Abstract
Purpose of Review
Ankylosing spondyloarthritis (AS) is a chronic inflammatory disease that involves the axial joints and entheses. Extra-spinal manifestations such as anterior uveitis, psoriasis, and colitis also occur frequently. This review on the pathogenesis of AS includes an update on the recent discoveries within the field.
Recent Findings
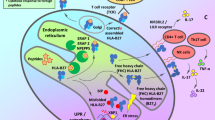
HLA-B*27 is still considered of major importance in the pathogenesis, and it has recently been shown to profoundly affect the gut microbiome and its metabolites and the handling of bacteria during infection. Biochemical and biophysical properties of HLA-B*27 influence its ability to misfold, to induce an endoplasmic reticulum stress response, and to promote autophagy/unfolded protein responses (UPR). HLA-B*27 free heavy chains may induce inflammation through T cells, NK cells, and myeloid cells. Induction of UPR genes results in release of tumor necrosis factor-α (TNF-α), interleukin-17 (IL-17), IL-23, and interferon-γ and increase in T helper (Th) 17 cells. Several other HLA-B and non-B molecules have been associated with AS, although their role in the pathogenesis is unknown.
Summary
Genotypes of endoplasmic reticulum aminopeptidases (ERAP) 1 and 2 have been associated with alterations in the antigenic pool expressed by HLA-B*27 molecules. In the gut, innate immune cells type 3 (ILC3) influence T cell expression of IL-17 and IL-22. Gamma-delta (γ/δ) T cells are induced by IL-23 to produce IL-17. IL-7 induces mucosa-associated invariant T (MAIT) cells to produce IL-17. Besides the microbiome, zonulin may be important through its effects on the permeability of tight junctions in the intestinal epithelial barrier.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, several studies have given information on the pathogenesis of ankylosing spondylitis (AS). Until the era of modern genetics, information on pathogenesis was primarily based on clinical studies of heredity, biopsy studies of joint synovium and gut mucosa, and animal models of AS. Simultaneously, the continuous achievements in genetics and development of novel targeted biological therapies have also added valuable information, even though the pathogenesis of AS is still not entirely recognized. The current review comprises a summary of our current understanding of the pathogenesis of AS, and it particularly focuses on advances in the last 2–3 years.
Epidemiology of Ankylosing Spondylitis and Its Relation to HLA-B*27
In the early 1970s, a strong connection between AS and the human leukocyte antigen (HLA)-B*27 was reported, which is also reflected in the worldwide occurrence of HLA-B*27 and AS [1]. HLA-B*27 and AS are common in native populations of Western Canada, Alaska, and Siberia (occurrence of HLA-B*27, 40–50%; occurrence of AS, 6–10%) and in the northern part of Scandinavia (15–25% and 1.1–1.8%, respectively). In Western Europe, the prevalence of HLA-B*27 and AS is lower (4–13% and 0.23–0.55%, respectively), and it declines further from Middle East/North Africa (2–5% and 0.07–0.14%, respectively) to Japan, where it is uncommon (1% and 0.0065%, respectively) [1]. AS also has a strong heredity background [2], which is not only connected to the presence of HLA-B*27, e.g., first-degree relatives of HLA-B*27-positive AS patients had a 6–16 times higher relative risk of the disease as compared with relatives of HLA-B*27-positive healthy subjects (HS) in the general population [3, 4]. In twin studies, the disease concordance of AS was higher in identical twins (50–63%) than in fraternal twins (13–20%) [5,6,7]. More than 160 subtypes of the HLA-B*27 allele have been discovered [8], but besides a few common subtypes, many of the individual subtypes are too rare to be investigated for disease associations. Not all subtypes are equally distributed in world populations, and thus, AS is relatively more often observed with HLA-B*27:05 in Caucasians (the ancestral subtype), HLA-B*27:02 in Mediterranean, HLA-B*27:03 in sub-Sahara/Middle Eastern populations, HLA-B*27:04 in Chinese and Asian populations, and HLA-B*27:07 in South East Asian populations [9]. Moreover, AS has been associated with HLA-B*27:08 and HLA-B*27:10, whereas HLA-B*27:06 and HLA-B*27:09 appear to be “disease neutral” or less strongly associated with AS [9].
The Pathobiology of HLA-B*27 Subtypes
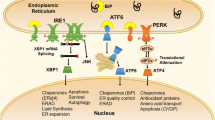
The physiological function of major histocompatibility complex (MHC) class I molecules is to present antigenic peptides to T cell receptors (TCR) of cytotoxic T cells. The HLA-B*27 molecules associated with AS differ only at a few amino acid residues as compared with HLA-B*27 molecules not associated with AS. Examples are HLA-B*27:04 and HLA-B*27:06, which only vary in the amino acids at positions 114 (histidine to aspartate) and 116 (aspartate to tyrosine) [10]. Additional examples are HLA-B*27:05 and HLA-B*27:09; they differ in a single amino acid at position 116 (aspartate to histidine) [10]. These amino acid residues are all located in the same area of the antigenic peptide-binding groove (the F pocket), and the change in amino acid has the potential to induce conformational changes in the pocket [11]. A recent study investigated the ability of HLA-B*27:05, HLA-B*27:06, and HLA-B*27:09 to assemble, dimerize, and interact with chaperones [12••]. The HLA-B*27:06 and HLA-B*27:09 molecules assembled more rapidly (30–90 min) as compared with HLA-B*27:05 (3.5 h) and demonstrated a reduced tendency to dimerize. HLA-B*27:05 formed endoplasmic reticulum (ER) resident heavy chain dimers and misfolded HLA-B*27 molecules. HLA-B*27:06 were only transiently bound to ERp57 and binding immunoglobulin protein (BiP), whereas the other two subtypes had more prolonged binding with ERp57 and BiP [12••]. Thus, subtype differences in HLA-B*27 clearly affect its biophysical properties.
Hypotheses on the Pathophysiological Role of HLA-B*27
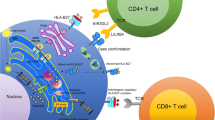
Numerous hypotheses have been suggested regarding the role of HLA-B*27 in the pathogenesis of AS. These are mainly based on animal models of rodents. One of the major hypotheses is the “the arthritogenic peptide” hypothesis [13], which proposes that alterations in the HLA-B*27 amino acid sequence change the specificity for peptides derived from certain bacterial proteins. This could result in cross-reactivity with peptides in the joints and/or entheses by induction of CD8+ T cell–mediated immune cross-reactivity. In the “ER stress model” [14], misfolded HLA-B*27 accumulates in the ER, where it induces a stress response that leads to an unfolded protein response (UPR) and autophagy [15]. In an animal model of AS, upregulation of UPR genes induced an increase in T helper (Th) 17 cells and in the proinflammatory cytokines IL-17, IL-23, and IFN-γ [16]. In the “HLA-B*27 homodimer” hypothesis [17], free heavy chains of B*27 form homodimers (B*272), which have other biologic functions not related to presentation of antigens such as abnormal interaction with killer cell immunoglobulin-like receptor 3DL2 (KIR3DL2) [18] and leucocyte immunoglobulin-like receptor molecules (LILR) [19]. This may increase expression of IL-17 and IL-23 through activation of CD8+ T cells and NK cells [20]. In the “mucosal immunodeficiency hypothesis” [21], HLA-B*27 fails to present bacterial peptides, which causes an increased invasion in the mucosa, which may lead to upregulation of the IL-23 pathway. Antoniou et al. [22••] investigated HLA-B*27:05 and UPR in relation to the life cycle of Salmonella in infected epithelial cells. In HLA-B*27:05-positive cells, Salmonella had augmented replication, which was not seen in HLA-B*35:01-positive cells. Salmonella activated UPR and induced misfolding of HLA-B*27, which was associated with the induction of an ER stress response, thereby facilitating the infection [22••].
MHC Genes Beyond HLA-B*27
Other MHC genes beyond HLA-B*27 have been associated with AS. Robinson et al. in 1989 first reported that HLA-Bw60, now renamed HLA-B*60 (which is the same as HLA-B*40:01), increases susceptibility to AS in HLA-B*27-positive subjects [23]. A subsequent study has confirmed the epistasis between HLA-B*27 and HLA-B*60 in Caucasian patients with AS [24]. In Han Chinese and African-American patients with AS, an increased frequency of HLA-B*40:01 (i.e., HLA-B*60) and decreased frequency of HLA-B*07 were observed [25••]. HLA-B*60 and HLA-B*61 have been associated with AS in HLA-B*27-negative Taiwanese-Chinese patients [26] and HLA-B*39 in HLA-B*27-negative Japanese patients [27]. A large cohort of Caucasian AS patients and controls were genotyped for MHC markers [28]. Regardless of their HLA-B*27 status, an increased risk for AS was observed for HLA-B*13, HLA-B*40, HLA-B*47, and HLA-B*51 subjects, whereas HLA-B*07 and HLA-B*57 decreased the risk for development of AS [28]. Non-HLA-B genes associated with AS comprise HLA-A*02:01 and the HLA-DRB1*01:03 and HLA-DPB1 alleles [28]. There was an earlier report of disease association of HLA-A2 (now renamed HLA-A*02:01) among HLA-B*27-positive patients with AS and acute anterior uveitis [29]. A recent study of HLA alleles in a large group of Caucasian HLA-B*27-negative AS patients and controls demonstrated that AS was positively associated with HLA-A*29, HLA-B*38, HLA-B*49, HLA-B*52, HLA-DRB1*11, and HLA-DPB1*03:01 and negatively associated with HLA-B*07, HLA-B*57, HLA-DRB1*15:01, HLA-DQB1*02:01, and HLA-DQB1*06:02 [25••]. This suggests an independent role of both MHC classes I and II alleles in predisposition to AS, but their further role in the development of the disease is currently not known.
The Pathobiology of HLA-B Molecules
The association of AS with HLA-B molecules is primarily via the amino acid located at position 97, where the presence of asparagine or threonine is related with AS, and the presence of serine or valine is not [28]. The amino acid at position 97 is also located in the peptide-binding groove, and recent mutagenesis experiments have demonstrated that a substitution at this position to threonine was associated with increased amount of free heavy chains on the cell surface [30], whereas presence of protective amino acid residues (serine or valine) and amino acids not related to AS (arginine or tryptophan) in position 97 did not alter the expression of free heavy chains. The AS-protective HLA-B*07 allele was associated with lower levels of free heavy chains than the AS associated HLA-B*27 or HLA-B*51, but when the amino acid at position 97 in HLA-B*07 was changed (from serine to asparagine), this increased the expression of free heavy chains [30]. Similarly, the HLA-DRB alleles are associated with AS through a change in amino acid residue at position 11 or 70, respectively, which are also situated in the peptide-binding groove [28].
Non-MHC Genes
Over 100 genes have been associated with the development of AS [31]. It has been estimated that 27.8% of the heritability of AS is known: 20.4% is considered associated with HLA-B*27 and only 7.4% with non-HLA-B*27 genes [32]. The first non-MHC genes clearly associated with AS were ERAP-1 (endoplasmic reticulum aminopeptidase 1) [33], ERAP-2, and LNPEP (leucyl/cysteinyl aminopeptidase) [34]. Moreover, genes associated with AS included the IL-23 pathway genes, e.g., IL-23 receptor (IL-23R) [33], IL-12B, IL-6 receptor (IL-6R), TYK2, IL-27 receptor (IL-27R), IL-1 receptor type 2 (IL-1R2) [35], IL-1 receptor type 1 (IL-1R1), and STAT-3 [33]. Genes associated with CD8+ T cell differentiation, such as RUNX3 (RUNX transcription factor 3), T-bet (T-box transcription factor/TBX21), EOMES (Eomesodermin)/TBR2 (T-box brain protein 2), IL-7R, and ZMIZ1 (zinc finger MIZ-type containing 1), have also been associated with AS [35, 36]. Two polymorphisms of RUNX3 have been associated with AS in UK patients (rs4648889 and rs4265380) [37•] and one in Korean patients (rs11249215) [38•], but RUNX3 alleles were not associated with AS in Han Chinese patients [39•]. RUNX3 is a central transcriptional factor for differentiation of innate lymphoid cells types 1 and 3 (ILC1 and ILC3) [40•], and it is also required for induction of the RAR (truncated retinoic acid receptor) and RORγt (related orphan nuclear receptor gamma t) transcriptional factors in ILC3 cells [40•].
ERAP-1, ERAP-2, and NPEPPS (puromycin-sensitive aminopeptidase) are also associated with AS. ERAP-1 trims peptides to a length of 10–16 amino acids so that they can be presented by MHC class I molecules [41]. ERAP-2 trims the N-terminal of precursors of antigenic peptides before they are bound to MHC class I molecules. Thus, aminopeptidases cleave the peptides before their binding to HLA-B*27, and such alterations in their function may change the pool of peptides (i.e., the peptidome) for presentation by HLA-B*27 [42, 43••]. There are significant interactions between the ERAP-1 non-synonymous SNP rs30187 and several HLA class I alleles, such as HLA-B*27 and HLA-B*40 in AS [26], HLA-Cw6 in psoriasis [44], and HLA-B*51 in Bechet’s disease [45]. The ERAP-1 allele variant rs30187 is only associated with AS in the presence of HLA-B*27 or HLA-B*40 [45]. When HLA-B*51 is present, the ERAP-1 allele variant rs30187 is associated with an increased risk of Bechet’s disease. In the ERAP-1 molecule, the amino acid variant of rs30187 is located at the enzymatically active site [46], which may therefore alter its function. This SNP has been associated with decreased trimming of peptides in vitro (a 40% reduction) [47]. The disease-associated ERAP-1 SNP rs10050860 has been shown to be functionally silent [47].
ERAP-1 and ERAP-2 Functional Aspects
In a large group of AS families, one ERAP-1 SNP (rs30187 [C]) out of a total of three investigated SNPs (rs27044 [C/G] and rs10050860 [C/T]) and a specific ERAP-1 and ERAP-2 haplotype (rs30187 [T] rs27044 [G] rs2549782 [T]) were significantly associated with AS [48]. ERAP-1 polymorphism has also been associated with differential expression of HLA-B*27. In patients with the ERAP-1 SNP rs27044 minor allele [G], HLA-B*27 expression was lower, whereas the SNP rs30187 did not affect the expression of HLA-B*27 and of HLA-B*27 free heavy chains [49]. The interaction between ERAP-1 and HLA-B*27 was functional, since inhibition of ERAP-1 resulted in changes in cells expressing HLA-B*27:04 or HLA-B*27:05, but not in cells expressing HLA-B*27:06 or HLA-B*27:09 [49]. The function of ERAP-2 was also investigated in AS patients homozygous for the ERAP-2 G allele [GG] (SNP rs2248374), which results in a non-functional ERAP-2 enzyme [50••]. Patients with the [GG] genotype had higher concentrations of HLA-B*27 free heavy chains on peripheral blood mononuclear cells as compared with patients that expressed other ERAP-2 alleles [50••]. Suppression of ERAP-2 induced numerous UPR molecules, e.g., BiP, C/EBP homologous protein (CHOP-10), and X-box binding protein 1 (XBP1) [50••]. Polymorphisms in ERAP-1 (SNP rs30187 and rs10050860) were associated with reduced/no peptide trimming activity and had significant impact on transcript and protein expression of ERAP-1 and ERAP-2 [51••]. The ERAP-1 SNP rs30187 changed the expression of the molecule, and SNP rs10050860 resulted in two other isoforms of the protein and a significant change in the concentration of the active ERAP-1 molecule [51••]. Thus, different alleles coding for ERAP-1 and ERAP-2 have a substantial impact on the function of the enzyme and on expression of different forms of HLA-B*27 on the cell surface and in this way may impact the pathogenesis of AS.
Gut Inflammation
AS and IBD are often seen in the same families, and relatives of AS patients develop IBD three times more often as compared with persons that are unrelated [52]. Studies based on endoscopy have revealed that up to 60% patients with spondyloarthropathy (SpA) have inflammation in the caecum and/or ileum [53] and up to 6% will develop IBD [52]. In genetic studies of AS patients with IBD, 20 of 31 genes associated with AS were also associated with IBD [54, 55, 56]. The most consistently associated genes are all related to the IL-23 pathway (IL-12B, IL-23R, IL-27, TYK-2, and JAK-2). Other genes are associated with other aspects of gut physiology such as mucosal immunity, differentiation and activation of gut lymphoid cells, and bacterial sensing. The hypothesis is that defective gut mucosal immunity is central in the pathogenesis of AS [54, 55, 56]. This hypothesis has been partly confirmed by Ciccia et al. [57•], who observed increased expression of zonulin in AS patients with bacterial ileitis. The patients also had an intestinal mucosal barrier and gut vascular barrier that were damaged. The concentration of zonulin and bacterial products in the blood was increased. Zonulin modifies tight junction permeability [58]. In vitro, zonulin can induce expansion of c-MAF+ CD163+ M2 polarized macrophages from blood mononuclear cells of AS patients [57•]. The presence of CD163+ M2 monocytes that produce IL-23 has been observed in peripheral blood, inflamed gut, and synovial tissues of AS patients [59, 60]. The HLA-B*27-transgenic rat, which is a model of SpA associated with IBD [61], does not develop inflammation in the intestine and peripheral joints if maintained in a germ-free environment [62]. This supports the hypothesis that exposure to microorganism(s) is important in the pathogenesis of AS.
Microbiome
The microbiome in axial SpA has been investigated in a few studies, which were all based on analyses of 16S ribosomal RNA. A study of biopsies from the terminal ileum of 9 AS patients and 9 healthy subjects (HS) showed that the microbiome in AS patients differed from HS [63]. AS patients had greater abundance of Bacteroidaceae, Lachnospiraceae, Porphyromonadaceae, Rikenellaceae, and Ruminococcaceae and lower abundance of Prevotellaceae and Veillonellaceae. HS had higher abundance of Actinomycetaceae, Gemellaceae, and Streptococcaceae. Tito et al. compared ileal and colonic biopsies of 27 SpA patients (14 with histologic bowel inflammation) and 15 HS [64]. They observed that SpA patients with versus those without bowel inflammation differed significantly in microbial composition. The amount of Dialister correlated with clinical disease activity [64]. Breban et al. compared the microbiome of 49 HLA-B*27-positive SpA patients, 17 rheumatoid arthritis (RA) patients, and 18 HS [65]. Compared with HS, SpA patients had more Lachnospiraceae, and SpA and RA patients had less Prevotellaceae and Paraprevotellaceae. When compared with 41 matched siblings, SpA patients had a significantly different bacterial composition, which however did not differ between HLA-B*27-positive versus HLA-B*27-negative siblings [65]. Finally, SpA patients had lower variety in the microbiome than HS, with the most notable change being a 2 to 3-fold increase in Ruminococcus gnavus, which correlated with disease activity [65].
HLA-B*27 and Dysbiosis
Rosenbaum et al. have studied the impact of HLA-B*27 on the intestinal microbiome of HLA-B*27/human beta-2-microglobulin-Tg rats and observed that the microbiome was significantly altered as compared with littermate controls [66]. Microbial and host metabolites in the gut were altered early in life such as the amino acid, carbohydrate, medium-chain fatty acid, and xenobiotic metabolites [67••]. This alteration was preceded by an inflammatory cytokine signature of Th17 and Th1 cells before development of dysbiosis and gut inflammation [68]. A similar inflammatory signature was seen in a study of three different HLA-B*27-transgenic rat strains that developed gut inflammation [69]. Surprisingly, the dysbiosis differed substantially between the rat strains, suggesting that numerous microbes influence the abnormal immune response, rather than a single or limited number of microbial genera [69]. In a recent study of HS, the presence of HLA-B*27 and HLA-DRB*1 RA-risk alleles for AS and RA, respectively, significantly influenced the composition of the intestinal microbiome [70••], and the intestinal dysbiosis associated with AS and RA is therefore not exclusively related to having the disease. Moreover, the microbiome on mucosal biopsies of the ileum differed substantially from biopsies from other parts of the gut and from the stools, and since patients with AS particularly have gut inflammation in the ilium, future studies in AS should ideally examine samples from the ileum [70••].
Innate Lymphoid Cells
Currently, innate lymphoid cells (ILCs) are considered important in the pathogenesis of AS. ILCs are divided into three categories: group 1 ILCs (ILC1s) that all have T-Bet as a transcriptional factor and produce Th1 cytokines such as INF-γ and TNF, ILC2s produce Th2-associated cytokines, and ILC3 is characterized by the transcriptional factor RORγt [71]. RORγt is crucial for lymphoid tissue inducer (LTi) cells [72]. In T cells, RORγt also has effects on the expression of IL-17 and IL-22 [73]. The most common type of ILC1 cells are NK cells, whose numbers are increased in ileal biopsies from AS patients [74]. ILC3 cells producing IL-17 and IL-22 are increased in the gut, peripheral blood, synovial fluid and bone marrow of AS patients [75], and in the skin of patients with psoriasis [76]. The ILC3 cells are also present in soft tissue and bone adjacent to human entheses, where they express IL-23R and where stimulation with IL-23 results in upregulation of IL-17 [77]. Finally, ILC3s also play a central role in gut homeostasis and in experimental animal models of IBD [78]. The number of ILC3s in the gut and peripheral blood is reduced after initiation of TNF-α inhibition [79].
Gamma-Delta (γ/δ) T Cells
Gamma-delta (γ/δ) T cells are a population of CD3+ T cells that express a T cell receptor (TCR) with γ and δ chains. In patients with Crohn’s disease and in SpA patients with ileitis, γ/δ T cells are abundant in the epithelium and mucosa [80]. Compared with HS, axial SpA and AS patients have an elevated number of γ/δ T cells that produce IL-17 and express IL-23R [81]. Kenna et al. have studied the relation between IL-23R signaling and IL-17 [82] and observed that in patients with AS, the number of γ/δ T cells was increased 3-fold in the peripheral blood as compared with HS, and 3–5 times as many γ/δ T cells in the patients expressed IL-23R as compared with HS. The frequency of γ/δ T cells producing IL-17 was 5-fold higher in AS patients, where only γ/δ T cells with IL-23R produced IL-17. Stimulation of γ/δ T cells with IL-23 resulted in a 6-fold induction of IL-17, and when anti-CD3/CD28 was added, this resulted in a 9-fold induction of IL-17 and a small amount of IFN-γ. In HS, INF-γ was induced 20-fold by T cell stimulation and a further 1.5-fold by IL-23 stimulation, whereas IL-17 only increased 1.5-fold. CD4+ T cells and IL-17+ CD8+ cells were not elevated. CD4+ T cells, CD8+ T cells, T reg cells, and NK receptor–positive T cells expressed IL-23R to the same extent [82]. Thus, IL-23 stimulates γ/δ T cells via the IL-23R to produce IL-17 in the peripheral blood of patients with AS but not in HS. Verken et al. recently discovered a novel innate-like T cell subset, RORγt+TbetloPLZF− invariant NK T cells (iNKT), and demonstrated that these cells, as well as γ/δ T cells, responded rapidly to stimulation with IL-23 with cytokine signatures like that of Th17 cells [83].
Mucosa-Associated Invariant T Cells
In the lamina propria of the intestinal mucosa, there are also T cells that express invariant forms of the TCR [84]. In humans, the mucosa-associated invariant T cells (MAIT cells) express a single TCR-α chain characterized by Vα7.2 and either Jα12, 20, or 33 and a restricted β chain repertoire characterized by Vβ2, Vβ8, Vβ13, or Vβ22 [85]. These TCRs are activated by cells expressing MHC class 1-like molecule (MR1) following infection with bacteria, and this results in production of cytokines and in cytolytic activity [84]. Five studies have investigated MAIT cells in patients with AS [86, 87, 88•, 89, 90]. In two studies, the amount of MAIT cells did not differ between SpA patients and control groups [86, 89], whereas other studies found lower proportion of MAIT cells in the circulation of AS patients than HC [88•, 90]. Gracey et al. studied the functional phenotype of MAIT cells in AS, HS, and RA patients [88•]. Compared with HS, AS patients had a significantly higher proportion of IL-17+ MAIT cells in the blood, whereas the proportion of IFN-γ+ MAIT cells was significantly lower while the proportion of TNF-α+ MAIT cells were similar. Male patients with AS but not female patients with AS had increased frequency of IL-17+ MAIT cells. In the synovial fluid of AS and RA patients, the frequency of IFN-γ+ and TNF-α+ MAIT cells was similar, but AS patients had a significantly lower frequency of IFN-γ+ and TNF-α+ MAIT cells in synovial fluid than in blood [88•]. Stimulation with IL-7, a cytokine involved in T cell activation and homeostasis, induced IL-7R on MAIT cells in both AS patients and HS. However, expression of IL-7R was significantly higher in AS patients. Induction of IL-17 in MAIT cells in AS patients was dependent on priming with IL-7 but not with IL-23. Moreover, the IL-7R SNP rs11742270 associated with AS did not change IL-7R expression or function [88•]. Toussirot et al. examined the functional capacity of MAIT cells to induce cytokines such as IL-17A, IL-22, and IFN-γ [90]. AS patients, independent of gender, had higher frequency of IL-17A+/IFN-γ− MAIT cells and IL-22+ MAIT cells, whereas only female patients had IL-17A+/IFN-γ− MAIT cells [90]. There was no association between disease activity and frequency of IL-17A+ or IL-22+ MAIT cells [90].
Interleukin-17
In AS patients, IL-17 is produced by cells from the innate immune system, such as neutrophils [91], mast cells [91], ILC3 [75, 91], γ/δ T cells, MAIT cells, Tc17 (CD8+ T cells that produce IL-17) cells, and Th17 cells [87, 88•]. The relation between IL-17, ILC3, MAIT cells, and γ/δ T cells is described in detail in the previous sections. Facet joint histopathology showed that the proportion of cells secreting IL-17 was significantly higher in the subchondral bone marrow in patients with AS compared with patients with osteoarthritis [91]. IL-17 has complex effects on bone metabolism. It is well established as a cytokine promoting enhanced activity of osteoclasts. In a murine model of collagen-induced arthritis (CIA) [92] and a murine model of antigen-induced arthritis (AIA) [93], IL-17 inhibition suppressed joint inflammation and reduced bone erosion. But IL-17A may also protect against generalized bone loss by inducing osteoblast generation from mesenchymal stem cells [94]. It suppresses adipogenesis and promotes osteoblast differentiation from mesenchymal stem cells but also inhibits bone regeneration in a rat model of calvarial defect [95••, 96]. The impact of IL-17A on differentiation of osteoblasts may depend on the stage of differentiation of the cell as well as the duration of exposure. Both prophylactic and therapeutic administrations of an antibody inhibiting IL-17A in HLA-B*27/hβ2m-transgenic rats led to diminished inflammation and new bone formation, the latter being clearer in peripheral versus axial joints [97••]. Thus, it is possible that inhibition of IL-17A could reduce new bone formation in axial SpA.
Interleukin-23
IL-23 is a heterodimeric cytokine composed of the IL-12B (IL-12p40) subunit that is shared with IL-12 and the IL-23A (IL-23p19) subunit that is specific to IL-23 [98]. Macrophages and dendritic cells produce IL-23 [99], and it is also secreted by the gut epithelium [100]. Studies have shown that IL-23 is present in the terminal ileum of AS patients with subclinical intestinal disease as compared with HS [101]. IL-23 can activate, expand, and maintain Th17 cells that express IL-17, and it can induce the immune-regulatory cytokine IL-22 [102]. IL-23 also regulates IL-17 and IL-22 expression in γ/δ T cells, NK cells, mast cells, and innate lymphoid cells (ILCs) [103, 104]. Further, IL-23 stimulates inflammation in entheses via a previously unidentified population of CD3+CD4−CD8− entheseal resident lymphocytes [105]. Neutralization of the IL-23p19 subunit resulted in reduced clinical disease activity and downregulation of mediators and genes involved in bone erosion, such as IL-6, IL-1b, matrix metalloproteinases, and RANKL [105]. This novel animal model of SpA also developed sacroiliitis, spondylitis, and aortic root inflammation [105, 106]. However, targeting IL-23 has not been a successful strategy in humans with AS. This may reflect the importance of IL-17A expressing cells that are not dependent on regulation by IL-23, such as gamma-delta T cells [107].
Conclusion and Perspectives
The expression of both TNF and IL-17 cytokines is pivotal to the development of axial SpA/AS, and it is now clear that immune cells of the innate and adaptive immune systems secrete these cytokines. Clinical trials in humans have revealed that IL-23 is not essential to the perpetuation of axial SpA/AS, implicating IL-17 expressing cells that are regulated by different mechanisms. A conundrum is that such cells may also be important for maintaining normal gut mucosal permeability. Key issues that remain unclear are the mechanism by which HLA-B*27 causes an inflammatory response, the role of IL-17 in bone remodeling, and mechanism(s) linking inflammation to the development of pathological new bone formation.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Stolwijk C, Boonen A, van Tubergen A, Reveille JD. Epidemiology of spondyloarthritis. Rheum Dis Clin N Am. 2012;38(3):441–76.
de Blecourt J, Polman A, Blecourt-Meindersma d. Hereditary factors in rheumatoid arthritis and ankylosing spondylitis. Ann Rheum Dis. 1961;20:215–20.
Calin A, Marder A, Becks E, Burns T. Genetic differences between B27 positive patients with ankylosing spondylitis and B27 positive healthy controls. Arthritis Rheum. 1983;26:1460–4.
van der Linden S, Valkenburg H, Cats A. The risk of developing ankylosing spondylitis in HLA-B27 positive individuals: a family and population study. Br J Rheumatol. 1983;22(4 Suppl 2):18–9.
Brown MA, Kennedy LG, MacGregor AJ, Darke C, Duncan E, Shatford JL, et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 1997;40:1823–8.
Jarvinen P. Occurrence of ankylosing spondylitis in a nationwide series of twins. Arthritis Rheum. 1995;38:381–3.
Pedersen OB, Svendsen AJ, Ejstrup L, Skytthe A, Harris JR, Junker P. Ankylosing spondylitis in Danish and Norwegian twins: occurrence and the relative importance of genetic vs. environmental effectors in disease causation. Scand J Rheumatol. 2008;37:120–6.
Khan MA. An update on the genetic polymorphism of HLA-B*27 with 213 alleles encompassing 160 subtypes (and still counting). Curr Rheumatol Rep. 2017;19:9.
Mathieu A, Paladini F, Vacca A, Cauli A, Fiorillo MT, Sorrentino R. The interplay between the geographic distribution of HLA-B27 alleles and their role in infectious and autoimmune diseases: a unifying hypothesis. Autoimmun Rev. 2009;8:420–5.
D’Amato M, Fiorillo MT, Carcassi C, Mathieu A, Zuccarelli A, Bitti PP, et al. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur J Immunol. 1995;25:3199–201.
Ostermeir K, Springer S, Zacharias M. Coupling between side chain interactions and binding pocket flexibility in HLA-B*44:02 molecules investigated by molecular dynamics simulations. Mol Immunol. 2015;63:312–9.
•• Guiliano DB, North H, Panayoitou E, Campbell EC, McHugh K, Cooke FG, et al. Polymorphisms in the F pocket of HLA-B27 subtypes strongly affect assembly, chaperone interactions, and heavy-chain misfolding. Arthritis Rheum. 2017;69:610–21. The study investigates the effects of polymorphism in the F pocket of HLA-B*27 on molecular characteristics, which differs among HLA-B*27 subtypes.
Benjamin R, Parham P. HLA-B27 and disease: a consequence of inadvertent antigen presentation? Rheum Dis Dis Clin North Am. 1992;18:11–21.
Mear JP, Schreiber KL, Munz C, Zhu X, Stevanovic S, Rammensee HG, et al. Misfolding of HLA-B27 as a result of its B pocket suggests a novel mechanism for its role in susceptibility to spondyloarthropathies. J Immunol (Baltimore, Md : 1950). 1999;163:6665–70.
Colbert RA, DeLay ML, Klenk EI, Layh-Schmitt G. From HLA-B27 to spondyloarthritis: a journey through the ER. Immunol Rev. 2010;233:181–202.
DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60:2633–43.
Allen RL, O’Callaghan CA, McMichael AJ, Bowness P. Cutting edge: HLA-B27 can form a novel beta 2-microglobulin-free heavy chain homodimer structure. J Immunol (Baltimore, Md : 1950) 1999;162:5045–5048.
Wong-Baeza I, Ridley A, Shaw J, Hatano H, Rysnik O, McHugh K, et al. KIR3DL2 binds to HLA-B27 dimers and free H chains more strongly than other HLA class I and promotes the expansion of T cells in ankylosing spondylitis. J Immunol (Baltimore, Md : 1950). 2013;190:3216–24.
Giles J, Shaw J, Piper C, Wong-Baeza I, McHugh K, Ridley A, et al. HLA-B27 homodimers and free H chains are stronger ligands for leukocyte Ig-like receptor B2 than classical HLA class I. J Immunol (Baltimore, Md : 1950). 2012;188:6184–93.
Bowness P, Ridley A, Shaw J, Chan AT, Wong-Baeza I, Fleming M, et al. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J Immunol (Baltimore, Md : 1950). 2011;186:2672–80.
Laitio P, Virtala M, Salmi M, Pelliniemi LJ, Yu DT, Granfors K. HLA-B27 modulates intracellular survival of Salmonella enteritidis in human monocytic cells. Eur J Immunol. 1997;27:1331–8.
•• Antoniou AN, Lenart I, Kriston-Vizi J, Iwawaki T, Turmaine M, McHugh K, et al. Salmonella exploits HLA-B27 and host unfolded protein responses to promote intracellular replication. Ann Rheum Dis. 2019;78:74–82. This study showed that HLA-B*27 misfolding is associated with enhanced Salmonella replication and that Salmonella exploits the UPR for replication.
Robinson WP, van der Linden SM, Khan MA, Rentsch HU, Cats A, Russell AS, et al. HLA Bw60 increases susceptibility to ankylosing spondylitis in HLA B27 positive individuals. Arthritis Rheum. 1989;32:1135–41.
van Gaalen FA, Verduijn W, Roelen DL, Bohringer S, Huizinga TW, van der Heijde DM, et al. Epistasis between two HLA antigens defines a subset of individuals at a very high risk for ankylosing spondylitis. Ann Rheum Dis. 2013;72:974–8.
•• Reveille JD, Zhou X, Lee M, Weisman MH, Yi L, Gensler LS, et al. HLA class I and II alleles in susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2019;78:66–73. This is a study investigating HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 alleles in AS to identify other risk alleles than HLA-B*27.
Wei JC, Tsai WC, Lin HS, Tsai CY, Chou CT. HLA-B60 and B61 are strongly associated with ankylosing spondylitis in HLA-B27-negative Taiwan Chinese patients. Rheumatology (Oxford, England). 2004;43:839–42.
Yamaguchi A, Tsuchiya N, Mitsui H, Shiota M, Ogawa A, Tokunaga K, et al. Association of HLA-B39 with HLA-B27-negative ankylosing spondylitis and pauciarticular juvenile rheumatoid arthritis in Japanese patients. Evidence for a role of the peptide-anchoring B pocket. Arthritis Rheum. 1995;38:1672–7.
Cortes A, Pulit SL, Leo PJ, Pointon JJ, Robinson PC, Weisman MH, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. 2015;6:7146.
Khan MA, Kushner I, Braun WE. Association of HLA A2 with uveitis in HLA-B27 positive patients with ankylosing spondylitis. J Rheumatol. 1981;8:295–8.
Chen L, Shi H, Yuan J, Bowness P. Position 97 of HLA-B, a residue implicated in pathogenesis of ankylosing spondylitis, plays a key role in cell surface free heavy chain expression. Ann Rheum Dis. 2017;76:593–601.
Costantino F, Breban M, Garchon HJ. Genetics and Functional Genomics of Spondyloarthritis. Front Immunol. 2018;18(9):2933.
Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–8.
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37.
Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–7.
Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–8.
Reveille JD, Sims AM, Danoy P, Evans DM, Leo P, Pointon JJ, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123– 7.
• Vecellio M, Cortes A, Roberts AR, Ellis J, Cohen CJ, Knight JC, et al. Evidence for a second ankylosing spondylitis-associated RUNX3 regulatory polymorphism. RMD Open. 2018;4(1):e000628. This study includes the identification of association between AS and polymorphism in transcription factor RUNX3.
• Cho SM, Jung SH, Chung YJ. A variant in RUNX3 is associated with the risk of ankylosing spondylitis in Koreans. Genomics Inform. 2017;15:65–8. This study includes the identification of association between AS and polymorphism in transcription factor RUNX3.
• Su W, Du L, Liu S, Deng J, Cao Q, Yuan G, et al. ERAP1/ERAP2 and RUNX3 polymorphisms are not associated with ankylosing spondylitis susceptibility in Chinese Han. Clin Exp Immunol. 2018;193:95–102. This study includes the investigation of RUNX3 in Chinese AS patients, where no association was found.
• Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immun. 2015;16:1124–33. This study investigates the role of RUNX3 in ILCs, and identifies it as a key transcription factor for lineage specific differentiation of ILC1 and ILC3.
Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc Natl Acad Sci U S A. 2005;102:17107–12.
Martin-Esteban A, Guasp P, Barnea E, Admon A, Lopez de Castro JA. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 2 with the HLA-B*27 peptidome in human cells. Arthritis Rheumatol. 2016;68:2466–75.
•• Martin-Esteban A, Sanz-Bravo A, Guasp P, Barnea E, Admon A, Lopez de Castro JA. Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome. J Autoimmun. 2017;79:28–38. ERAP1 and ERAP2 influence the HLA-B27 peptidome, i.e., the pool of peptides processed by HLA-B*27.
Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat genet. 2010;42:985–90.
Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Tugal-Tutkun I, Seyahi E, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat genet. 2013;45:202–7.
Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–13.
Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci U S A. 2011;108:7745–50.
Tsui FW, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW, et al. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis. 2010;69:733–6.
Haroon N, Tsui FW, Uchanska-Ziegler B, Ziegler A, Inman RD. Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann Rheum Dis. 2012;71:589–95.
•• Zhang Z, Ciccia F, Zeng F, Guggino G, Yee K, Abdullah H, et al. Brief report: functional interaction of endoplasmic reticulum aminopeptidase 2 and HLA-B27 activates the unfolded protein response. Arthritis Rheumatol. 2017;69:1009–15. Lack of ERAP2 in HLA-B*27-positive cells increased FHC and UPR.
•• Hanson AL, Cuddihy T, Haynes K, Loo D, Morton CJ, Oppermann U, et al. Genetic variants in ERAP1 and ERAP2 associated with immune-mediated diseases influence protein expression and the isoform profile. Arthritis Rheumatol. 2018;70:255–65. AS-associated gene polymorphism in ERAP1 and ERAP2 changed not only the gene expression but also the handling of the transcript and the products of the ERAP1 enzyme.
Edmunds L, Elswood J, Kennedy LG, Calin A. Primary ankylosing spondylitis, psoriatic and enteropathic spondyloarthropathy: a controlled analysis. J Rheumatol. 1991;18(5):696–8.
Mielants H, Veys EM, Cuvelier C, de Vos M. Ileocolonoscopic findings in seronegative spondylarthropathies. Br J Rheumatol. 1988;27(Suppl 2):95–105.
De Vos M, Mielants H, Cuvelier C, Elewaut A, Veys E. Long-term evolution of gut inflammation in patients with spondyloarthropathy. Gastroenterology. 1996;110:1696–703.
Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24.
Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73.
• Ciccia F, Guggino G, Rizzo A, Alessandro R, Luchetti MM, Milling S, et al. Dysbiosis and zonulin upregulation alter gut epithelial and vascular barriers in patients with ankylosing spondylitis. Ann Rheum Dis. 2017;76:1123–32. This study investigated the difference in gut intestinal barrier in patient with AS with ileitis versus healthy subjects.
Sturgeon C, Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue barriers. 2016;4:e1251384.
Baeten D, Moller HJ, Delanghe J, Veys EM, Moestrup SK, De Keyser F. Association of CD163+ macrophages and local production of soluble CD163 with decreased lymphocyte activation in spondylarthropathy synovitis. Arthritis Rheum. 2004;50:1611–23.
Ciccia F, Alessandro R, Rizzo A, Accardo-Palumbo A, Raimondo S, Raiata F, et al. Macrophage phenotype in the subclinical gut inflammation of patients with ankylosing spondylitis. Rheumatology (Oxford, England). 2014;53:104–13.
Milia AF, Ibba-Manneschi L, Manetti M, Benelli G, Messerini L, Matucci-Cerinic M. HLA-B27 transgenic rat: an animal model mimicking gut and joint involvement in human spondyloarthritides. Ann N Y Acad Sci. 2009;1173:570–4.
Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64.
Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief report: intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheum. 2015;67:686–91.
Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, et al. Brief report: Dialister as a microbial marker of disease activity in spondyloarthritis. Arthritis Rheum. 2017;69:114–21.
Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76:1614–22.
Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, et al. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One. 2014;9:e105684.
•• Asquith M, Davin S, Stauffer P, Michell C, Janowitz C, Lin P, et al. Intestinal metabolites are profoundly altered in the context of HLA-B27 expression and functionally modulate disease in a rat model of spondyloarthritis. Arthritis Rheumatol. 2017;69:1984–95. This study investigates the influence of HLA-B*27 on intestinal metabolites.
Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT. Perturbed mucosal immunity and dysbiosis accompany clinical disease in a rat model of spondyloarthritis. Arthritis Rheum. 2016;68:2151–62.
Gill T, Asquith M, Brooks SR, Rosenbaum JT, Colbert RA. Effects of HLA-B27 on gut microbiota in experimental spondyloarthritis implicate an ecological model of dysbiosis. Arthritis Rheum. 2018;70:555–65.
•• Asquith M, Sternes PR, Costello ME, Karstens L, Diamond S, Martin TM, et al. HLA alleles associated with risk of ankylosing spondylitis and rheumatoid arthritis influence the gut microbiome. Arthritis Rheumatol. 2019. https://doi.org/10.1002/art.40917. This study investigates the impact of HLA alleles on the content of the gut microbiome.
Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–9.
Cherrier M, Eberl G. The development of LTi cells. Curr Opin Immunol. 2012;24(2):178–83.
Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33.
Ciccia F, Accardo-Palumbo A, Alessandro R, Rizzo A, Principe S, Peralta S, et al. Interleukin-22 and interleukin-22-producing NKp44+ natural killer cells in subclinical gut inflammation in ankylosing spondylitis. Arthritis Rheum. 2012;64:1869–78.
Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A, et al. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis. 2015;74:1739–47.
Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol. 2014;134:984–91.
Cuthbert RJ, Fragkakis EM, Dunsmuir R, Li Z, Coles M, Marzo-Ortega H, et al. Brief report: group 3 innate lymphoid cells in human enthesis. Arthritis Rheum. 2017;69:1816–22.
Forkel M, Mjosberg J. Dysregulation of group 3 innate lymphoid cells in the pathogenesis of inflammatory bowel disease. Curr Allergy Asthma Rep. 2016;16:73.
Paustian AMS, Paez-Cortez J, Bryant S, Westmoreland S, Waegell W, Kingsbury G. Continuous IL-23 stimulation drives ILC3 depletion in the upper GI tract and, in combination with TNFalpha, induces robust activation and a phenotypic switch of ILC3. PLoS One. 2017;12:e0182841.
Cuvelier CA, De Wever N, Mielants H, De Vos M, Veys EM, Roels H. Expression of T cell receptors alpha beta and gamma delta in the ileal mucosa of patients with Crohn’s disease and with spondylarthropathy. Clin Exp Immunol. 1992;90:275–9.
Jansen DT, Hameetman M, van Bergen J, Huizinga TW, van der Heijde D, Toes RE, et al. IL-17-producing CD4+ T cells are increased in early, active axial spondyloarthritis including patients without imaging abnormalities. Rheumatology (Oxford, England). 2015;54:728–35.
Kenna TJ, Davidson SI, Duan R, Bradbury LA, McFarlane J, Smith M, et al. Enrichment of circulating interleukin-17-secreting interleukin-23 receptor-positive gamma/delta T cells in patients with active ankylosing spondylitis. Arthritis Rheum. 2012;64:1420–9.
Venken K, Jacques P, Mortier C, Labadia ME, Decruy T, Coudenys J, et al. RORgammat inhibition selectively targets IL-17 producing iNKT and gammadelta-T cells enriched in Spondyloarthritis patients. Nat Commun. 2019;10:9.
Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–85.
Chiba A, Murayama G, Miyake S. Mucosal-associated invariant T cells in autoimmune diseases. Front Immunol. 2018;9:1333.
Sugimoto C, Konno T, Wakao R, Fujita H, Fujita H, Wakao H. Mucosal-associated invariant T cell is a potential marker to distinguish fibromyalgia syndrome from arthritis. PLoS One. 2015;10:e0121124.
Hayashi E, Chiba A, Tada K, Haga K, Kitagaichi M, Nakajima S, et al. Involvement of mucosal-associated invariant T cells in ankylosing spondylitis. J Rheumatol. 2016;43(9):1695–703.
• Gracey E, Qaiyum Z, Almaghlouth I, Lawson D, Karki S, Avvaru N, et al. IL-7 primes IL-17 in mucosal-associated invariant T (MAIT) cells, which contribute to the Th17-axis in ankylosing spondylitis. Ann Rheum Dis. 2016;75:2124–32. This study investigates IL-7 in AS patients and associates IL-7 with IL-17 in MAIT cells.
Cho YN, Kee SJ, Kim TJ, Jin HM, Kim MJ, Jung HJ, et al. Mucosal-associated invariant T cell deficiency in systemic lupus erythematosus. J Immunol (Baltimore, Md : 1950). 2014;193(8):3891–901.
Toussirot E, Laheurte C, Gaugler B, Gabriel D, Saas P. Increased IL-22- and IL-17A-producing mucosal-associated invariant T cells in the peripheral blood of patients with ankylosing spondylitis. Front Immunol. 2018;9:1610.
Appel H, Maier R, Wu P, Scheer R, Hempfing A, Kayser R, et al. Analysis of IL-17(+) cells in facet joints of patients with spondyloarthritis suggests that the innate immune pathway might be of greater relevance than the Th17-mediated adaptive immune response. Arthritis Res Ther. 2011;13(3):R95.
Lubberts E, Joosten LA, Oppers B, van den Bersselaar L, Coenen-de Roo CJ, Kolls JK, et al. IL-1-independent role of IL-17 in synovial inflammation and joint destruction during collagen-induced arthritis. J Immunol (Baltimore, Md : 1950). 2001;167:1004–13.
Koenders MI, Lubberts E, Oppers-Walgreen B, van den Bersselaar L, Helsen MM, Di Padova FE, et al. Blocking of interleukin-17 during reactivation of experimental arthritis prevents joint inflammation and bone erosion by decreasing RANKL and interleukin-1. Am J Pathol. 2005;167:141–9.
Huang H, Kim HJ, Chang EJ, Lee ZH, Hwang SJ, Kim HM, et al. IL-17 stimulates the proliferation and differentiation of human mesenchymal stem cells: implications for bone remodeling. Cell Death Differ. 2009;16:1332–43.
•• Ono T, Okamoto K, Nakashima T, Nitta T, Hori S, Iwakura Y, et al. IL-17-producing gammadelta T cells enhance bone regeneration. Nat Commun. 2016;7:10928. This is a study of the impact of gamma-delta T cells and IL-17A in bone remodeling.
Goswami J, Hernandez-Santos N, Zuniga LA, Gaffen SL. A bone-protective role for IL-17 receptor signaling in ovariectomy-induced bone loss. Eur J Immunol. 2009;39:2831–9.
•• van Tok M, van Duivenvoorde LM, Kramer I, Ingold P, Pfister S, Roth L, et al. Interleukin-17a inhibition diminishes inflammation and new bone formation in experimental spondyloarthritis. Arthritis Rheum. 2019;71:612–25. This is a study of inhibition of IL-17A in a rat model of SpA.
Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25.
Tan ZY, Bealgey KW, Fang Y, Gong YM, Bao S. Interleukin-23: immunological roles and clinical implications. Int J Biochem Cell Biol. 2009;41:733–5.
Becker C, Wirtz S, Blessing M, Pirhonen J, Strand D, Bechthold O, et al. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J Clin Invest. 2003;112:693–706.
Ciccia F, Bombardieri M, Principato A, Giardina A, Tripodo C, Porcasi R, et al. Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis Rheum. 2009;60:955–65.
Benham H, Rehaume LM, Hasnain SZ, Velasco J, Baillet AC, Ruutu M, et al. Interleukin-23 mediates the intestinal response to microbial beta-1,3-glucan and the development of spondyloarthritis pathology in SKG mice. Arthritis Rheum. 2014;66:1755–67.
Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol (Baltimore, Md : 1950). 2009;182:5904–8.
Kenna TJ, Brown MA. The role of IL-17-secreting mast cells in inflammatory joint disease. Nat Rev Rheumatol. 2013;9:375–9.
Sherlock JP, Joyce-Shaikh B, Turner SP, Chao CC, Sathe M, Grein J, et al. IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nat Med. 2012;18:1069–76.
Reinhardt A, Yevsa T, Worbs T, Lienenklaus S, Sandrock I, Oberdorfer L, et al. Interleukin-23-dependent gamma/delta T cells produce interleukin-17 and accumulate in the enthesis, aortic valve, and ciliary body in mice. Arthritis Rheum. 2016;68:2476–86.
Cuthbert R, Fragkakis EM, Bridgewood C, Dunsmuir R, Watad A, Rao A, et al. The Vδ2 subset of Γδt-cells are present at healthy human enthesis and have transcriptional and functional characteristics consistent with a capacity for IL-17A production in response to IL-23 [abstract]. Arthritis Rheum. 2018;70(suppl.)):1833.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
WPM has received research support and/or consultancy/speaker fees from Abbvie, Boehringer, Celgene, Eli-Lilly, Galapagos, Janssen, Merck, Novartis, Pfizer, and UCB. SJP has received honoraria for speaking from MSD, Pfizer, AbbVie, Novartis, and UCB, has been an advisory board member for AbbVie and Novartis, and has received research support from AbbVie, MSD, and Novartis.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Spondyloarthritis
Rights and permissions
About this article
Cite this article
Pedersen, S.J., Maksymowych, W.P. The Pathogenesis of Ankylosing Spondylitis: an Update. Curr Rheumatol Rep 21, 58 (2019). https://doi.org/10.1007/s11926-019-0856-3
Published:
DOI: https://doi.org/10.1007/s11926-019-0856-3