Abstract
This study aimed to develop biocontrol Bacillus and explore bacterial biocontrol substances. According to the blood agar test, strain FJAT-14262 was screened as a biosurfactant-producer. The biosurfactant-producing ability of FJAT-14262 was further confirmed by the oil spreading tests because of its amphipathic character. Furthermore, its fermentation supernatant could decrease the surface tension from 74.1 to 32.7 mN m−1. Fourier transform infrared spectroscopy (FT-IR) analysis indicated that the biosurfactant produced by the strain FJAT-14262 was a kind of lipopeptides. Reverse-phase high-performance liquid chromatography (RP-HPLC) and liquid chromatography-mass spectrometry (LC-MS) analysis demonstrated that this lipopeptide contained surfactin with polar amino acids and hydrophobic fatty acid chains. Moreover, bioinformatic analysis revealed that the nonribosomal peptide synthetases genes srfAA, srfAB, and srfAC were structurally conserved in the FJAT-14262 genome. Importantly, the crude surfactant exhibited strong inhibitory activities against Fusarium oxysporum, suggesting that strain FJAT-14262 could be a potential biological control agent against Fusarium wilt.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biosurfactants are variety of microbial compounds that exhibit noticeable surface and emulsifying activities. They are divided into glycolipids, lipopeptides, polysaccharide-protein complexes, phospholipids, fatty acids and neutral lipids based on their chemical structures [1, 2]. Biosurfactants have potential applications in pharmaceutical, mining, bioremediation, cosmetics, and food industries of human daily life. Some bacteria, yeasts, and fungi can produce biosurfactants as secondary metabolites [3]. For example, Bacillus species produced a broad spectrum of lipopeptide biosurfactants as bacterial biocontrol agents [4, 5]. Comparing to chemical surfactants, biosurfactants are considered to be less toxic and eco-friendly. In order to produce biosurfactants economically, much effort has been carried out [6]. Vecino et al. [7] extracted biosurfactants from corn steep, which significantly decreased their production cost.
Bacillus species are well known as safe microorganisms and have an outstanding ability of sporulation, which assures their prevalence in varied environments [8]. After the cyclic lipopeptide surfactin was first isolated from Bacillus subtilis ATCC 21332 by Arima et al. [9], an increasing number of biosurfactants produced by Bacillus sp. had been discovered. Many Bacillus strains can produce a range of antimicrobial cyclic lipopeptides with commercial and therapeutic importance. Notably, biosurfactants can be produced by Bacillus strains from many inexpensive substrates [10]. Noah et al. [11] used potato process effluent as substrates for surfactin production by B. subtilis in an airlift reactor.
Most lipopeptide biosurfactant from Bacillus can be classified into three families: surfactin, fengycin, and iturin [1, 12]. The biosynthesis of these lipopeptides is mediated by nonribosomal peptide synthase (NRPSs), and NRPSs of surfactin composed of three conserved genes srfAA, srfAB, and srfAC [3]. Surfactin is a heptapeptide with a β-amino fatty acid, resulting from the condensation of peptide and lipid moieties that are independently synthesized [13]. It has the strongest surfactant activity which can significantly reduce surface tension of water from 72 to 27 mN m−1 [9] and is inhibitory to fungi, bacteria, mycoplasmas, and viruses [14, 15]. Fengycin is cyclic lipopeptide with moderate surfactant activities. It exhibit strong antifungal ability to filamentous fungi and capable of inhibiting biofilm formation of bacteria [16]. NRPSs of fengycin composed of five subunits: FenC, FenD, FenE, FenA, and FenB, while NRPSs of iturin composed of four subunits. Iturin is cyclic lipopeptides linked by a β-amino acid residue. Members of this family have strong antibiotic activity and moderate surfactant activity [6].
In this study, the biosurfactant-producing Bacillus strain FJAT-14262 was isolated from rhizosphere soil of the medicinal plant Anoectochilus roxburghii and identified as a member of the species Bacillus. The results demonstrated that the biosurfactant produced by strain FJAT-14262 displayed strong inhibitory activities against plant pathogenic fungus Fusarium oxysporum. Therefore, the strain FJAT-14262 would be a potential biocontrol agent against Fusarium wilt. The aim of the present work was to find biocontrol Bacillus and exploring bacterial biocontrol substances.
Material and Methods
Cultural Conditions of Bacteria
The strain FJAT-14262 isolated from the rhizosphere soil of medicinal plant A. roxburghii in Guangze county, Fujian province, China. The soil sample was suspended in sterilized water, serially diluted, spread on Luria-Bertani (LB) agar (1% tryptone, 0.5% yeast tract, 0.5% NaCl, pH 7.0–7.2) incubated at 30 °C for 48 h. Pure cultures were obtained by several successive single colony isolations. When cultured on blood agar media at 30 °C for 72 h, FJAT-14262 formed a transparent zone around itself. Due to the strong hemolytic activity [17], the strain was selected to further evaluate the biosurfactant-producing ability by both the oil displacement activity and the surface tension tests [18, 19]. In the hemolytic activity test, the blood agar media (Biosune, Shanghai, China) were sterilized, and 5% sheep blood was added after cooling to 55 °C. The fermentation medium in oil displacement test was comprised of 0.5% glucose, 0.3% beef extract, 1% tryptone, 0.02% MgSO4·7H2O, pH 7.0–7.2. After centrifugation at 12,000×g for 20 min, the surface tension of the culture broth of the strain FJAT-14262 was measured through the bubble pressure method using the tensiometer SITA science line t100 (SITA, Berlin, Germany). All the measurements were performed in triplicate.
Phenotypic and Physiological Characterization
After the strain FJAT-14262 was grown for 2 days (d) at 30 °C on LB, the colonial morphology was characterized and the cellular morphology was further viewed by a transmission electron microscopy (Hitachi, HT7700, Tokyo, Japan). Growth at different temperatures (20o–50 °C, at increments of 5 °C) was tested. Growth was also assessed at different pHs (i.e., 5, 6, 7, 8, 9, and 10) and different salt concentrations (in LB medium supplemented with 0, 2, 4, 6, 8% (w/v) NaCl) at 30 °C. Motility was examined on motility agar. Physiological and biochemical characterization of the strain FJAT-14262 was also analyzed using API 20E and API 50 CH biochemical strips (bioMérieux, Marcy, France) at 37 °C for 48 h.
Phylogenetic Analysis
Genomic DNA was obtained by DNA extraction kit (Generay, Shanghai, China). The 16S ribosomal ribonucleic acid (rRNA) gene was amplified by PCR. According to Liu et al. [20], the primes are 27F (5′-ACTCAAAGGAATTGACGG-3′) and 1492R (5′-TACGGCTACCTTG TTACGACTT-3′). The program was as follows: initial denaturation at 94 °C for 3 min; 30 cycles of 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 5 min. The partial 16S rRNA gene sequences of the strain FJAT-14262 and related taxa (obtained from GenBank) were edited with BioEdit version 7.2.5. Multiple alignments were performed using CLUSTALX. The phylogenetic tree was constructed by the neighbor-joining methods using the MEGA 5.0 software with 1000 bootstrap replicates.
Preparation of Crude Biosurfactants
After fermentation at 30 °C for 48 h, the culture broth of the strain FJAT-14262 was centrifuged at 10,000×g for 10 min. The supernatant was adjusted to pH 2.0 with 6 M HCl and stored at 4 °C for 12 h. Then, it was centrifuged at 10,000×g for 20 min to obtain the precipitate. The precipitate was washed twice with deionized water (pH 2.0) and extracted three times with methanol [5]. The extracts were dried with nitrogen evaporators (N-EVAP-111, Organomation, Boston, MA) to obtaining crude biosurfactants.
Characterization of the Biosurfactants
To characterize the biosurfactants produced by the strain FJAT-14262, the resulted crude biosurfactants were analyzed by FT-IR and reverse-phase high-performance liquid chromatography (RP-HPLC), respectively. For FT-IR analysis, 0.1 g dried sample was mixed with 1.0 g KBr and scanned in the range of 500–4000 cm−1 at a resolution of 4 cm−1 by Nicolet 380 spectrometer (Thermo Fisher, Waltham, MA) [21]. For RP-HPLC analysis, an Agilent Technologies 1100 HPLC (Palo Alto, CA) equipped with a variable wavelength detector (VWD) was used. The sample and standard surfactin (Sigma, St. Louis, MO) were dissolved in methanol, and then passed through a 0.22-μm pore filter. The filtrate was subjected to the reversed-phase HPLC C18 column (4.6 × 250 mm, 5 μm, welch, Shanghai China). The column was eluted with solutions of acetonitrile and Milli-Q water (containing 0.05% trifluoroacetic acid) in the ratio of 90:10 (v/v) at a flow rate of 1 mL min−1 [7]. The eluate was monitored at 210 nm.
Mass Q-TOF Spectrometry
Liquid chromatography-mass spectrometry (LC-MS) and LC-tandem MS (LC-MS/MS) were performed by Agilent Technologies 6250 Accurate-Mass quadrupole time of flight (Q-TOF) LC-MS system (Palo Alto, CA) with Agilent ZORBAX Extend C18 column (2.1 μm × 50 mm, 1.8 μm). The LC-MS operating parameters were as follows: temperature was 350 °C, gas flow 8.0 L·min−1, nebulizer pressure was 30 psig, capillary voltage was 3500 V, and in the positive ion mode [22, 23]. To ensure full coverage of the m/z range of interest for biosurfactant studies, data were recorded in full MS mode along the m/z 100 to 3000 range. The target molecular ion mass identified by LC-MS analysis was further subjected to LC-MS/MS analysis by adding a voltage of 35 V in targeted MS/MS mode.
Bioinformatics Analysis of the Surfactin Peptide Biosynthesis Genes srfAA, srfAB, and srfAC
The genome sequencing of strain FJAT-14262 was performed via the Illumina HiSeq 2500 system and annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) utilizing GeneMark, Glimmer, and BLASTx. The lipopeptide synthesis genes were analyzed with the ORF Finder platform (http://www.bioinformatics.org/sms/orf_find.html). Prediction of the protein structure from the biosurfactant operon was examined using the antiSMASH version 3.0.5.
Amplification of sfp genes was carried out by PCR, and specific primers were designed according to Sen et al. [15]. The amplification reaction was performed using the primers sfp F (5′-ATG AAG ATT TAC GGA ATT TA-3′) and sfp R (5′-TTA TAA AAG CTC TTC GTA-3′) for the sfp fragment, with the following program: initial denaturation at 94 °C for 3 min; 30 cycles of 94 °C for 1 min, annealing at 50 °C for 1 min, and extension at 72 °C for 1 min; and a final extension at 72 °C for 5 min.
Assay for the Antifungal Activity of the Biosurfactants
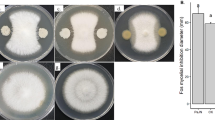
The crude biosurfactants of the strain FJAT-14262 were dissolved in methanol and diluted with phosphate buffered saline (PBS) (pH 8.5) to a concentration of 1 mg mL−1. Surfactin from Sigma was dissolved and diluted in the same method. Subsequently, the ability of the crude biosurfactants to inhibit fungal growth was determined using the dual culture technique on potato dextrose agar (PDA) plates [21]. Firstly, the agar overlays were prepared by mixing 50 mL of melted agar (0.7% agar at 45 °C) with 1 mL of fungal pathogen cultures (108 CFU mL−1) covering the surface of a PDA agar plate. Four fungal phytopathogen of F. oxysporum f. sp. Cubense, F. oxysporum f. sp. Melonis, F. oxysporum f. sp. Melongenae, and F. oxysporum f. sp. lycopersici kept in our lab were selected for this test. Secondly, the Oxford cups were put carefully on the agar overlays. Finally, 5, 10, 20, or 30 μL of crude biosurfactants was separately added to each Oxford cup, and the plates were incubated at 30 °C for 3 to 5 days (d), depending on the fungal pathogens. PBS buffer was used as a blank control and surfactin (1 mg mL−1) as positive control. Each experiment was repeated three times. The size of inhibition zone was used to assess the antifungal activity of the biosurfactants.
Results and Discussion
The Strain FJAT-14262 Is a Biosurfactant-Producing Bacterium
The blood agar test is a convenient technology to detect the hemolytic activity of the biosurfactants [17, 24]. Therefore, the strain which was isolated from the rhizosphere soil of the medicinal plant, A. roxburghii, was subjected to the hemolytic activity test to screen preliminarily biosurfactant-producing bacteria. The strain FJAT-14262 appeared to have the strong hemolytic activity. Thus, Bacillus sp. strain FJAT-14262 was selected to further test and verify its biosurfactant-producing ability. Oil spreading test indicated that the culture supernatants of the strain FJAT-14262 had high oil displacement activity. The diameter of the oil-displaced circle could reach up to 7.5 cm, suggesting a high concentration of the biosurfactants in the culture supernatants [5]. FJAT-14262 can significantly displace the oil layer, showing a displacement zone (Fig. 1S). Biosurfactant can trigger the reduction of surface tension or interfacial tension of liquids in which it is dissolved [14, 15]. Consequently, the surface tension of the culture broths of the strain FJAT-14262 was measured through the bubble pressure method. These results demonstrated that the surface tension could be reduced from 74.1 to 32.7 mN m−1. Taken together, these results consistently confirmed that the strain FJAT-14262 could produce biosurfactants.
The Strain FJAT-14262 Is a Member of Genus Bacillus
The colony characteristics and cell morphology of the strain FJAT-14262 were very similar to those of Bacillus. The colonies grown on LB agar for 2 days were white, dry, and flat. The cells of strain FJAT-14262 were motile rods that were 0.76–0.94 μm in diameter and 1.76–2.94 μm in length after 2-day culture on LB agar (Fig. 1). Growth of the strain FJAT-14262 was observed in the temperature range of 30° to 50 °C (optimum 35 °C), pH range of 5.0 to 9.0 (optimum pH 8.0), and in the presence of 0–6% (w/v) NaCl (optimum 0%). The oxidase and catalase tests were positive; this strain could decompose lysine and ornithine, but not urea; this strain could use arabinose, cellobiose, melibiose, raffinose, glucose, and lactose as carbon sources, but not affinose, melibiose, xylose, inositol, and mannitol (Table 1). Finally, a phylogenetic analysis based on 16S rRNA gene sequence showed that the strain FJAT-14262 was closely related to Bacillus tequilensis KCTC 13622T, B. subtilis 168T, and B. subtilis TU-B-10T with 99.9, 99.7, and 99.8% sequence similarity, respectively (Fig. 2).
Chemical Characteristic of Biosurfactant from Strain FJAT-14262
To characterize the biosurfactant produced by Bacillus sp. FJAT-14262, its fermentation broth was dried. Subsequently, 0.1 g of the dried sample was mixed with 1.0 g KBr, and the mixed samples were analyzed by FT-IR. Five specific absorbance modes were observed at 1378, 1545, 1650, 2930, and 3325 cm−1, respectively (Fig. 3). The wavenumbers of these vibration modes were very similar with those of a lipopeptide (surfactin) produced by Bacillus safensis CCMA-560 [25]. The stretching and vibration modes observed at 2930 and 1378 cm−1 were separately indicative of aliphatic chains –CH3 and –CH2–, suggesting the presence of alkyl chains in the compound from Bacillus sp. FJAT-14262. Similar stretching for the hydrocarbon chain (−C–H–) was also found in a lipopeptide from B. tequilensis CH [21]. According to the FT-IR spectra of a glycolipopeptide from Lactobacillus pentosus and the fengycin from B. subtilis LSFM-05, the stretching absorption at 3325 and bending mode 1650 cm−1 represented the chemical bonds N–H and –CO–NH–, respectively [26, 27]. The vibration mode at 1545 cm−1 suggested the presence of the chemical bond amide II, being typical of a carbon-containing compound with amino group [28]. According to these results (particularly, the comparison of the FT-IR spectra of the tested compound with those of the lipopeptides produced by B. tequilensis CH and B. safensis CCMA-560), the biosurfactants produced by Bacillus sp. FJAT-14262 was identified as a kind of lipopeptides. Through extraction by methanol, the maximum yield of crude lipopeptides could reach 576 mg in 1 L fermentation broth of Bacillus sp. FJAT-14262 under the present cultural conditions.
In order to further confirm the lipopeptide produced by strain FJAT-14262, the crude lipopeptides from Bacillus sp. FJAT-14262 and the standard substances of surfactin were further analyzed and compared by RP-HPLC. The result demonstrated that the lipopeptide sample displayed a very similar RP-HPLC profile to the standard substances of surfactins (Fig. 4) [7]. In the RP-HPLC profile of the standard surfactins, four major peaks/fractions could be discriminated at the retention times of 8.85, 11.07, 11.68, and 13.98 min, respectively (Fig. 4a). In the RP-HPLC profile of the lipopeptide sample, three similar peaks/fractions appeared at the retention times at 8.45, 11.18, and 14.01 min, respectively (Fig. 4b). Consequently, the lipopeptides produced by Bacillus sp. FJAT-14262 could contain surfactin.
The structure of the lipopeptide was investigated by mass spectrometry analysis. Verification of the molecular mass of lipopeptide from strain FJAT-14262, the ESI (+)-MS showed ions of m/z 1036 as the most predominant in this sample (Fig. 5a). The detailed LC-MS/MS analysis of m/z 1036 proved the presence of fragmented masses of 923, 810, 695, 596, 484, and 370.3 (Fig. 5b). The fragmentation pattern was checked by the Metlin database. According the LC-MS/MS result, MS/MS fragmentation pattern of m/z 1036 was found to be highly matching to surfactin with seven amino acids and a fatty acid chain with 15 carbons [29–31].
The Surfactin Peptide Biosynthesis Genes Are Structurally Conserved in the Strain FJAT-14262 Genome
Surfactin was synthesized by a linear nonribosomal peptide synthetase. The corresponding coding genes are organized in an operon in the genomes of Bacillus sp. The genome of strain FJAT-14262 had been sequenced and deposited in the GenBank under the accession no. LGRW00000000 [32]. Bioinformatic analysis revealed that the surfactin peptide biosynthesis genes srfAA, srfAB, and srfAC were organized in the srf operon in the FJAT-14262 genome. The genes srfAA, srfAB, and srfAC were 10,692, 10,677, and 3831 bp in length, coding the corresponding proteins with 3563, 3558, and 1276 amino acids, respectively. The structural prediction indicated that the proteins SrfAA and SrfAB had similar structure and both possessed 10 modules, including three AMP-binding domains (A), three condensation sites (C), three peptidyl carrier domains (PCP), and one epimerization domain (E) (Fig. 6). The protein SrfAC contained one AMP-binding domain (A), one condensation site (C), one peptidyl carrier domain (PCP), and one (TE) domain. All the surfactin peptide biosynthesis genes of strain FJAT-14262 had high structurally similarity with Bacillus species [1].
Downstream to this operon is a sfp gene, encoding sfp protein [27]. The sfp protein belonged to the 4′-phosph-opantetheinyl transferase family and the structure of sfp host highly conserved regions of the alignment involved in binding the magnesium ion [33, 34]. The sfp gene of FJAT-14262a is 675 bp in length, and codes for a protein of 224 amino acids showed a structurally conserved region. The amino acid sequences are very similar between FJAT-14262a, tequilensis, over 90%, while lower amino acid similar with Salinibacillus aidingensis, Bacillus axarquiensis, Bacillus mojavensis, Bacillus atrophaeus, Bacillus amyloliquefaciens, and Bacillus siamensis (Fig. 7). A single nucleotide insertion in sfp gene of B. subtilis 168T led to an incompletion of the surfactin biosynthetic pathway [35]. Although B. subtilis 168T has a complete srf operon, the strain cannot produce surfactin. Strain FJAT-14262 containing both complete srf operon and sfp gene has real potential to producing surfactin.
The Biosurfactant of Bacillus sp. FJAT-14262 Possesses Strong Inhibition Activities Against F. oxysporum
The antifungal activity of lipopeptide from strain FJAT-14262 was tested against the phytopathogen F. oxysporum. The dual cultural tests illustrated that the crude lipopeptide from strain FJAT-14262 exhibited significantly inhibitory activities against all the four forma specialis of F. oxysporum, which can specifically invade four different host plants (Fig. 2S). Moreover, the inhibitory activities were increased with increasing dosage of the crude lipopeptide, appearing a dose-dependent manner (Table 2). But surfactin from sigma had no effect on four forma specialis of F. oxysporum. Surfactin was found to have effective antimicrobial properties such as antibacterial, antiviral, antifungal, antimycoplasma, and hemolytic activities [5]. Snook et al. [36] found surfactin from endophytic bacterium B. mojavensis RRC 101 is toxic to Fusarium verticillioides. The HPLC and LC-MS analyses revealed that the main biosurfactant from strain FJAT-14262 was surfactin. Thus, it could be concluded that surfactin in the crude compounds might play an important role in inhibition of fungal. In order to confirm the antifungal activity of surfactin, removing the srfA operon merits further study.
Conclusions
The strain FJAT-14262 that was isolated from the rhizosphere soil of the medicinal plant A. roxburghii was confirmed to be able to produce biosurfactants. It was also identified as a member of the species Bacillus sp. by the polyphasic taxonomy analyses. Furthermore, biosurfactants produced by Bacillus sp. FJAT-14262 could be a kind of surfactin. Bioinformatic analysis revealed that the surfactin peptide biosynthesis genes srfAA, srfAB, and srfAC were structurally conserved in the FJAT-14262 genome. Importantly, the crude lipopeptide exhibited strong inhibitory activities against the four forma specializes of F. oxysporum. Thus, it can be suggested that Bacillus sp. FJAT-14262 could be a potential biological control agent against Fusarium wilt.
References
Ron, E. Z., & Rosenberg, E. (2001). Natural roles of biosurfactants. Environmental Microbiology, 3, 229–236.
Sachdev, D. P., & Cameotra, S. S. (2013). Biosurfactants in agriculture. Applied Microbiology and Biotechnology, 97(3), 1005–1016.
Roelants, S. L., De Maeseneire, S. L., Ciesielska, K., Van Bogaert, I. N., & Soetaert, W. (2014). Biosurfactant gene clusters in eukaryotes: regulation and biotechnological potential. Applied Microbiology and Biotechnology, 98(8), 3449–3461.
Sharma, R. R., Singh, D., & Singh, R. (2009). Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: a review. Biological Control, 50, 205–221.
Shoda, M. (2000). Bacterial control of plant diseases. Journal of Bioscience and Bioengineering, 89, 515–521.
Rodrigues, L., Banat, I. M., Teixeira, J., & Oliveira, R. (2006). Biosurfactants: potential applications in medicine. The Journal of Antimicrobial Chemotherapy, 26, 609–618.
Vecino, X., Barbosa-Pereira, L., Devesa-Rey, R., Cruz, J. M., & Moldes, A. B. (2015). Optimization of liquid-liquid extraction of biosurfactants from corn steep liquor. Bioprocess and Biosystems Engineering, 38(9), 1629–1637.
Liu, B., Qiao, H., Huang, L., Buchenauer, H., Han, Q., Kang, Z., & Gong, Y. (2009). Biological control of take-all in wheat by endophytic Bacillus subtilis E1R-j and potential mode of action. Biological Control, 49, 277–285.
Arima, K., Kakinuma, A., & Tamura, G. (1968). Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochemical and Biophysical Research Communications, 31, 488–494.
Bonmatin, J. M., Laprévote, O., & Peypoux, F. (2003). Diversity among microbial cyclic lipopeptides: iturins and surfactins. Activity-structure relationships to design new bioactive agents. Combinatorial Chemistry & High Throughput Screening, 6(6), 541–556.
Noah, K. S., Fox, S. L., Bruhn, D. F., Thompson, D. N., & Bala, G. A. (2002). Development of continuous surfactin production from potato process effluent by Bacillus subtilis in an airlift reactor. Applied Biochemistry and Biotechnology, 98, 803–813.
Raaijmakers, J. M., Irene, D. B., Ole, N., et al. (2010). Natural functions of lipopeptides from Bacillus, and Pseudomonas: more than surfactants and antibiotics[J]. South African Journal of Science, 82(4), 172–172.
Singh, P., & Cameotra, S. S. (2004). Potential applications of microbial surfactants in biomedical sciences. Trends in Biotechnology, 22, 142–146.
Zeriouh, H., de Vicente, A., Perez-Garcia, A., & Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environmental Microbiology, 16, 2196–2211.
Sen, R. (2010). Surfactin: biosynthesis, genetics and potential applications. Advances in Experimental Medicine and Biology, 672, 316–323.
Haniyavarn, J., Roongsawang, N., Kameyama, T., et al. (2003). Production and characterization of biosurfactants from Bacillus licheniformis F2.2.[J]. Bioscience Biotechnology and Biochemistry, 67(6), 1239–1244.
Mulligan, C. N., Cooper, D. G., & Neufeld, R. J. (1984). Selection of microbes producing biosurfactants in meida without hydrocarbons. Journal of Fermentation Technology, 62(4), 311–314.
Morikawa, M., Daido, H., Takao, T., Murata, S., Shimonishi, Y., & Imanaka, T. (1993). A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. Journal of Bacteriology, 175, 6459–6466.
Vater, J., Wilde, C., & Kell, H. (2009). In situ detection of the intermediates in the biosynthesis of surfactin, a lipoheptapeptide from Bacillus subtilis OKB 105, by whole-cell cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in combination with mutant analysis. Rapid Communications in Mass Spectrometry, 23(10), 1493–1498.
Liu, B., Liu, G. H., Cetin, S., Schumann, P., Wang, M. K., Tang, J. Y., & Chen, M. C. (2014). Bacillus cihuensis sp. nov., isolated from rhizosphere soil of a plant in the Cihu area of Taiwan. Anton Leeuw., 106, 1147–1155.
Pradhan, A. K., Pradhan, N., Mall, G., Panda, H. T., Sukla, L. B., Panda, P. K., & Mishra, B. K. (2013). Application of lipopeptide biosurfactant isolated from a halophile: Bacillus tequilensis CH for inhibition of biofilm. Applied Biochemistry and Biotechnology, 171, 1362–1375.
Xin, B., Zheng, J., Xu, Z., Li, C., Ruan, L., Peng, D., & Sun, M. (2015). Three novel lantibiotics, ticins A1, A3, and A4, have extremely stable properties and are promising food biopreservatives. Applied and Environmental Microbiology, 81(20), 6964–6972.
Yoshida, S., Hiradate, S., Tsukamoto, T., Hatakeda, K., & Shirata, A. (2001). Antimicrobial activity of culture filtrate of Bacillus amyloliquefaciens RC-2 isolated from mulberry leaves. Phytopathol., 91, 181–187.
Mulligan, C. N., Yong, R. N., & Gibbs, B. F. (2001). Heavy metal removal from sediments by biosurfactants. Journal of Hazardous Materials, 85, 111–125.
Domingos, D. F., de Faria, A. F., de Souza Galaverna, R., Eberlin, M. N., Greenfield, P., Zucchi, T. D., Melo, I. S., Tran-Dinh, N., Midgley, D., & de Oliveira, V. M. (2015). Genomic and chemical insights into biosurfactant production by the mangrove-derived strain Bacillus safensis CCMA-560. Applied Microbiology and Biotechnology, 99, 3155–3167.
Faria, A. F., Stefani, D., Vaz, B. G., & Silva, I. S. (2011). Purification and structural characterization of fengycin homologues produced by Bacillus subtilis LSFM-05 grown on raw glycerol. Journal of Industrial Microbiology & Biotechnology, 38, 863–871.
Moldes, AB., Paradelo, R., Vecino, X. (2013). Partial characterization of biosurfactant from Lactobacillus pentosus and comparison with sodium dodecyl sulphate for the bioremediation of hydrocarbon contaminated soil. Biomed Res Int. Epub: 961842.
Silverstein, R. N., Webster, F. X., Kiemle, D. J., & Bryce, D. L. (2014). Spectrometric identification of organic compounds. 8th ed. New York: Wiley.
Jasim, B., Sreelakshmi, K. S., Mathew, J., & Radhakrishnan, E. K. (2016). Surfactin, iturin, and fengycin biosynthesis by endophytic Bacillus sp. from Bacopa monnieri. Microbial Ecology, 72(1), 106–119.
Roongsawang, N., Washio, K., & Morikawa, M. (2010). Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. International Journal of Molecular Sciences, 12(1), 141–172.
Ma, Z., & Hu, J. (2015). Production and characterization of surfactin-type lipopeptides as bioemulsifiers produced by a pinctada martensii -derived bacillus mojavensis, b0621a. Appl Biochem Biotech., 177(7), 1520–1529.
Chen, Q. Q., Liu, B., Liu, G. H., Wang, J. P., & Che, J. M. (2015). Draft genome sequence of Bacillus tequilensis strain FJAT-14262. Genome Announcements, 12, e01317–e01315.
Nakano, M. M., Marahiel, M. A., & Zuber, P. (1988). Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. Journal of Bacteriology, 170(12), 5662–5668.
Quadri, L. E., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., & Walsh, C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. The Biochemist, 37(6), 1585–1595.
Barbe, V., Cruveiller, S., Kunst, F., Lenoble, P., Meurice, G., Sekowska, A., Vallenet, D., Wang, T., Moszer, I., Médigue, C., & Danchin, A. (2009). From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiol., 155, 1758–1775.
Snook, M. E., Mitchell, T., Hinton, D. M., et al. (2009). Isolation and characterization of Leu7-surfactin from the endophytic bacterium Bacillus mojavensis RRC 101, a biocontrol agent for Fusarium verticillioides[J]. J Agr Food Chem., 57(10), 4287–4292.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (grant #31370059), the Natural Science Foundation of Fujian Province (grant #2013 J01106), the Fujian Key Science and Technology Special Projects—Key Agricultural Science and Technology Special Project (grant #2015NZ0003-1), the Scientific Research Foundation for Returned Scholars, Fujian Academy of Agricultural Sciences (grant #YJRC2014-1), and the Seed industry innovation project of Fujian Province—“ Fujian Resource Preservation Center of the Bacillus-like Bacteria” in the Seed Industry innovation and industrialization of project of Fujian Province (grant #FJZZZY-1544).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Bo Liu and Xiong Guan contributed to the paper equally.
Rights and permissions
About this article
Cite this article
Chen, Q., Liu, B., Wang, J. et al. Antifungal Lipopeptides Produced by Bacillus sp. FJAT-14262 Isolated from Rhizosphere Soil of the Medicinal Plant Anoectochilus roxburghii . Appl Biochem Biotechnol 182, 155–167 (2017). https://doi.org/10.1007/s12010-016-2317-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2317-z