Abstract
In the present study, we first reported a cold-active xylanase of glycosyl hydrolase family 11, Xyn11, from the filamentous fungus Bispora antennata. The coding gene (xyn11) was cloned and successfully expressed in Pichia pastoris. Deduced Xyn11 exhibited the highest identity of 65 % with a family 11 endo-β-1,4-xylanase from Alternaria sp. HB186. Recombinant Xyn11 exhibited maximal activity at 35 °C and remained 21 % of the activity at 0 °C. Sequence alignment showed that the N-terminal sequence of Xyn11 is distinct from those of thermophilic xylanases of family 11. To determine its effect on enzyme properties, the Xyn11 mutant without the N-terminal sequence, t-Xyn11, was then constructed, expressed in P. pastoris, and compared with Xyn11. Both enzymes showed optimal activities at 35 °C and pH 5.5 and were stable at pH 2.0–12.0. Compared with truncated mutant t-Xyn11, Xyn11 retained more activity after 20-min incubation at 40 °C (Xyn11:28 % vs. t-Xyn11:4 %) and degraded xylan substrates more completely. Thus, a new factor affecting the thermostability of cold-active xylanase of family 11 was identified.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hemicelluloses are the major constituents of plant cell walls. Based on their backbone composition, hemicelluloses can be classified as xylans, mannans, arabinogalactans, and arabinans [1]. Of all enzymes required for efficient hemicellulose degradation, endo-β-xylanase (EC 3.2.1.8) is the most critical component that catalyzes the hydrolysis of β-1,4-xylosidic linkages of xylan. Xylanases have been classified into glycosyl hydrolase (GH) families (http://www.cazy.org/fam/acc_GH.html) 5, 7, 8, 10, 11, 30, and 43 [2–4] according to the catalytic domain sequences.
There seems to be an ongoing interest to discover novel xylanases that will be used in various industries [2, 5–7]. Compared with thermoactive enzymes that have received much attention from research and development [8–10], cold-active xylanases are less known in amount and seldom concerned. In comparison to mesophilic (40–60 °C) and thermophilic (>60 °C) counterparts, cold-active enzymes exhibit optimal activities at 0 to 40 °C and maintain a constant enzyme reaction rate at such a low temperature [11, 12]. To our knowledge, there are only 15 cold-active xylanases that have been characterized, including two xylanases A and B from Euphausia superba Dana [13], three GH8 xylanases from Pseudoalteromonas haloplanktis TAH 3a [14], Pseudoalteromonas arctica [15], and an environmental sample [16], and ten GH10 xylanases from Bacillus sp. SN5 [17], Flavobacterium johsoniae [18], Glaciecola mesophila [19], Penicillium sp. FS010 [20], Flavobacterium sp. MSY2 [21], Cryptococcus adeliae [22], goat rumen contents [23], Sorangium cellulosum [24], Bacillus sp. HJ2 [25], and Zunongwangia profunda [26].
Many industrial processes require low temperatures to stabilize product, reduce microbial pollution and/or fermentation, and avoid product denaturation [2, 11]. For example, the psychrophilic xylanases from P. haloplanktis TAH3A, Flavobacterium sp. MSY-2, and unknown bacterial origin have effects on the improvement of dough property and bread volume (up to 28 %) [27]. In our preliminary screening test, Bispora antennata CBS 126.38 grown at 15 °C showed significant xylanase activity in malt extract medium, suggesting the possible existence of cold-active xylanase. Here, we report the cloning and expression of the first cold-active xylanase of GH11, Xyn11, from the filamentous fungus B. antennata. It had a temperature optimum of 35 °C and exhibited 21 % activity even at 0 °C. Sequence analysis indicated that Xyn11 has a distinct N-terminal sequence in comparison with thermophilic counterparts. Functions of this sequence were determined by construction of an N-terminal deletion mutant.
Materials and Methods
Strains and Vectors
B. antennata CBS 126.38 isolated from a beech stump was purchased from the Centraalbureau voor Schimmelcultures (CBS, Utrecht, Netherlands) and cultivated in malt extract medium containing 130 g/l malt extract (Fortune Biotech, Shanghai, China) and 100 mg/l chloramphenicol (BioDee, Beijing, China), pH 5.6 ± 0.2 at 28 °C. The host-vector system (TransGen, Beijing, China) for gene cloning contained Escherichia coli Trans1-T1 competent cells and the plasmid pEASY-T3. The heterologous protein expression system (Invitrogen, Carlsbad, CA, USA) was composed of Pichia pastoris GS115 and pPIC9. Medium preparation and heterologous expression followed the protocol described in the Pichia expression manual (Invitrogen).
Chemicals and Reagents
Birchwood xylan, beechwood xylan, wheat arabinoxylan, carboxymethyl cellulose sodium (CMC-Na), Avicel, p-nitrophenyl cellobioside, and p-nitrophenyl xylopyranoside were purchased from Sigma (St. Louis, MO, USA). The DNA purification kit, LA Taq DNA polymerase, and restriction endonucleases were purchased from TaKaRa (Otsu, Japan). T4 DNA ligase was purchased from New England Biolabs (Ipswich, MA, USA). All other chemicals were of reagent grade or better.
Cloning of the Xylanase Encoding Gene (xyn11)
Mycelia of B. antennata were collected after 20-h growth in 200 ml of malt extract medium at 25 °C. The genomic DNA and total RNA were isolated and purified as described previously [4]. Using the degenerate primers dF and dR (Table S1) specific for GH11 xylanase genes, a PCR product was amplified from the genomic DNA of B. antennata and cloned into pEASY-T3 vector for sequencing and BLAST analysis. Six special primers (usp1–3 and dsp1–3) (Table S1) were designed based on the known sequences and used to obtain the 5′ and 3′ flanking regions by thermal asymmetric interlaced (TAIL)-PCR [28]. The PCR products were sequenced and then assembled with the known fragment. The full-length DNA of xyn11 was directly amplified from the genomic DNA of B. antennata with specific primers SF and SR (Table S1). To obtain the complementary DNA (cDNA) sequence of xyn11, a ReverTra Ace kit (Toyobo, Osaka, Japan) was used to synthesize the first-strand cDNAs and a PCR amplification was performed using primers FF and RR (Table S1) with an annealing temperature of 60 °C.
Sequence Analysis
Vector NTI 7.0 software was used for sequence assembly and analysis. The signal peptide sequence was predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP/). The BLAST server was used for homology searches in GenBank. Multiple alignments of deduced Xyn11 and those mesophilic or thermophilic GH11 xylanases from Chaetomium thermophilum [29, 30], Trichoderma reesei [31], Paecilomyces varioti bainier [32], and Aspergillus niger [33], respectively, were carried out using the ClustalW program (http://www.ebi.ac.uk/clustalW). Homology modeling and electrostatic interaction analysis of key residues were performed using Accelrys Discovery Studio software (DS 2.5, http://www.accelrys.com) with the GH11 β-xylanase from T. reesei (PDB 1XYP; 68 % identity) as the template [34]. The credibility of the putative tertiary model was evaluated with Ramanchandran plot [35].
Enzyme Assay
Xylanase activity assay was performed using 3,5-dinitrosalicylic acid (DNS) method [36]. Standard assay mixture was composed of 900 μl of 100 mM sodium citrate-phosphate buffer (pH 5.5) containing 1 % (w/v) beechwood xylan and 100 μl of appropriately diluted enzyme. The amount of enzyme releasing 1 μmol of reducing sugar per min at given assay conditions was defined as one unit (U) of xylanase activity.
Expression of Xyn11 and t-Xyn11 in P. pastoris
Sequence analysis indicated that deduced mature Xyn11 had an extra eleven N-terminal residues when compared to its mesophilic or thermophilic counterparts. To verify its function, the cDNAs encoding mature Xyn11 without the signal peptide coding sequence and N-terminal deletion mutant t-Xyn11 were amplified by PCR with primers FF and RR and tF and RR, respectively (Table S1). The PCR products were digested with SnaBI and NotI and cloned into pPIC9 vector, respectively, downstream of the α-factor signal peptide sequence. The recombinant plasmids, pPIC9-xyn11 and pPIC9-t-xyn11, were linearized with PmeI, and then transformed into P. pastoris GS115 competent cells by electroporation, respectively. The transformed cells were cultured on minimal dextrose base agar plates and incubated at 30 °C for about 2 days. His + transformants were transferred to minimal dextrose agar plates and 3 ml of buffered methanol complex medium for 2-day growth at 30 °C. The cells of buffered glycerol-complex medium were pelleted by centrifugation (5000×g, 5 min) and resuspended in 1 ml of buffered methanol complex medium. After 2-day induction with 0.5 % methanol at 30 °C, the culture was pooled, and the culture supernatant was collected by centrifugation (12,000×g, 4 °C, 3 min) and subjected to xylanase activity assay. The transformant having the highest xylanase activity was fermented in 1-l flasks and used for subsequent analysis.
Purification of Recombinant Xyn11 and t-Xyn11
The induced cultures were centrifuged at 12,000×g for 10 min at 4 °C, and the cell-free culture supernatants were desalted against 20 mM Tris-HCl (pH 7.0) with a GE Healthcare desalting column. The crude enzymes were loaded onto a HiTrap Q Sepharose XL 5 ml FPLC column (GE Healthcare, Uppsala, Sweden) that was equilibrated with the same buffer. A linear gradient of NaCl (0–1.0 M) at a flow rate of 3.0 ml/min was used to elute proteins. Fractions with enzyme activity were pooled for characterization. Protein purification profiles were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) to confirm the purity [37]. The protein concentration was measured by Bradford method [38] with bovine serum albumin as the standard. To identify the proteins, the bands were excised from the SDS-PAGE gel and analyzed utilizing liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) at the Tianjin Biochip Corp., Tianjin, China.
Characterization of Purified Recombinant Xylanases Xyn11 and t-Xyn11
The pH and temperature optima, pH stability, thermal stability, and the effect of chemicals on the activities of purified recombinant xylanases were determined using the methods described previously [4].
Substrate Specificity and Kinetic Parameters
The substrate specificity of purified recombinant Xyn11 was tested by measuring its enzyme activity against birchwood xylan, beechwood xylan, wheat arabinoxylan, CMC-Na, Avicel, p-nitrophenyl cellobioside, and p-nitrophenyl xylopyranoside in 100 mM citric acid-Na2HPO4 (pH 5.5), respectively. The K m and V max values of Xyn11 were determined by using 1–10 mg/ml birchwood xylan and beechwood xylan as the substrates. The K m and V max values of t-Xyn11 were determined by using 1–10 mg/ml beechwood xylan as the substrates. The data were plotted to construct Lineweaver-Burk plots. Each experiment was repeated three times.
Analysis of Hydrolysis Products
Reactions containing 100 U of Xyn11 and 500 μl of 100 mM citric acid-Na2HPO4 (pH 5.5) containing 0.2 % (w/v) birchwood or beechwood xylan were incubated at 37 °C for 12 h. After removal of the extra enzyme by 3 kDa cutoff membrane ultrafiltration tube (Pall, USA), the hydrolysis products were assayed by using high-performance anion-exchange chromatography (HPAEC) with a model 2500 HPAEC system (Dionex, Sunnyvale, CA, USA) [39].
Nucleotide Sequence Accession Number
The nucleotide sequence for the GH11 xylanase gene (xyn11) of B. antennata was deposited in the GenBank database under accession number JQ685507.
Results
Cloning and Sequence Analysis of xyn11
A 200-bp xylanase gene fragment was amplified from the genomic DNA of B. antennata using degenerate primers specific for GH11 xylanases. The 5′ and 3′ flanking regions, 370 and 160 bp, respectively, were obtained by TAIL-PCR and assembled with the core region to generate a DNA sequence of 730 bp. The full-length DNA and cDNA sequences of xyn11 were determined to be 674 and 603 bp, respectively (Fig. S1). One intron, 71 bp in length, was identified in the genomic sequence of xyn11. SingalP predicted the presence of a putative N-terminal signal peptide at residues 1–19 of the deduced amino acids sequence of xyn11. The mature protein was composed of 181 residues with a calculated molecular mass of 21.7 kDa and exhibited the highest identity of 74 % with a family 11 endo-β-1,4-xylanase from Alternaria sp. HB186. Sequence alignment analysis indicated that deduced mature Xyn11 has 11 extra residues (APSEVLVERGG) at the N-terminus in comparison with mesophilic and thermophilic counterparts (Fig. 1). Using GH11 β-xylanase from T. reesei (1XYP; 68 % identity) as the template [34], a thumb region conserved amongst GH11 xylanases was shown and the single catalytic domain of Xyn11 contains two twisted anti-parallel β-sheets and one α-helix. The two catalytic glutamates, Glu95 and Glu187, are located at the concave site of the palm. Modeled t-Xyn11 has similar structure to that of Xyn11 except for the significant difference in the N-terminal areas (Fig. S2). The consensus sequence PSIDG was also identified.
Heterologous Expression and Purification of Xyn11 and t-Xyn11
The mature forms of Xyn11 and t-Xyn11 were successfully expressed in P. pastoris GS115. Xyn11 and t-Xyn11 in the culture supernatants were purified to electrophoretic homogeneity (Fig. 2). t-Xyn11 migrated a single band on SDS-PAGE with an apparent molecular mass of 21 kDa, in good agreement with its calculated value. Xyn11 showed two bands on the gel with apparent molecular masses of 22 kDa (similar to the calculated molecular weight) and 19 kDa, respectively. Both bands were identified to be Xyn11 by LC-ESI-MS/MS (Fig. S1). The loss of molecular weight might be ascribed to protein truncation during expression.
Properties of the Purified Recombinant Xyn11 and t-Xyn11
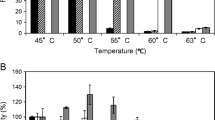
Xyn11 and t-Xyn11 had maximal activities at pH 5.5 (35 °C) (Fig. 3a). Both enzymes displayed high activities over a wider pH range, remaining more than 30 % activity at pH 3.0–8.0. The enzymes retained stable after 1-h incubation at 20 °C over a broad pH range of 2.0 to 12.0 (Fig. 3b). Xyn11 and t-Xyn11 had similar temperature optima of 35 °C (Fig. 3c). Xyn11 was relatively stable at 30 °C, retaining 80 % relative activity after 60-min incubation and 28 % activity at 40 °C for 20 min (Fig. 3d). t-Xyn11 had worse stability than Xyn11, retaining 60 % activity after incubation at 30 °C for 60 min and 4 % activity at 40 °C for 20 min. These results suggest that the distinct N-terminal sequence of Xyn11 is important for enzyme thermostability.
Characterization of the purified recombinant Xyn11and t-Xyn11. a The effect of pH on xylanase activity. The activity assays were performed at 35 °C in buffers of pH 2.0–9.0 for 10 min. b pH stability of Xyn11 and t-Xyn11. After preincubating the enzymes at 20 °C for 1 h at pH 2.0–11.0 without substrate, the residual activities were measured in 100 mM citric acid-Na2HPO4 (pH 5.5, 35 °C, 10 min). c The effect of temperature on xylanase activities measured in 100 mM citric acid-Na2HPO4 (pH 5.5) for 10 min. d Thermostability of purified Xyn11 and t-Xyn11. The enzymes were preincubated at 30 or 40 °C in 100 mM citric acid-Na2HPO4 (pH 5.5) without substrate. Aliquots were removed at specific time points for measurement of residual activities in the same buffer at 35 °C. Each value in the panel represents the mean ± SD (n = 3)
Substrate Specificity and Kinetic Parameters of Xyn11
The same as the cold-active xylanase Xyn8 from P. arctica [15], XYL from Penicillium sp. FS010 [20] and XynGR40 from goat rumen contents [21], Xyn11 exhibited the highest activity towards birchwood xylan (100 %). It also had high activities against beechwood xylan (99.5 %) and wheat arabinoxylan (96.1 %). No activity could be detected on CMC-Na, Avicel, p-nitrophenyl cellobioside, and p-nitrophenyl xylopyranoside. These results indicated that Xyn11 has high but relatively narrow substrate specificity.
The kinetic parameters of Xyn11 and t-Xyn11 for xylan substrates were determined at the optimal temperature (35 °C). With birchwood xylan as substrate, the K m and V max values of Xyn11 were 1.65 mg/ml and 236.3 μmol/min/mg, respectively. With beechwood xylan as substrate, the K m and V max values of Xyn11 and t-Xyn11 were 1.73 mg/ml and 276.6 μmol/min/mg and 1.14 mg/ml and 100.3 μmol/min/mg, respectively.
Analysis of Hydrolysis Products
The hydrolysis products of birchwood xylan and beechwood xylan by Xyn11 were determined by HPAEC. Like XynA from S. cellulosum and XynGR40 from goat rumen contents (Table 1), Xyn11 catalyzed the hydrolysis of beechwood and birchwood xylan to produce mainly xylose and xylobiose. The hydrolysis products of birchwood xylan were comprised of 68.79 % xylose, 30.39 % xylobiose, and 0.82 % other xylan polymers. The composition of the hydrolysis products of beechwood xylan was 40.21 % xylose, 57.92 % xylobiose, and 1.87 % other xylan polymers. The hydrolysis products of beechwood xylan by t-Xyn11 mainly consisted of xylose (25.86 %) and xylobiose (74.14 %). These results indicate that Xyn11 can hydrolyze xylan substrates more completely.
Discussion
In this study, we selected B. antennata as the source strain and identified in it a GH11 endo-1,4-β-xylanase (Xyn11). Xyn11 has the typical characteristics of cold-active xylanases [2], such as a low temperature optimum, high catalytic activity at low temperatures, and poor thermostability. It is the first reported cold-active GH11 xylanase known so far.
Like most cold-active xylanases that have acid to neutral pH optima (Table 1), Xyn11 had maximal activity at pH 5.5 (35 °C). However, Xyn11 displayed high activity over a wider pH range, remaining more than 30 % activity at pH 3.0–8.0. By comparison with Xyn11, Xyn8 from P. arctica lost all activity at pH 4.0, XynA from G. mesophila had less than 10 % activity at pH 3.0, and xylanases A and B from E. superba had less than 20 % relative activity at pH 4.0. Moreover, Xyn11 had the greatest pH stability of all known cold-active xylanases (Table 1). It remained stable after 1-h incubation at 20 °C over a broad pH range of 2.0 to 12.0. On the other hand, cold-active xylanases known so far generally have temperature optima of 35 °C or below (Table 1). Xyn11 had a similar temperature optimum (35 °C). It remained more activity (21 %) at 0 °C than most known GH10 cold-active xylanases but less than Xyn8 from P. arctica (60 % activity). Its thermostability at 40 °C is much worse than that of almost all of the counterparts. The result indicated that Xyn11 may have a more flexible structure. Further removal of the N-terminal sequence makes Xyn11 less stable. Thus, the probable mechanisms intrigue us for deeper analysis.
It has been reported that cold-active enzymes are characterized by high flexibility that allows the molecular motions necessary for activity at low temperatures and thermolability at moderate temperatures [40, 42]. Amino acid composition and secondary structure are the key factors of enzyme thermostability. When compared with GH11 thermophilic xylanases that have an increased occurrence of Thr, Tyr, and Arg and decreased Ser and Asn [29], Xyn11 showed the reverse trend in amino acid composition (Table 2). Arg and Tyr may form both short and long range interactions due to their large side chains. The guanidinium group in Arg and the hydroxyl group in Tyr can form salt bridges and hydrogen bonds, respectively. Hence, they appear to function in both binding and folding at high temperatures, consequently contributing towards protein stability [41, 43]. For example, Turunen et al. introduced five Arg residues into T. reesei xylanase II and improved the enzyme thermotolerance significantly [44]. On the other hand, Ser forms mostly local interactions due to its short side chain [45]. An interesting phenomenon has shown that the ratio of Thr:Ser in thermophilic xylanases is higher than that in mesophilic proteins. The reason might be that more Thr and less Ser improve the β-strand forming propensities, since over half of Thr and Ser of GH11 xylanases are located in the β-strands [29]. Asn has a low forming propensity and is easily deamidated at high temperature, and thus might be avoided in the strands of thermophilic xylanases [29]. These amino acid composition characteristics may combine altogether to result in the thermolability of Xyn11 (retaining only 28 % relative activity at 40 °C after 20 min).
Sequence alignment analysis indicated that the N-terminal sequence APSEVLVERGG of deduced Xyn11 is absent from thermophilic GH11 xylanases. The effect of N-terminus on the thermostability of xylanases has been reported previously [46–49]. For example, the thermostability of a mesophilic xylanase from Steptomyces olivaceovirdis has been significantly improved by substituting the N-terminal residues with a corresponding sequence of a thermophilic Thermomonospora fusca xylanase [50]. Then it is interesting to clarify whether the distinct N-terminal sequence of cold-active Xyn11 exerts influence on enzyme thermostability. Thus, its functions on enzyme properties were further studied by constructing a truncated mutant. Xyn11 and t-Xyn11 had similar temperature optima of 35 °C but varied in thermostability and hydrolysis capability. Moreover, they have similar tertiary structures except for the N-terminal regions (Fig. S2). It might be the reason why t-Xyn11 was less thermostable than Xyn11 at 40 °C. It is speculated that there are some interactions between the extra N-terminal residues and other residues in close proximity, and deletion of these residues may destroy their interactions and further influence the enzyme stability.
In comparison with t-Xyn11, Xyn11 showed decreased substrate binding affinity but increased catalytic activity. However, it showed higher binding affinity and catalytic activity towards birchwood xylan when compared with other cold-active xylanases (Table 1). Moreover, Xyn11 showed higher activity (203.8 U/mg) than XynAs from S. cellulosum and G. mesophila, but less than Xyn8 from P. arctica, XynGR40 from goat rumen and XYL from P. chrysogenum. Its hydrolysis products are simple, only consist of xylose and xylobiose. These properties make Xyn11 potential for use in some low-temperature industries.
References
Hilge, M., Gloor, S. M., Rypniewski, W., Sauer, O., Heightman, T. D., Zimmermann, W., Winterhalter, K., & Piontek, K. (1998). High-resolution native and complex structures of thermostable β-mannanase from Thermomonospora fusca-substrate specificity in glycosyl hydrolase family 5. Structure, 6, 1433–1444.
Collins, T., Gerday, C., & Feller, G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiology Reviews, 29, 3–23.
Haegeman, A., Vanholme, B., & Gheysen, G. (2009). Characterization of a putative endoxylanase in the migratory plant-parasitic nematode Radopholus similis. Molecular Plant Pathology, 10, 389–401.
Luo, H., Yang, J., Li, J., Shi, P., Huang, H., Bai, Y., Fan, Y., & Yao, B. (2010). Molecular cloning and characterization of the novel acidic xylanase XYLD from Bispora sp. MEY-1 that is homologous to family 30 glycosyl hydrolases. Applied Microbiology and Biotechnology, 86, 1829–1839.
Biely, P. (1991). Biotechnological potential and production of xylanolytic systems free of cellulases. In G. F. Leatham & M. E. Himmel (Eds.), Enzymes in biomass conversion (Vol. 460, pp. 408–416). Washington D.C: ACS Symposium.
Paës, G., Berrin, J. G., & Beaugrand, J. (2011). GH11 xylanases: structure/function/properties relationships and applications. Biotechnology Advances, 30, 564–592.
Subramaniyan, S., & Prema, P. (2002). Biotechnology of microbial xylanases: enzymology, molecular biology, and application. Critical Reviews in Biotechnology, 22, 33–64.
Andrews, S. R., Taylor, E. J., Pell, G., Vincent, F., Ducros, V. M., Davies, G. J., Lakey, J. H., & Gilbert, H. J. (2004). The use of forced protein evolution to investigate and improve stability of family 10 xylanases. The Journal of Biological Chemistry, 279, 54369–54379.
Sunna, A., & Bergquist, P. L. (2003). A gene encoding a novel extremely thermostable 1,4-β-xylanase isolated directly from an environmental DNA sample. Extremophiles, 7, 63–70.
Turunen, O., Janis, J., Fenel, F., & Leisola, M. (2004). Engineering the thermotolerance and pH optimum of family 11 xylanases by site-directed mutagenesis. Methods in Enzymology, 388, 156–167.
Gerday, C., Aittaleb, M., Bentahir, M./., Chessa, J. P., Claverie, P., Collins, T., D’Amico, S., Dumont, J., Garsoux, G., Georlette, D., Hoyoux, A., Lonhienne, T., Meuwis, M. A., & Feller, G. (2000). Cold-adapted enzymes: from fundamentals to biotechnology. Trends in Biotechnology, 18, 103–107.
Siddiqui, S. K., & Cavicchioli, R. (2006). Cold-adapted enzymes. Annual Review of Biochemistry, 75, 403–433.
Turkiewicz, M., Kalinowska, H., Zielińska, M., & Bielecki, S. (2000). Purification and characterization of two endo-1,4-β-xylanases from Antarctic krill, Euphausia superba Dana. Comparative Biochemistry and Physiology. Part B, Biochemistry and Molecular Biology, 127, 325–335.
Collins, T., Meuwis, M. A., Stals, I., Claeyssens, M., Feller, G., & Gerday, C. (2002). A novel family 8 xylanase, functional and physicochemical characterization. The Journal of Biological Chemistry, 277, 35133–35139.
Elleuche, S., Piascheck, H., & Antranikian, G. (2011). Fusion of the OsmC domain from esterase EstO confers thermolability to the cold-active xylanase Xyn8 from Pseudoalteromonas arctica. Extremophiles, 15, 311–317.
Lee, C. C., Kibblewhite-Accinelli, R. E., Wagschal, K., Robertson, G. H., & Wong, D. W. (2006). Cloning and characterization of a cold-active xylanase enzyme from an environmental DNA library. Extremophiles, 10, 295–300.
Bai, W., Xue, Y., Zhou, C., & Ma, Y. (2012). Cloning, expression and characterization of a novel salt-tolerant xylanase from Bacillus sp. SN5. Biotechnology Letters, 34, 2093–2099.
Chen, S. C., Kaufman, M. G., Miazgowicz, K. L., Bagdasarian, M., & Walker, E. D. (2013). Molecular characterization of a cold-active recombinant xylanase from Flavobacterium johnsoniae and its applicability in xylan hydrolysis. Bioresource Technology, 128, 145–155.
Guo, B., Chen, X., Sun, C., Zhou, B., & Zhang, Y. (2009). Gene cloning, expression and characterization of a new cold-active and salt-tolerant endo-β-1,4-xylanase from marine Glaciecola mesophila KMM 241. Applied Microbiology and Biotechnology, 84, 1107–1115.
Hou, Y., Wang, T., Long, H., & Zhu, H. (2006). Novel cold-adaptive Penicillium strain FS010 secreting thermo-labile xylanase isolated from Yellow Sea. Acta Biochimica et Biophysica Sinica, 38, 142–149.
Lee, C. C., Smith, M., Kibblewhite-Accinelli, R. E., Williams, T. G., Wagschal, K., Robertson, G. H., & Wong, D. W. (2006). Isolation and characterization of a cold-active xylanase enzyme from Flavobacterium sp. Current Microbiology, 52, 112–116.
Petrescu, I., Lamotte-Brasseur, J., Chessa, J. P., Ntarima, P., Claeyssens, M., Devreese, B., Marino, G., & Gerday, C. (2000). Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles, 4, 137–144.
Wang, G., Luo, H., Wang, Y., Huang, H., Shi, P., Yang, P., Meng, K., Bai, Y., & Yao, B. (2011). A novel cold-active xylanase gene from the environmental DNA of goat rumen contents: direct cloning, expression and enzyme characterization. Bioresource Technology, 102, 3330–3336.
Wang, S., Hu, W., Lin, X., Wu, Z., & Li, Y. (2012). A novel cold-active xylanase from the cellulolytic myxobacterium Sorangium cellulosum So9733-1: gene cloning, expression, and enzymatic characterization. Applied Microbiology and Biotechnology, 93, 1503–1512.
Zhou, J., Dong, Y., Tang, X., Li, J., Xu, B., Wu, Q., Gao, Y., Pan, L., & Huang, Z. (2012). Molecular and biochemical characterization of a novel intracellular low-temperature-active xylanase. Journal of Microbiology and Biotechnology, 22, 501–509.
Liu, X., Huang, Z., Zhang, X., Shao, Z., & Liu, Z. (2014). Cloning, expression and characterization of a novel cold-active and halophilic xylanase from Zunongwangia profunda. Extremophiles, 18, 441–450.
Dornez, E., Verjans, P., Arnaut, F., Delcour, J. A., & Courtin, C. M. (2011). Use of psychrophilic xylanases provides insight into the xylanase functionality in bread making. Journal of Agricultural and Food Chemistry, 59, 9553–9562.
Liu, Y. G., & Whittier, R. F. (1995). Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 25, 674–681.
Hakulinen, N., Turunen, O., Janis, J., Leisola, M., & Rouvinen, J. (2003). Three-dimensional structures of thermophilic β-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. European Journal of Biochemistry, 270, 1399–1412.
Janis, J., Hakanpaa, J., Hakulinen, N., Ibatullin, F. M., Hoxha, A., Derrick, P. J., Rouvinen, J., & Vainiotalo, P. (2005). Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography. FEBS Journal, 272, 2317–2333.
Törrönen, A., Harkki, A., & Rouvinen, J. (1994). Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO Journal, 13, 2493–2501.
Kumar, P. R., Eswaramoorthy, S., Vithayathil, P. J., & Viswamitra, M. A. (2000). The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier. Journal of Microbiology and Biotechnology, 295, 581–593.
Krengel, U., & Dijkstra, B. W. (1996). Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. Journal of Microbiology and Biotechnology, 263, 70–78.
Törrönen, A., & Rouvinen, J. (1995). Structural comparison of two major endo-1,4-xylanases from Trichoderma reesei. Biochemistry, 34, 847–856.
Ramachandran, G. N., Ramakrishnan, C., & Sasisekharan, V. (1963). Stereochemistry of polypeptide chain configurations. Journal of Microbiology and Biotechnology, 7, 95–99.
Miller, G. L., Blum, R., Glennon, W. E., & Burton, A. L. (1960). Measurement of carboxymethylcellulase activity. Analytical Biochemistry, 1, 127–132.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Yang, P., Shi, P., Wang, Y., Bai, Y., Meng, K., Luo, H., Yuan, T., & Yao, B. (2007). Cloning and overexpression of a Paenibacillus β-glucanase in Pichia pastoris: purification and characterization of the recombinant enzyme. Journal of Microbiology and Biotechnology, 17, 58–66.
Collins, T., Roulling, F., Piette, F., Marx, J. C., Feller, G., Gerday, C., & D’Amico, S. (2008). Fundamentals of cold-adapted enzymes. In R. Margesin, F. Schinner, J. C. Marx, & C. Gerday (Eds.), Psychrophiles: from biodiversity to biotechnology (pp. 211–227). London: Springer.
Querol, E., Perez-Pons, J. A., & Mozo-Villarias, A. (1996). Analysis of protein conformational characteristics related to thermostability. Protein Engineering, 9, 265–271.
Spiwok, V., Lipovová, P., Skálová, T., Dušková, J., Dohnálek, J., Hašek, J., Russell, N. J., & Králová, B. (2007). Cold-active enzymes studied by comparative molecular dynamics simulation. Journal of Molecular Modeling, 13, 485–497.
Kumar, S., Tsai, C. J., & Nussinov, R. (2000). Factors enhancing protein thermostability. Protein Engineering, 13, 179–191.
Turunen, O., Vuorio, M., Fenel, F., & Leisola, M. (2002). Engineering of multiple arginines into the Ser/Thr surface of Trichoderma reesei endo-1,4-beta-xylanase II increases the thermotolerance and shifts the pH optimum towards alkaline pH. Protein Engineering, 15, 141–145.
Jeffrey, G. A., Saenger, W. (1994). Hydrogen bonding in biological structures. Berlin and New York
Fenel, F., Leisola, M., Jänis, J., & Turunen, O. (2004). A de novo designed N-terminal disulphide bridge stabilizes the Trichoderma reesei endo-1,4-β-xylanase II. Journal of Biotechnology, 108, 137–143.
Sung, W. L., Yaguchi, M., & Ishikawa, K. (1998). Modification of xylanase to improve thermophilicity, alkophilicity and thermostability for pulp bleaching. Patent US-575984. Canada: National Research Council of Canada.
Wakarchuk, W. W., Sung, W. L., Campbell, R. L., Cunningham, A., Watson, D. C., & Yaguchi, M. (1994). Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Engineering, 7, 1379–1386.
Liu, L., Zhang, G., Zhang, Z., Wang, S., & Chen, H. (2011). Terminal amino-acids disturb xylanase thermostability and activity. The Journal of Biological Chemistry, 286, 44710–44715.
Zhang, S., Zhang, K., Chen, X., Chu, X., Sun, F., & Dong, Z. (2010). Five mutations in N-terminus confer thermostability on mesophilic xylanase. Biochemical and Biophysical Research Communications, 395, 200–206.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31201829), the National High-Tech Researchand Development Program (863 Program, No. 2012AA022208), and the National “948” Project (2014-S1).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table S1
(DOC 1908 kb)
Fig. S1
The cDNA nucleotide and deduced amino acid sequences of xyn11 from B. attennata. The putative signal peptide is boxed. The stop codon is indicated with an asterisk. The internal peptides of Xyn11 and t-Xyn11 identified by LC-ESI-MS/MS are underlined in red. (DOC 1908 kb)
Fig. S2
The tertiary structures of Xyn11 (a), t-Xyn11 (b) and their fitness (c) predicted with Accelrys Discovery Studio 8 software using the GH11 xylanase from Trichoderma reesei (68% identity; PDB: 1XYP) as the template. The putative catalytic residues are indicated. (DOC 1908 kb)
Rights and permissions
About this article
Cite this article
Liu, Q., Wang, Y., Luo, H. et al. Isolation of a Novel Cold-Active Family 11 Xylanase from the Filamentous Fungus Bispora antennata and Deletion of its N-Terminal Amino Acids on Thermostability. Appl Biochem Biotechnol 175, 925–936 (2015). https://doi.org/10.1007/s12010-014-1344-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1344-x