are the averages of three reactions. The standard deviations were calculated using the “Descriptive Statistics Std. deviation” function in IBM SPSS Statistic 19. A fitting equation between zone diameter and antimicrobial activity was established using Origin 9.0. The relative activity was defined as the ratio of post-treatment activity to untreated activity. The activities of all positive controls were set as 100%.
Time-Killing Kinetics Assay, Growth Inhibition, and Cellular Content Leakage
The time-killing kinetics assays of recombinant plantaricins were carried out using M. luteus as described previously [15, 41]. The initial concentration of the indicator bacteria was 1 × 106 CFU/mL. One MIC of plantaricins was added after culturing at 37 °C for 2 h, and cells were then cultured for an additional 6 h. The OD600 was detected every hour using a microplate reader. The viable cells of all samples were counted by dilution plate counting.
To study the effects of recombinant plantaricins on the cell membrane, the leakage of intercellular contents (DNA and protein) was examined. Overnight cultures were washed twice in PBS (pH 7.4). One MIC of recombinant plantaricins was added, and cells were then cultured at 37 °C for an additional 1 h. Citrate buffer (0.1 M, pH 5.0) was added as the negative control. The values of OD260 and OD280 were detected to measure leakage of protein and DNA, respectively, from the indicator bacteria.
Electron Microscopy Study
To investigate whether recombinant plantaricins affected the overall cell morphology and to visualize the bacterial membrane, we examined cell morphology after treatment with one MIC of plantaricins by using scanning electron microscopy (SEM). The indicator bacterium (M. luteus) was grown at 37 °C for 8 h. Cultures were then washed twice in PBS (pH 7.4) and adjusted to 1 × 106 CFU/mL using fresh LB medium. One MIC of plantaricins was added, and cells were then cultured for an additional 2 h. Next, cells were harvested and washed twice in PBS (pH 7.4). Finally, samples were treated for SEM as previously described [37].
Results and Discussion
Analysis of plnE and Its Mutant
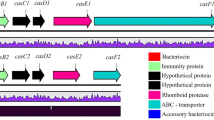
The synthetic gene of plantaricin E was mutated in L. plantarum 163 compared with that in L. plantarum WCFS1, C11, and NC8 [6, 19, 33] (gene ID of plnE 1064171). There were five codons missing in plnEm, resulting in the formation of a termination codon (TAG). Therefore, the length of mature PlnEm was only 14 amino acids (FNRGGYNFGKSVRH) compared with the 33-amino acid sequence (FNRGGYNFGKSVRHVVDAIGSVAGIRGILKSIR) of PlnE (Fig. 1b).
Pln locus in Lactobacillus plantarum 163 and analysis of plnE and its mutant. a The pln locus in L. plantarum 163. This locus contained five operons: pln123, plnRL, plnEFI, plnABD, and plnGHSTUVW. PlnE in plnEFI was missing 19 amino acids. b Sequence of the signal peptide (MLQFEKLQYSRLPQKKLAKISGG). The peptide had a typical double-glycine leader and a net charge of +4. The alpha helix and hydrophobic domain are indicated in burgundy and orange, respectively
PlnEm, which had only one α-helix domain and one hydrophobic domain, also exhibited antimicrobial activity, as shown below. Additionally, PlnE and PlnEm had a positive net charge, as described in Fig. 1b; this could play an important role during the initial reaction when plantaricin bound to the cell membrane.
Further analyses showed that the signal peptide (23 amino acids) had a positive charge and a hydrophobic domain that could bind and insert into the lipid bilayer and guide the secretion of plantaricin E.
Expression and Purification of Recombinant Protein
Because several reports have described heterologous expression of plantaricin E [24, 42], we decided to further clone plnE and its mutant amplicons, followed by expression and purification of the cloned products. For this purpose, the total gene and mature region of plnE and plnEm were cloned into pET32a (+) and sequenced. The data showed that the constructs showed 100% similarity to the template sequence.
For heterologous expression, the cells were disrupted after induced by IPTG, and the supernatants were analyzed by SDS-PAGE. A substantial band was observed at 18.9–24.4 kDa (Fig. 2a). The theoretical masses of recombinant PlnEm, PlnE, SP-PlnEm, and SP-PlnE (with fusion proteins) were 19.6, 21.7, 22.1, and 24.2 kDa, respectively. These results suggested that four recombinant plantaricins were successfully expressed in E. coli BL21 (DE3).
Expression and purification of target genes. a Protein expression results for plnEm, plnE, sp-plnEm, and sp-plnE. Lane 1 indicates supernatants from ultrasonic disruption of positive clones before induction by IPTG and lane 2 indicates supernatants from ultrasonic disruption of positive clones after induced by IPTG. b Cleavage and purification results of PlnEm, PlnE, SP-PlnEm, and SP-PlnE, respectively, in tricine-SDS-PAGE. Lane 1 indicates recombinant proteins that were digested by enterokinase and lane 2 indicates recombinant plantaricins that were purified by cation exchange column (Hitrap SPFF). c Results of PlnEm, PlnE, SP-PlnEm, and SP-PlnE analyses by RP-HPLC. Analysis of the peak areas showed that more than 92.19, 75.17, 88.78, and 85.74% of the proteins were objective proteins, respectively
For further purification, the supernatants were subjected to Ni-NTA affinity chromatography. The bound proteins were eluted in 200 mM imidazole. The proteins were detected by Tricine-SDS-PAGE after enterokinase cleavage (Fig. 2b). The cleavage mixture was then subjected to Ni-NTA affinity chromatography again to remove fusion protein and further purified using a cation exchange column (SP FF column). The proteins were analyzed by Tricine-SDS-PAGE after desalting by dialysis in citrate buffer (0.1 M, pH 5.0).
For further purification, target proteins were purified by RP-HPLC; the target proteins were eluted at about 6 min. The percentages of target protein yields were analyzed by peak calculus and found to be 92.19, 75.17, 88.78, and 85.74% for PlnEm, PlnE, SP-PlnEm, and SP-PlnE, respectively (Fig. 2c).
Antimicrobial Activities of Plantaricin E and Its Mutant
The results of our study showed that PlnEm, PlnE, SP-PlnEm, and SP-PlnE had different antimicrobial activities against gram-positive bacteria (Table 2). All target proteins had activity against M. luteus CMCC 63202, S. epidermidis, L. lactis NZ3900, L. paracasei CICC 20241, and L. innocua CICC 10417 at different concentrations. PlnE and SP-PlnE had activity against S. aureus ATCC29213 and MRSA, whereas PlnEm and SP-PlnEm did not. Additionally, PlnEm also showed no activity against MRSE.
Overall, PlnE had better activity than PlnEm, indicating that the mutant had a significant influence on the activity of PlnE, which may be explained by the length of the alpha helix and the positive charge. Compared with the positive control (nisin), PlnEm and SP-PlnEm had reduced activity for all indicator bacteria. However, PlnE and SP-PlnE had better activities against M. luteus, S. aureus, S. epidermidis, MRSE, and MRSA than nisin. This difference may be associated with the distinct structure and positive net charge of the mutants. For example, plnE had a greater positive net charge than plnEm, which may affect the interaction between peptides and the bacterial phospholipid bilayer during the initial stage. Moreover, plnE had a more hydrophobic region than plnEm, altering hydrophobic interactions and binding strength. Therefore, the missing amino acids of plnE decreased the efficiency and strength of the interaction between plantaricins and the cell membrane.
In previous studies [24, 42], almost all of the heterologous expressed bacteriocins were analyzed without consideration of the influence of the signal peptide. It is commonly thought that the signal peptide has no effect or a negative effect on bacteriocin activity. However, the signal peptide of plnE has a positive effect on the activity of plnEm. We found that SP-plnEm had higher activity than plnEm. The role of the signal peptide may be attributed to its large hydrophobic domain, which could bind to the lipid bilayer, and the +4 positive charge, which could play a role in initial membrane binding to enhance the binding stability of PlnEm [5].
Stability of Plantaricin E and Its Mutant
The antimicrobial activities of PlnEm, PlnE, SP-PlnEm, and SP-PlnE were reduced when exposed to acidic/alkaline conditions or heat. Additionally, stability was enhanced as the peptide length increased. SP-PlnE showed the best stability, whereas PlnEm showed the worst stability. All target proteins had more than 87% relative activity when treated in an acidic environment (pH 1.0), demonstrating good stability under these conditions. However, relative activity was reduced when exposed to alkaline conditions; specifically, the activity of PlnEm was only around 78% when the protein was exposed to an alkaline environment at pH 14.0 for 2 h. The activities of PlnEm, PlnE, SP-PlnEm, and SP-PlnE were rapidly reduced after heat treatment at temperatures above 100 °C but were stable at below 80 °C. Indeed, PlnEm had only 68% of the original activity after processing at 120 °C for 1 h (Fig. 3). Additionally, the proteins did not have activity after treatment with proteolytic enzymes, such as proteinase E, proteinase K, pepsin, or papain.
Protein stability at varying pH values and temperatures. All assays were carried out against Micrococcus luteus CMCC 63202. The relationship between zone diameter and antimicrobial activity was analyzed to establish a fitting equation using Origin 9.0. The relative activity was defined as the ratio of post-treatment activity to untreated activity. Y = 0.395X + 1.881 and r 2 = 0.996; Y is log C, C is the concentration of target protein (μM/mL), and X is the ring diameter of the inhibition zone (mm). The activity of all positive controls was set to 100%
Time-Killing Kinetics Assays and SEM Results
Some models for antimicrobial peptide membrane lysis have been proposed [40, 43]. Based on the variability of the microbial membrane ultrastructure, a specific peptide may have different mechanisms under different membrane environments. Many models have shown that antimicrobial peptides result in the formation of membrane pores, leakage of ions and intercellular substances, changes in metabolism, and cell death [40].
In order to study the activities of plantaricins against indicator bacteria, time-killing kinetics assays and growth inhibition assays were conducted. The results showed that four plantaricins could inhibit the growth of the indicator bacteria when the plantaricin was added (Fig. 4b). The time-killing kinetics assays showed that the number of viable cells of indicator bacteria did not decrease when treated with plantaricins (Fig. 4a). These results indicated that the four recombinant plantaricins exerted their inhibitory effects by inhibiting bacterial growth.
Time-killing kinetics assays and SEM results. a Growth inhibition of recombinant plantaricins to Micrococcus luteus CMCC 63202. The plantaricins were added after 2 h of culture. The OD600 was detected every 2 h using a microplate reader. b The CFU were counted by diluted coating in plates at the same time. c Results of leakage of intercellular contents following treatment with plantaricins. A260 and A280 represent the amounts of protein and nucleic acids, respectively, that leaked from the indicator bacteria. d SEM images after plantaricin treatment. The acceleration voltage was 7.0 kV, and the magnification was ×5000
The leakage of intercellular contents and SEM were conducted to evaluate the destruction of the cell membrane. The OD260 and OD280 values were significantly different between treated samples and the negative control (Fig. 4c). These data indicated that proteins and DNA leaked from the cell; thus, the cell membrane was damaged. However, SEM images showed that the cell morphology was not substantially altered (Fig. 4d). We could not find “holes” or specific occurrences of leakage.
Cell growth was inhibited when cells were treated with the four plantaricins. The number of dividing cells (dumbbell-shaped cells) was reduced significantly, as shown in our SEM images. Notably, the better the bacteriocin, the greater the inhibitory effect on cell division (Fig. 4d). The cells barely divided when treated with SP-PlnE.
Overall, the bacteriostasis mechanisms of these four plantaricins caused irreversible breakage of the cell membranes. Peptides may create membrane pores, leading to ion and metabolite leakage, membrane depolarization, pH gradient (ΔpH) and electrical potential (ΔΨ) dissipation, and ultimately cell death [21, 43]. Recent studies have shown that plantaricins E and F have complementary effects; plantaricins E and F interact in an antiparallel manner and the C-terminal of plantaricin E and N-terminal of plantaricin F are localized to the outer part of the target cell membrane, whereas the N-terminal of plantaricin E and C-terminal of plantaricin F are localized to the inner part of the membrane. The preference for an aromatic residue at position 6 (tyrosine) in plantaricin E suggests positioning of this residue in or near the membrane interface inside the cells [8].
PlnEm had the main G5 × × × G9 motif, which is critical for the antimicrobial activity of the protein, but showed loss of the G20 × × × G24 motif, which has a secondary role in antimicrobial activity. Therefore, although PlnEm had antimicrobial activity, the activity was poorer than that of PlnE. When the indicator bacteria were treated with PlnE or PlnEm alone (without PlnF), the ability to create pores was weakened; many small pores may have been created, allowing leakage of ions and metabolites. However, these changes did not cause cell death, and the cell morphology was not changed significantly. Additionally, these plantaricins may influence cell metabolism, thereby affecting the growth of the indicator bacteria.
Conclusion
Currently, staphylococcal infections and other resistant bacteria account for a significant proportion of hospital-acquired infections, with S. epidermidis infections being the most common cause of bacteremia in hospitals [32, 44]. Many antimicrobial peptides had activities against resistant bacteria, providing new approaches for the treatment of resistant bacterial infections. Moreover, the potential use of purified plantaricins as food biopreservatives may result in the replacement of synthetic chemical preservatives in the future. Currently, nisin is the only approved bacteriocin that can be used as a food additive [38]. In this study, PlnE and SP-PlnE had better activities against M. luteus, S. aureus, S. epidermidis, MRSE, and MRSA than nisin. Thus, these antimicrobial peptides may have applications as new food additives and may provide new approaches to overcoming antibiotic resistance in food production.
Abbreviations
- plnE :
-
synthesized plantaricin E gene
- plnEm :
-
mutant of plnE in Lactobacillus plantarum 163
- sp-plnE :
-
plnE gene with the signal peptide
- sp-plnEm :
-
plnEm gene with the signal peptide
References
Anderssen, E. L., Diep, D. B., Nes, I. F., et al. (1998). Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Applied and Environmental Microbiology, 64, 2269–2272.
Andersson, R. E., Daeschel, M. A., & Hassan, H. M. (1988). Antibacterial activity of plantaricin SIK-83, a bacteriocin produced by Lactobacillus plantarum. Biochimie, 70, 381–390.
Baym, M., Stone, L. K., & Kishony, R. (2016). Antibiotic resistance multidrug evolutionary strategies to reverse antibiotic resistance. Science, 351, 40–U46.
Chen, Y. S., Wang, Y. C., Chow, Y. S., et al. (2014). Purification and characterization of plantaricin Y, a novel bacteriocin produced by Lactobacillus plantarum 510. Archives of Microbiology, 196, 193–199.
Dathe, M., Nikolenko, H., Meyer, J., et al. (2001). Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Letters, 501, 146–150.
Diep, D. B., Havarstein, L. S., & Nes, I. F. (1996). Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. Journal of Bacteriology, 178, 4472–4483.
Diep, D. B., Havarstein, L. S., Nissen-Meyer, J., et al. (1994). The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Applied and Environmental Microbiology, 60, 160–166.
Ekblad, B., Kyriakou, P. K., Oppegard, C., et al. (2016). Structure-function analysis of the two-peptide bacteriocin plantaricin EF. Biochemistry, 55, 5106–5116.
Enan, G., El-Essawy, A. A., Uyttendaele, M., et al. (1996). Antibacterial activity of Lactobacillus plantarum UG1 isolated from dry sausage: characterization, production and bactericidal action of plantaricin UG1. International Journal of Food Microbiology, 30, 189–215.
Fimland, N., Rogne, P., Fimland, G., et al. (2008). Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF. Biochimica et Biophysica Acta, 1784, 1711–1719.
Gonzalez, B., Arca, P., Mayo, B., et al. (1994). Detection, purification, and partial characterization of plantaricin C, a bacteriocin produced by a Lactobacillus plantarum strain of dairy origin. Applied and Environmental Microbiology, 60, 2158–2163.
Gupta, A., & Tiwari, S. K. (2014). Plantaricin LD1: a bacteriocin produced by food isolate of Lactobacillus plantarum LD1. Applied Biochemistry and Biotechnology, 172, 3354–3362.
Hata, T., Tanaka, R., & Ohmomo, S. (2010). Isolation and characterization of plantaricin ASM1: a new bacteriocin produced by Lactobacillus plantarum A-1. International Journal of Food Microbiology, 137, 94–99.
Hu, M., Zhao, H., Zhang, C., et al. (2013). Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. Journal of Agricultural and Food Chemistry, 61, 11676–11682.
Jacob, B., Kim, Y., Hyun, J. K., et al. (2014). Bacterial killing mechanism of sheep myeloid antimicrobial peptide-18 (SMAP-18) and its Trp-substituted analog with improved cell selectivity and reduced mammalian cell toxicity. Amino Acids, 46, 187–198.
Jimenez-Diaz, R., Ruiz-Barba, J. L., Cathcart, D. P., et al. (1995). Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Applied and Environmental Microbiology, 61, 4459–4463.
Lee, J. H., Park, K. S., Karim, A. M., et al. (2016). How to minimise antibiotic resistance. The Lancet Infectious Diseases, 16, 17–18.
Lopes, J. L., Nobre, T. M., Siano, A., et al. (2009). Disruption of Saccharomyces cerevisiae by plantaricin 149 and investigation of its mechanism of action with biomembrane model systems. Biochimica et Biophysica Acta, 1788, 2252–2258.
Maldonado, A., Ruiz-Barba, J. L., & Jimenez-Diaz, R. (2004). Production of plantaricin NC8 by Lactobacillus plantarum NC8 is induced in the presence of different types of gram-positive bacteria. Archives of Microbiology, 181, 8–16.
Meng, F., Zhao, H., Zhang, C., et al. (2016). Expression of a novel bacteriocin—the plantaricin Pln1—in Escherichia coli and its functional analysis. Protein Expression and Purification, 119, 85–93.
Moll, G. N., Van Den Akker, E., Hauge, H. H., et al. (1999). Complementary and overlapping selectivity of the two-peptide bacteriocins plantaricin EF and JK. Journal of Bacteriology, 181, 4848–4852.
Navarro, L., Rojo-Bezares, B., Saenz, Y., et al. (2008). Comparative study of the pln locus of the quorum-sensing regulated bacteriocin-producing L. plantarum J51 strain. International Journal of Food Microbiology, 128, 390–394.
Nishie, M., Nagao, J. I., & Sonomoto, K. (2012). Antibacterial peptides “bacteriocins”: an overview of their diverse characteristics and applications. Biocontrol Science, 17, 1–16.
Pal, G., & Srivastava, S. (2014). Cloning and heterologous expression of plnE, -F, -J and -K genes derived from soil metagenome and purification of active plantaricin peptides. Applied Microbiology and Biotechnology, 98, 1441–1447.
Pal, G., & Srivastava, S. (2015). Scaling up the production of recombinant antimicrobial plantaricin E from a heterologous host, Escherichia coli. Probiotics and antimicrobial proteins, 7, 216–221.
Rekhif, N., Atrih, A., & Lefebvre, G. (1995). Activity of plantaricin SA6, a bacteriocin produced by Lactobacillus plantarum SA6 isolated from fermented sausage. The Journal of Applied Bacteriology, 78, 349–358.
Rogne, P., Haugen, C., Fimland, G., et al. (2009). Three-dimensional structure of the two-peptide bacteriocin plantaricin JK. Peptides, 30, 1613–1621.
Rumjuankiat, K., Perez, R. H., Pilasombut, K., et al. (2015). Purification and characterization of a novel plantaricin, KL-1Y, from Lactobacillus plantarum KL-1. World journal of microbiology & biotechnology, 31, 983–994.
Sabo, S. D., Vitolo, M., Gonzalez, J. M. D., et al. (2014). Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Research International, 64, 527–536.
Sambrook, J., Maniatis, T., & Fritsch, E. F. (1989). Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory.
Sand, S. L., Oppegard, C., Ohara, S., et al. (2010). Plantaricin A, a peptide pheromone produced by Lactobacillus plantarum, permeabilizes the cell membrane of both normal and cancerous lymphocytes and neuronal cells. Peptides, 31, 1237–1244.
Sandiford, S., & Upton, M. (2012). Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrobial Agents and Chemotherapy, 56, 1539–1547.
Siezen, R. J., Francke, C., Renckens, B., et al. (2012). Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. Journal of Bacteriology, 194, 195–196.
Song, D. F., Zhu, M. Y., & Gu, Q. (2014). Purification and characterization of plantaricin ZJ5, a new bacteriocin produced by Lactobacillus plantarum ZJ5. PloS One, 9, e105549.
Tenover, F. C. (2006). Mechanisms of antimicrobial resistance in bacteria. The American Journal of Medicine, 119, S3–10 discussion S62-70.
Tiwari, S. K., & Srivastava, S. (2008). Purification and characterization of plantaricin LR14: a novel bacteriocin produced by Lactobacillus plantarum LR/14. Applied Microbiology and Biotechnology, 79, 759–767.
Todokoro H, Ezumi M, Ose Y et al. (2003) Scanning electron microscope. In: Google Patents.
Umer Abdullah, S., Badaruddin, M., Ali, R., et al. (2010). Effect of elementary and advanced glycation products of nisin on its preservative efficacy and digestibility. Food Chemistry, 122, 1043–1046.
Van Reenen, C. A., Van Zyl, W. H., & Dicks, L. M. (2006). Expression of the immunity protein of plantaricin 423, produced by Lactobacillus plantarum 423, and analysis of the plasmid encoding the bacteriocin. Applied and Environmental Microbiology, 72, 7644–7651.
Wimley, W. C. (2010). Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chemical Biology, 5, 905–917.
Xie, J., Gou, Y., Zhao, Q., et al. (2014). Antimicrobial activities and membrane-active mechanism of CPF-C1 against multidrug-resistant bacteria, a novel antimicrobial peptide derived from skin secretions of the tetraploid frog Xenopus clivii. Journal of peptide science : an official publication of the European Peptide Society, 20, 876–884.
Yang Fang, Y. Z.-Q., & Ri-Jun, Z. (2010). Heterologly expression of plantaricin genes PlnE and PlnF in E. coli. China Animal Husbandry & Yeterinary Medicine, 37, 55–59.
Yeaman, M. R., & Yount, N. Y. (2003). Mechanisms of antimicrobial peptide action and resistance. Pharmacological Reviews, 55, 27–55.
Zhang, J., Yang, Y. L., Teng, D., et al. (2011). Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expression and Purification, 78, 189–196.
Zhu, X., Zhao, Y., Sun, Y., et al. (2014). Purification and characterisation of plantaricin ZJ008, a novel bacteriocin against Staphylococcus spp. from Lactobacillus plantarum ZJ008. Food Chemistry, 165, 216–223.
Acknowledgments
This project was supported by the Food Biotechnology and Enzyme Engineering Laboratory and the National Natural Science foundation of China (No. 31271936). We thank all of our colleagues for their support and suggestions. We would like to thank Editage (http://www.editage.cn/) for English language editing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meng, F., Zhu, X., Lu, F. et al. Functional Analysis of Plantaricin E and Its Mutant by Heterologous Expression in Escherichia coli . Appl Biochem Biotechnol 182, 311–323 (2017). https://doi.org/10.1007/s12010-016-2328-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2328-9