Abstract
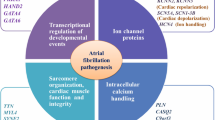
Atrial fibrillation (AF) is a common clinical arrhythmia that appears to be highly heritable, despite representing a complex interplay of several disease processes that generally do not manifest until later in life. In this manuscript, we will review the genetic basis of this complex trait established through studies of familial AF, linkage and candidate gene studies of common AF, genome wide association studies (GWAS) of common AF, and transcriptomic studies of AF. Since AF is associated with a five-fold increase in the risk of stroke, we also review the intersection of common genetic factors associated with both of these conditions. Similarly, we highlight the intersection of common genetic markers associated with some risk factors for AF, such as hypertension and obesity, and AF. Lastly, we describe a paradigm where genetic factors predispose to the risk of AF, but which may require additional stress and trigger factors in older age to allow for the clinical manifestation of AF.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atrial fibrillation (AF) is the most common sustained arrhythmia seen in clinical practice. The etiology of AF is multifactorial, but includes a genetic predisposition to the arrhythmia. The road towards discovering the genetic basis of AF has progressed from identification of genes associated with familial AF to clinical observational studies demonstrating hereditability of common AF and to genome wide association studies (GWAS) that have identified to date 14 genetic loci associated with AF (Table 1) [1, 18, 30, 35, 43, 44, 46•, 47, 48•, 49–52, 53•, 54, 55]. However, identification of risk loci is only the start of a long process to discover the mechanisms by which these variants increase AF risk. Moreover, despite the identification of inherent genetic risk, we do not fully understand why AF typically does not manifest until later decades of life, nor why certain clinical factors, such as hypertension, obesity, male sex, or Caucasian ancestry, predispose to AF. Here, we will review the published evidence for the genetic basis of AF and discuss the results of recent genomic and transcriptomic analyses that may yield insights into some clinical observations of AF incidence.
Heritability of Atrial Fibrillation
The lifetime risk for developing AF is one in four for people older than 40 years [56]. Traditional risk factors for AF include older age, obesity, alcohol consumption, hypertension, left ventricular hypertrophy, heart failure, prolonged PR interval, and coronary artery disease [57]. The prevalence of AF has increased as a result of the aging and increased prevalence of obesity in the general population and in response to an increased awareness and diagnosis of the arrhythmia [57].
Despite the complex and aging-related clinical risk factors for the disease, AF is heritable. In the Framingham study, parental AF increased the risk of AF in offspring [58]. The odds ratio was 1.85 for AF in offspring who had one parent with AF and 3.23 when the sample was restricted to patients with parents who developed AF when they were <75 years old. A more recent study involving the Framingham cohort reported that new onset AF occurred more frequently in subjects with compared to without familial AF (adjusted hazard ratio 1.40; 95 % CI 1.13–1.74), independent of traditional AF risk factors [59]. In Iceland, Arnar et al. studied 5269 patients diagnosed with AF and age- and sex-matched controls [49]. The relative risk of AF in first-degree relative pairs was 1.77. When the analysis was restricted to first-degree relatives of patients with early onset AF (diagnosed <60 years of age), relative risk of AF was 4.67. Ellinor and colleagues reported that lone AF (i.e., AF in the absence of structural heart disease) greatly increases the risk of AF in family members. A family history of AF was found in 38 % of patients with lone AF; the relative risk for sons was 8.1, daughters 9.5, brothers 70, and sisters 34 [60]. Lastly, a Danish twin study reported 22.0 % concordance rates for monozygotic pairs compared to 11.6 % for dizygotic pairs (p < 0.0001) with heritability of AF estimated to be 62 % [61]. Of note, such twin studies are of particular interest because they are less likely to lead to inflated estimates of heritability as a consequence of shared environmental risk factors.
Familial Monogenic AF
Families exhibiting a very high prevalence of AF have been reported as early as 1936 [62–64]. Traditional approaches to identifying genes responsible for AF in these presumably monogenic (or Mendelian) forms of AF include linkage studies and candidate gene searches. These studies have identified several ion channel gene mutations that were associated with familial AF, including potassium channel mutations (e.g., in KCNQ1, KCNE2, KCNH2, KCNJ2, KCNA5) for which gain-of-function mutations might be expected to promote AF by shortening of action potential duration and effective refractory periods, as well as in the sodium channel mutations (in SCN5A and SCN4B), which have also been associated with dilated cardiomyopathy [2–11].
Several non-ion channel genes have also been associated with familial monogenic AF, including PRKAG2, associated with familial Wolff-Parkinson-White syndrome and AF [12], and NPPA, which encodes for atrial natriuretic peptide [13]. Further, some developmental genes have been implicated, including genes encoding transcription factors involved in cardiogenesis from the GATA family [20–22] and NKX2-5, a homeobox-containing transcription factor involved in cardiac development and septation with mutations associated with atrial septal defect and AV conduction abnormalities [65]. Structural genes have also been associated with familial AF, including GJA5, which encodes for connexin 40 [66], a gap junction protein that is expressed in the atria and the conduction system and that is crucial for action potential propagation through myocytes, and RYR2, a gene which encodes the ryanodine receptor responsible for releasing intracellular stores of calcium [15, 16]. Finally, genes encoding nuclear membrane-associated proteins have been linked to familial AF. These include LMNA, which encodes for lamin expressed in the inner nuclear lamina. Mutations in LMNA have also been associated with muscular dystrophy, dilated cardiomyopathy, risk of sudden cardiac death [18, 19], and progeria syndrome. NUP155 has also been associated with AF [17, 67]. This gene encodes for a nucleoporin that is involved in the assembly and functioning of the nuclear pore complex that regulates transport of macromolecules across the nuclear envelope between the cytoplasm and the nucleus [17, 67].
Polygenic Non-familial Common AF
Genetic Architecture
Although familial AF studies have contributed much to our understanding of the genes associated with monogenic forms of AF, they represent only a small fraction of the overall burden of disease. Overall, the genetics of common AF pathogenesis is complex, involving modest contributions to the risk of disease from genetic variations in many genes. The genetic architecture of a disease depends on the number of variants that influence a given pathology, the effect size of each variant on the phenotype, and the frequency of the variant. Complex diseases such as AF follow the “common disease common variant” hypothesis of genetic architecture where common variants with small effects appear to be responsible for a majority of the heritability of the trait [68].
Linkage and Candidate Gene Studies
Initial strategies to identify polymorphisms predisposing to common polygenic AF included linkage analyses and candidate gene-based association studies. Linkage analyses attempted to apply the same analytic techniques used to identify monogenic forms of disease to common polygenic forms of disease but proved to be inadequately powered to detect the modest effects of polymorphisms contributing to the risk of common AF [69].
Failed linkage studies were followed by candidate gene association studies, which relied on prior knowledge to generate hypotheses about possible polymorphisms that could have a causal relationship with a given disease and were limited by the potential bias of studying only what is already known [70]. Since these studies concentrated on only a small region of the genome, their pretest probability of identifying a real signal was low. The data published was further influenced by publication bias. A meta-analysis of 370 studies reporting on 36 different genetic associations illustrated this point well. The study found that, in 25 of 36 cases, the first study overestimated the replicated effect [69]. Further, 39 % of the genetic associations studied had significant heterogeneity between results [69]. Lack of replication for candidate gene association studies is common and can be a result of differences in study design, differences in studied populations with variable allele frequencies or relative risks of a certain variant, or due to real biological differences [70]. Furthermore, the polygenic nature of common and complex diseases involving many variants with small relative risks resulted in many studies being substantially underpowered leading to a high rate of false-positive reports [69].
While as a whole candidate gene association studies have been disappointing, such studies nevertheless yielded some valuable insights into the potential mechanisms leading to AF. To begin, such studies implicated ion channel genes as contributants to risk of common AF, including those identified in familial studies. Potassium channel genes, including KCNQ1, KCNH2, KCNJ5, KCNJ8, KCND3, and KCNE 1 through 5, have been associated with AF risk [23–30]. The KCNE genes encode the beta subunits of voltage gated channels: KCNE1 and KCNQ1 contribute to the IKs current [24–26, 71]. Gain of function of IKs [24, 72] results in a faster repolarizing current and shorter effective refractory period, increasing the excitability of cells and susceptibility to arrhythmia. Loss of function prolongs the action potential and increases the likelihood of early after depolarizations [73]. KCNA5 encodes for the α-subunit of the voltage sensor of a channel that generates the ultra-rapid potassium current, IKur, which is abundant in atria and detectable in failing ventricles. Hayashi et al. found a novel gain of function variant and a previously described KCNA5 loss of function variant in a cohort of 72 patients with lone AF [30, 74].
Mutations in sodium channel subunit genes have also been associated with AF. The voltage-gated sodium channel consists of a pore-forming α subunit, encoded by the SCNA genes, and 4 different β subunits encoded by SCN1B-4B. This channel is responsible for the rapid influx of sodium ions that underlies the upstroke (phase 0) of the cardiac action potential. Several variants of SCNA5, which encodes the cardiac α-subunit of Nav1.5, have been reported in patients with common polygenic AF [75]. Gain-of-function mutations decrease the threshold voltage for the action potential and increase cellular excitability [76]. β-Subunits modulate the gating and voltage dependence of the channels, impact the trafficking of sodium channel to and from the cell membrane, and aid in cell adhesion. Mutations in the four β-subunits have been reported to attenuate sodium currents and shift the voltage dependence of sodium channel gating [31, 32]. A single nucleotide polymorphism (SNP) in SCN10A, which encodes Nav1.8, a voltage-gated sodium channel that participates in the late sodium current, was recently described to be associated with early onset AF [33].
Candidate gene analyses have also identified polymorphisms in non-ion channel genes with familial AF. Implicated genes include those encoding connexin 40, GJA5 [35–37, 77], the transcription factor gene NKX2.5 [38, 39], and the LMNA gene [78]. Genes involved in the renin-angiotensin-aldosterone pathway, including the angiotensin-converting enzyme, angiotensin gene promoter, and angiotensinogen [40–42, 79], have also been associated with AF.
Identification of AF-Associated Genetic Loci Through Genome Wide Association Studies
Genome wide association studies (GWAS) represent a relatively new but important study design to overcome the inherent bias of candidate gene analyses. By comparing the frequencies of common genetic variants among subjects with and without a disease or level of quantitative trait, GWAS has been able to identify hundreds of novel genetic associations over the last 10 years involving common single nucleotide polymorphisms (SNPs). GWAS analyze hundreds of thousands to millions of SNPs concurrently. To correct for multiple testing, thresholds for significance typically need to be adjusted to a p value of <5X10−8, which effectively corrects for one million independent tests. Once a SNP or a set of SNPs in a region reaches genome wide significance, the next important step is to link the SNP association to a causal gene. Generally, this process begins with an assessment of the genes that are in proximity or within the susceptibility locus and should not be deemed certain until a functional mechanism has been fully worked out in detail through multiple additional experiments [80].
Developmental Genes
In 2007, the first GWAS for AF identified a susceptibility locus at chromosome 4q25 in a population of European descent and replicated this association in an Asian population [43]. This association has been further replicated in all AF GWAS, to date [81–83]. Further, this locus was associated with recurrence of AF after ablation and new onset AF in the postoperative period after coronary bypass surgery [50, 81].
Additional GWAS have identified at least 4 independent loci at chromosome 4q25 for AF [84]. As is often the case, the lead SNPs in this region are in non-coding regions. Nevertheless, the closest gene is the paired-like homeodomain 2 PITX2 gene, located in a linkage disequilibrium (LD) block upstream to the susceptibility locus. PITX2 is involved in embryogenesis and left-right differentiation of the heart. PITX2 encodes 3 different isoforms of the protein by alternative splicing. Importantly, the Pitx2c isoform is mainly found in the left atrium and pulmonary veins [85, 86]. In Pitx2c conditional knockout mice, Pitx2c-deficient mice do not develop a pulmonary myocardial sleeve, and Pitx2 appears to suppress a left atrial sinus node program [87]. Compared to wild-type mice, Pitx2null+/− and Pitx2null−/− mice heterozygous and homozygous for a Pitx2-null allele that removes all Pitx2 isoform function showed step-wise increased expression of HCN4, which encodes a hyperpolarization-activated cyclic nucleotide-gated channel contributing to pacemaker currents, as well as TBX3 and SHOX2 that are also required for sinoatrial node development [86]. Expression of KCNQ1 was also increased [87], which would result in a shorter effective refractory period, facilitating action potential triggering and conduction. Mouse PITX2 conditional or heterozgygote mutation models have shown alterations in ion channel, calcium handling, gap and tight junction gene expression [88–91], PITX2 regulation of miRNAs [90–92], and atrial arrhythmia inducibility [87, 88, 90]. Thus, PITX2 is a biologically attractive candidate causative gene in the region, as it appears to be involved in the regulation of several ion channel genes that plausibly may predispose to atrial arrhythmia, it directs asymmetric morphogenesis of the heart, is involved in the formation of pulmonary veins, the putative site of AF initiation targeted by clinical AF ablation [93], and appears to repress a left atrial sinus node program, genetic variation of which might contribute to electrical triggers recorded within pulmonary vein ostia.
The expression of PITX2 has been shown to markedly decrease postnatally. The mechanisms leading to postnatal inactivation of PITX2 are actively being researched and include possible mediation by various risk factors, such as pressure and volume overload and oxidative stress that come with age. Tao et al. showed that postnatal inactivation of PITX2 leads to abnormal R-R intervals and to differential expression of genes involved in cell junction organization, including the intercalated disk, ion channels, and caveolae [94].
GWAS of AF have also identified several other loci associated with transcription factors that appear to be important for cardiac development. One of these is located on chromosome 16q22 intronic to the zinc finger homeobox 3 gene (ZFHX3), known as AT motif-binding transcription factor 1(ATBF1) [44], which is involved in regulation of growth and differentiation of skeletal muscle and neuronal tissues [44]. ZFHX3 is required for the transcriptional activation of the gene POU class I homebox 1 (POU1F1), which interacts with PITX2, enabling DNA binding and transcriptional activity. This might be a mechanism by which it predisposes to AF [83]. Later, another GWAS described PRRX1 which encodes for a homeodomain transcription factor expressed in connective tissue in the developing heart [46•]. In animal models, knockout of PRRX1 led to abnormalities in the pulmonary vasculature [95].
Ion Channels and Pores
Thus far, two loci in ion channel genes have been significantly associated with AF in GWAS studies. First, a locus was identified on 1q22 within an intron of the KCNN3 gene, which encodes for the calcium-activated potassium channel SK3 [47]. SK3 contributes to the repolarization phase of the action potential [96]. Second, a locus on chromosome 15q24 within an intron of the hyperpolarization-activated cyclic nucleotide-gated 4 (HCN4) gene was described [46•]. HCN4 is expressed in the sinoatrial (SA) node and underlies the If current normally responsible for the pacemaker current in nodal myocytes. Mutations of this channel may lead to diminished action potential frequency (heart rate slowing) and delayed after depolarizations that might trigger AF [97]. As discussed above, Pitx2 represses Hcn4 expression in the left atrium [86, 87].
Structural Genes
A meta-analysis of GWAS in 2010 identified several new AF loci, including three in/near genes involved in cellular structural components. Cell membranes contain invaginations, known as caveolae, that participate in cell transport. A locus on chromosome 7q31 within an intron of the gene CAV1, which encodes caveolin-1, was associated with AF [46•]. CAV1 is co-expressed with a potassium channel gene (KCNH2), which is involved in the repolarization phase of the action potential. The risk allele and AF are associated with decreased expression of CAV1 [98]. Further, caveolin-1 has been postulated as an anti-fibrotic element in atrial tissue since down-regulation of Cav-1 by siRNA leads to increased TGF-β1-induced activation of the Smad signal pathway and collagen production [99].
An intronic SNP located on chromosome 14q23 in the gene SYNE2, which encodes for nesprin-2, has also been associated with AF. Nesprin-2 is located in the outer nuclear membrane and sarcomere and anchors nuclei to the cytoskeleton. A large meta-analysis of GWAS by Ellinor, et al. identified a susceptibility locus between two genes, SYNPO2L and MYOZ1, on chromosome 10 and another locus in the an open-reading frame on chromosome 9 (C9orf3) [46•]. Myozenin-1 is a protein involved in stabilizing the sarcomere, encoded by MYOZ1. C9orf3 region is associated with three coding genes, including FBP1 and FBP2, which are involved in gluconeogenesis [46•].
Gene mRNA Expression studies
Although GWAS studies identify regions in the genome that are associated with a phenotype, the lead SNP within a susceptibility locus is often not the causal variant. Furthermore, the lead SNP is often not within a coding region of a gene and usually near several genes, making it challenging to determine which gene in the region is the causal gene. One way to identify with more certainty the causal gene is to leverage gene expression studies to determine whether a lead SNP is preferentially associated with the level of mRNA of one or more genes nearby. SNPs that are associated with mRNA are often termed expression quantitative trait loci (eQTLs) [100]. As the control of gene expression is often tissue-specific, assessment of relationships between SNPs and mRNA expression among circulating cells in blood may be inadequate to assess the effects of SNPs on the expression of genes predisposing to AF. Gene expression studies involving more relevant target tissues, such as the left atrium itself, may substantially increase the yield of identifying causal genes among susceptibility loci of AF.
Genetic variants can change the expression of mRNA and proteins by cis or trans mechanisms. Cis-acting variants affect expression of genes nearby on the same chromosome as the gene variants, while trans-acting variants can act on genes that are far from them. Such transcriptional studies can help guide the identification of the genes associated with genetic variants identified by GWAS.
eQTL analysis of the AF-associated locus in between SYNPO2L and MYOZ1 has demonstrated an association between the high risk alleles in this region and decreased expression of MYOZ1, implicating MYOZ1 as the most likely causal gene associated with this AF locus [98, 101]. In another eQTL study predominantly involving right atrial appendage tissues, AF risk alleles were associated with increased expression of PITX2a and decreased expression of MYOZ1, CAV1, C9orf3, and FANCC [98].
Despite the finding of cis-eQTLs for genes noted above, significant eQTLs for the top GWAS locus from AF on chromosome 4q25 were not found for PITX2c in human adult left atrial appendage tissues [102]. Recently, we performed RNA sequencing on human left/right atrial tissue pairs and reported the presence of an intergenic long non-coding RNA sequence adjacent to the PITX2 gene (PANCR) differentially expressed in the left atria and eye, but no other heart chambers. Expression of PANCR RNA correlated with PITX2c mRNA expression. In human embryonic stem cells, both PANCR and PITX2c RNAs were coordinately expressed during early differentiation of the stem cells towards cardiomyocytes. siRNA-mediated PANCR knockdown decreased PITX2c expression and resulted in similar RNA expression profiles as PITX2c knockdown cells, whereas PITX2c knockdown did not affect PANCR expression, suggesting that PANCR is an important regulator of PITX2c expression [103]. However, similar to PITX2c in human adult left atrial appendage tissue, chromosome 4q25 risk SNPs for AF were not associated with PANCR or PITX2c expression [102, 103]. These findings may be a consequence of PITX2 and PANCR being most active during embryologic development in the left atrium or in the tissue within the pulmonary vein–left atrial junction which may have some distinct differences with the tissue making up the adult left atrial appendage.
A combination of large-scale GWAS, cis-eQTL, and functional evaluation led to the identification of 5 novel loci associated with AF near the genes NEURL, GJA1, TBX5, CAND2, and CUX2 [48•]. AF-associated cis-eQTLs were identified for CAND2, GJA1, and TBX5. NEURL encodes for an E3 ubiquitin ligase that was found to co-localize with PITX2. Another signal was found in a SNP intronic to TBX5, a transcription factor implicated in conduction abnormalities, and in CAND2 [48•], which encodes for a TATA-binding protein transcription factor involved in myogenesis [104]. eQTL analysis of CAND2 and TBX5 showed that the risk alleles were associated with an increased expression of these genes. Additional functional studies demonstrated that NEURL and CAND2 knockout zebrafish models had longer atrial action potential duration [48•]. Propagation of the action potential through myocytes requires a balanced expression of two gap junction proteins: connexin 40 and 43 [105]. Cis-eQTL associations showed the AF risk allele at the GJA1 locus was associated with lower expression of GJA1, which encodes connexin 43 [48•]. Connexin abnormalities can lead to reduced conduction velocity and action potential propagation. In the same paper, a novel SNP located within an intron of the CUX2 gene was found to be associated with AF only in a Japanese population [48•]. CUX2 was recently associated with increased oxidative DNA damage in embryonic neurons [106]. However, a significant cis-eQTL was not identified for the risk allele.
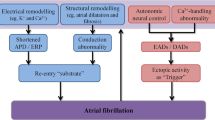
Transcriptomic Analyses and the Age Paradox in AF
Despite the association of several developmental genes with AF risk, AF typically does not manifest until decades after birth. To gain insight on this paradox, we recently performed a gene expression study of left atrial appendage tissue from 207 adults with AF and 32 adults without AF to compare gene expression in 3 groups based on AF history and rhythm at the time of resection of the atrial appendage: (1) no AF history and in sinus rhythm (SR/SR), (2) AF history and in sinus rhythm (AF/SR), and (3) AF history and in AF rhythm (AF/AF) [53•]. We identified differentially coexpressed gene modules between the SR/SR and AF/SR groups (“AF susceptibility” comparison) and between the AF/SR and AF/AF groups (“AF activity or persistence” comparison). Gene set enrichment analyses showed that AF susceptibility was associated with decreased transcriptional responses to cellular stress, inflammation, oxidation, and unfolded proteins, while AF persistence was associated with changes in ion channel expression and decreased stress responses. These findings may help explain the AF age paradox, whereby AF does not manifest until there is impaired responsiveness to persistent cellular stress and aging, decades after the GWAS-identified AF loci exert their activity during cardiac and pulmonary vein development.
The reduced cellular stress response to protein unfolding is of particular interest, as this geneset includes targets of several transcription factors such as the CREB/ATF family, HSF1, ATF6, SRF, E2F1, and SP1 [53•]. Alterations in many of these transcription factors can lead to abnormal proteostasis and deposition of unfolded amyloid proteins. Deposition of isolated atrial amyloid is well described and associated with older age and the presence of AF [107, 108]. Interestingly, atrial amyloid has also been reported to be more frequent in left than in right atrial appendages [108]. Isolated atrial amyloid protein has been shown to be due to aggregates of atrial natriuretic peptide (ANP) [107, 109], and expression levels of NPPA, which encodes for ANP, are high in adult human atrial appendage tissues. Expression of NPPA was up-regulated in Pitx2 heterozygote and null mice [86]. Further, a recent study in a Chinese population suggested that TBX5 mutations increases the expression of ANP and connexin 40 [110]. Senile accumulation of the protein aggregates may contribute to the predisposition to AF with aging.
AF Genetics and Stroke
One of the most devastating complications of AF is stroke. Conventional risk factors account for only 60 % of the variation in the risk of stroke [111]. Similar to AF, stroke has a genetic predisposition. Further, risk factors differ depending on the etiology of the stroke, which includes hemorrhagic, ischemic, cardioembolic, large artery due to atherosclerosis, small vessel, and cryptogenic forms [112]. Genetic studies have attempted to elucidate new stroke prediction, prevention, and treatment approaches and have led to the discovery of common genetic variants associated with both AF and stroke (Table 2, Fig. 1).
Soon after publication of the first GWAS of AF implicating SNPs in the 4q25 region, the same SNPs were also identified as risk factors for ischemic stroke. Gretarsdottir et al. showed that rs2200733, one of the first 4q25 SNPs demonstrated to be strongly associated with AF risk, was associated with ischemic stroke, and even more strongly with cardioembolic stroke [115]. This association was further replicated in a targeted sequencing study of a Polish population [114]. Lastly, another study showed a different locus in the same region was associated only with cardioembolic stroke [113].
Large-scale studies have found similar genetic associations between AF and stroke. Gudbjartsson et al. reported a case control genomic study, finding that the SNP rs7193343 at the ZFHX3 locus was associated with ischemic and cardioembolic stroke [45]. Several GWAS studies have confirmed this association, including the METASTROKE study, which incorporated about 70,000 subjects. These studies confirmed that the PITX2 and ZFHX3 SNPs were associated with ischemic stroke and more specifically with cardioembolic stroke, even though only half of their population was sub classified by type of stroke [116]. A recent exome sequencing study replicated the ZFHX3 locus association with ischemic stroke [119]. Using the METASTROKE collaboration data, an association between ischemic stroke with an AF-risk SNP intronic to CUX2 was found, though the AF risk allele was paradoxically associated with a decreased the risk of stroke (OR 0.95; 95 % CI 0.91–0.98). Sub-analysis by type of stroke found an association of GJA1 and TBX5 AF-risk SNPs with cardioembolic stroke [48•].
The mentioned associations are likely due to an enrichment of AF cases among ischemic and cardioembolic stroke cases. They might be markers of AF as a predominant cause of embolic rather than non-embolic stroke. Studies have not consistently classified strokes, and the lack of differentiation by subtype of stroke reduces power for stroke subtype analyses. Without a large study of isolated cardioembolic stroke, signals attributed to ischemic stroke are an aggregate of multiple mechanisms leading to stroke. Nevertheless, these genetic variants might ultimately prove useful in differentiating the etiology of an ischemic stroke and in identifying patients with cryptogenic stroke who are most likely to have underlying undiagnosed AF. These patients may warrant more monitoring and potential anticoagulation for secondary prevention of stroke.
Current stroke prevention scores rely on clinical data to determine therapy, but adding genetic markers may improve the predictive value of these tools. Tada et al. constructed a 12 SNP genetic risk score (including SNPs near PITX2, KCNN3, PRRX1, CAV1, C9orf3, SYNPO2L, SYNE2, HCN4, ZFHX3) to identify people at higher risk for AF and found that the score in addition to the CHADS2 score led to better prediction of ischemic stroke in AF patients [120•]. The renin angiotensin aldosterone (RAAS) system plays an important role in inflammation, hypertension, tissue remodeling, and fibrosis. A preliminary association of the G6 polymorphism (a polymorphism located in the promoter region of the angiotensin gene leading to increased angiotensinogen production) has been found in stroke in two candidate gene studies [117, 121] and in one candidate gene study of AF patients [42]. Tsai et al. showed that the addition of the G6 polymorphism to the CHADS2 score improved the ability to predict stroke in an AF population [122]. Other elements of the RAAS system, such as the angiotensinogen gene and angiotensinogen-converting enzyme gene, have also been associated with AF and stroke in candidate gene analyses [40, 41, 118, 123].
Inflammation likely plays a role in both stroke and AF. Ischemic brain injury is characterized by inflammation. However, data regarding a polymorphism in the CRP gene, which has been reported in candidate gene analysis as an independent risk factor for stroke, have been conflicting [124, 125]. Other components of the inflammatory cascade studied include IL-1a, TNFα, IL-10, ICAM-1, though evidence has again been inconsistent [126–130] with studies limited by heterogeneity and small sample size.
Risk factors for AF: Hypertension, Obesity, and Common Genetic Variants
Hypertension is an important modifiable risk factor for AF [131]. While GWAS for hypertension have not identified common SNPs that have also been associated with AF, several genetic variants associated with hypertension may contribute to AF.
The genetic architecture of blood pressure appears to be extremely complex with ∼100 independent variants accounting for only around 2 % of the variance in blood pressure [132]. These include loci near NPPA and NPPB, which encode precursors of the natriuretic peptides. Polymorphisms in these loci correlate with the level of circulating natriuretic peptide and blood pressure [132, 133]. AF has been associated with variants in the NPPA gene [134, 135], causing resistance to ANP breakdown and increasing its half-life [13]. Atrial amyloid is largely derived from ANP. Transcriptomic differential gene expression analysis (discussed above) showed that AF susceptibility is associated with decreased targets of cellular stress response to protein unfolding [53•], leading to abnormal proteostasis and deposition of unfolded amyloid protein.
Several genes involved in the RAAS, including those for the angiotensin-converting enzyme, angiotensin and angiotensin receptor, have been associated with essential hypertension by candidate gene analysis [136–138]. Some studies of patients with essential hypertension have identified RAAS gene variants that are related to increased risk of AF [42, 79, 139]. Polymorphisms in this system have also been linked to stroke. However, the RAAS system is involved in multiple pathologies and attributing some variants to certain disease states might be challenging in the setting of so many confounders. Nevertheless, the possibility that some variants contribute to multiple pathologies through pleiotropy seems plausible, as AF, obesity, and hypertension are highly intertwined conditions and such pleiotropy has been well documented in other settings (Fig. 1).
Other clinical risk factors for AF that have been associated with common genetic variants include obesity and the metabolic syndrome. Obesity increases the risk of atrial fibrillation by almost 50 % [140]. In a GWAS meta-analysis of waist–hip ratio, TBX15 was associated with increased waist–hip ratio. Unlike the other SNP hits in this study, TBX15 did not differ by gender [141]. Candidate gene studies have shown that CAV1 polymorphisms, resulting in decreased expression of the gene, are associated with insulin resistance and the metabolic syndrome in Hispanic and Caucasian cohorts [142, 143].
Conclusions
Genomic analyses have provided important insights into the pathogenesis of AF, though much still remains unknown. While GWAS have identified common genetic variants associated with AF, identification of the causative genetic variants and their functional mechanisms remains challenging. For example, the top replicated AF-associated locus on chromosome 4q25 is in an intergenic region, but the functional variants and the direct mechanisms by which these variants increase AF susceptibility is still poorly understood. The nearest gene, PITX2, is a biologically attractive candidate causative gene, as it directs asymmetric morphogenesis of the heart, is involved in the formation of pulmonary veins, and represses a left atrial sinus node program that might contribute to pulmonary vein triggers. However, the connections between the AF-associated SNPs and gene expression or function of PITX2 remain elusive. Transcriptional studies in atrial tissue have made connections between SNPs and genes in several other AF risk loci, including for MYOZ1, CAV1, C9orf3, CAND2, GJA1, and TBX5. Analyses also suggest that the delay in AF manifestation until later in life may be associated with impaired transcriptional responses to cellular stress. Future GWAS of AF with much larger sample sizes will undoubtedly increase the number of susceptibility loci that are significantly associated with AF risk. Further studies are needed to determine functional mechanisms by which these genetic loci contribute to AF risk, stroke, and/or other common co-morbid conditions. Understanding these mechanisms will set the stage for the development of new therapeutic agents that target the causative genes.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–4.
Otway R, Vandenberg JI, Guo G, et al. Stretch-sensitive KCNQ1 mutation A link between genetic and environmental factors in the pathogenesis of atrial fibrillation? J Am Coll Cardiol. 2007;49:578–86.
Das S, Makino S, Melman YF, Shea MA, et al. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm. 2009;6:1146–53.
Hong K, Piper DR, Diaz-Valdecantos A, et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433–40.
Yang Y, Xia M, Jin Q, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905.
Hong K, Bjerregaard P, Gussak I, et al. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16:394–6.
Xia M, Jin Q, Bendahhou S, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–9.
Olson TM, Alekseev AE, Liu XK, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–91.
Makiyama T, Akao M, Shizuta S, et al. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–34.
Benito B, Brugada R, Perich RM, et al. A mutation in the sodium channel is responsible for the association of long QT syndrome and familial atrial fibrillation. Heart Rhythm. 2008;5:1434–40.
Li RG, Wang Q, Xu YJ, et al. Mutations of the SCN4B-encoded sodium channel beta4 subunit in familial atrial fibrillation. Int J Mol Med. 2013;32:144–50.
Gollob MH, Green MS, Tang AS, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–31.
Hodgson-Zingman DM, Karst ML, Zingman LV, et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–65.
Olson TM, Alekseev AE, Moreau C, et al. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med. 2007;4:110–6.
Di Pino A, Caruso E, Costanzo L, et al. A novel RyR2 mutation in a 2-year-old baby presenting with atrial fibrillation, atrial flutter, and atrial ectopic tachycardia. Heart Rhythm. 2014;11:1480–3.
Kazemian P, Gollob MH, Pantano A, et al. A novel mutation in the RYR2 gene leading to catecholaminergic polymorphic ventricular tachycardia and paroxysmal atrial fibrillation: dose-dependent arrhythmia-event suppression by beta-blocker therapy. Can J Cardiol. 2011;27:870.e7–10.
Zhang X, Chen S, Yoo S, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–27.
Beckmann BM, Holinski-Feder E, Walter MC, et al. Laminopathy presenting as familial atrial fibrillation. Int J Cardiol. 2010;145:394–6.
Pan H, Richards AA, Zhu X, et al. A novel mutation in LAMIN A/C is associated with isolated early-onset atrial fibrillation and progressive atrioventricular block followed by cardiomyopathy and sudden cardiac death. Heart Rhythm. 2009;6:707–10.
Wang J, Sun YM, Yang YQ. Mutation spectrum of the GATA4 gene in patients with idiopathic atrial fibrillation. Mol Biol Rep. 2012;39:8127–35.
Posch MG, Boldt LH, Polotzki M, et al. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur J Med Genet. 2010;53:201–3.
Wang XH, Huang CX, Wang Q, et al. A novel GATA5 loss-of-function mutation underlies lone atrial fibrillation. Int J Mol Med. 2013;31:43–50.
Li L, Shen C, Yao Z, et al. Genetic variants of potassium voltage-gated channel genes (KCNQ1, KCNH2, and KCNE1) affected the risk of atrial fibrillation in elderly patients. Genet Test Mol Biomarkers. 2015;19:359–65.
Ravn LS, Aizawa Y, Pollevick GD, et al. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart Rhythm. 2008;5:427–35.
Mao T, Miao HJ, Xu GJ, et al. Association of single nucleotide polymorphism of KCNE1 and KCNE4 gene with atrial fibrillation in Xinjiang Uygur and Han population. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:916–21.
Lundby A, Ravn LS, Svendsen JH, et al. KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cell Physiol Biochem. 2008;21:47–54.
Jabbari J, Olesen MS, Holst AG, et al. Common polymorphisms in KCNJ5 [corrected] are associated with early-onset lone atrial fibrillation in Caucasians. Cardiology. 2011;118:116–20.
Delaney JT, Muhammad R, Blair MA, et al. A KCNJ8 mutation associated with early repolarization and atrial fibrillation. Europace. 2012;14:1428–32.
Olesen MS, Refsgaard L, Holst AG, et al. A novel KCND3 gain-of-function mutation associated with early-onset of persistent lone atrial fibrillation. Cardiovasc Res. 2013;98:488–95.
K. Hayashi, T. Konno, H. Tada, et al. “Functional characterization of rare variants implicated in susceptibility to lone atrial fibrillation,” Circ. Arrhythm Electrophysiol., Jun 30, 2015.
Watanabe H, Darbar D, Kaiser DW, et al. Mutations in sodium channel beta1- and beta2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268–75.
Olesen MS, Jespersen T, Nielsen JB, et al. Mutations in sodium channel beta-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786–93.
Savio-Galimberti E, Weeke P, Muhammad R, et al. SCN10A/Nav1.8 modulation of peak and late sodium currents in patients with early onset atrial fibrillation. Cardiovasc Res. 2014;104:355–63.
Jabbari J, Olesen MS, Yuan L, et al. Common and rare variants in SCN10A modulate the risk of atrial fibrillation. Circ Cardiovasc Genet. 2015;8:64–73.
Christophersen IE, Holmegard HN, Jabbari J, et al. Rare variants in GJA5 are associated with early-onset lone atrial fibrillation. Can J Cardiol. 2013;29:111–6.
Wirka RC, Gore S, Van Wagoner DR, et al. A common connexin-40 gene promoter variant affects connexin-40 expression in human atria and is associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2011;4:87–93.
Juang JM, Chern YR, Tsai CT, et al. The association of human connexin 40 genetic polymorphisms with atrial fibrillation. Int J Cardiol. 2007;116:107–12.
Xie WH, Chang C, Xu YJ, et al. Prevalence and spectrum of Nkx2.5 mutations associated with idiopathic atrial fibrillation. Clinics (Sao Paulo). 2013;68:777–84.
Yu H, Xu JH, Song HM, et al. Mutational spectrum of the NKX2-5 gene in patients with lone atrial fibrillation. Int J Med Sci. 2014;11:554–63.
Ravn LS, Benn M, Nordestgaard BG, et al. Angiotensinogen and ACE gene polymorphisms and risk of atrial fibrillation in the general population. Pharmacogenet Genomics. 2008;18:525–33.
Zhao LQ, Wen ZJ, Wei Y, et al. Polymorphisms of renin-angiotensin-aldosterone system gene in Chinese Han patients with nonfamilial atrial fibrillation. PLoS ONE. 2015;10, e0117489.
Tsai CT, Lai LP, Lin JL, et al. Renin-angiotensin system gene polymorphisms and atrial fibrillation. Circulation. 2004;109:1640–6.
Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7.
Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81.
Gudbjartsson DF, Holm H, Gretarsdottir S. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8.
Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–5. Large GWAS meta-analysis identified new AF risk loci.
Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4.
Sinner MF, Tucker NR, Lunetta KL, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130:1225–35. Combines transcriptional analysis and functional analysis to give some information regarding the possible mechanism by which a risk SNP could lead to AF.
Arnar DO, Thorvaldsson S, Manolio TA, et al. Eur Heart J. 2006;27:708–12.
Body SC, Collard CD, Shernan SK, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506.
Brugada R, Tapscott T, Czernuszewicz GZ, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–11.
I. E. Christophersen and P. T. Ellinor, “Genetics of atrial fibrillation: from families to genomes,” J. Hum. Genet., May 21, 2015.
Deshmukh A, Barnard J, Sun H, et al. Left atrial transcriptional changes associated with atrial fibrillation susceptibility and persistence. Circ Arrhythm Electrophysiol. 2015;8:32–41. Differential gene expression by atrial rhythm status showed AF was associated with decreased transcriptional responses to cellular stress.
Lee KT, Yeh HY, Tung CP, et al. Association of RS2200733 but not RS10033464 on 4q25 with atrial fibrillation based on the recessive model in a Taiwanese population. Cardiology. 2010;116:151–6.
Thibodeau IL, Xu J, Li Q, et al. Paradigm of genetic mosaicism and lone atrial fibrillation: physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122:236–44.
Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation. 2004;110:1042–6.
Schnabel RB, Yin X, Gona P, et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. Lancet. 2015;386:154–62.
Fox CS, Parise H, D’Agostino S, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–5.
Lubitz SA, Yin X, Fontes JD, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304:2263–9.
Ellinor PT, Yoerger DM, Ruskin JN, et al. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–84.
Christophersen IE, Ravn LS, Budtz-Joergensen E, et al. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol. 2009;2:378–83.
Bertram H, Paul T, Beyer F, et al. Familial idiopathic atrial fibrillation with bradyarrhythmia. Eur J Pediatr. 1996;155:7–10.
Brugada R. Atrial fibrillation: is the solution in the genes? Rev Esp Cardiol. 1997;50:367.
Levy R. Paroxysmal auricular fibrillation and flutter without signs of organic cardiac disease in two brothers. J Mt Sinai Hosp. 1942;8:765–70.
Huang RT, Xue S, Xu YJ, et al. A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2013;31:1119–26.
Gollob MH, Jones DL, Krahn AD, et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–88.
Oberti C, Wang L, Li L, et al. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation. 2004;110:3753–9.
Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53.
Ioannidis JP, Ntzani EE, Trikalinos TA, et al. Replication validity of genetic association studies. Nat Genet. 2001;29:306–9.
Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–7.
Nielsen JB, Bentzen BH, Olesen MS, et al. Gain-of-function mutations in potassium channel subunit KCNE2 associated with early-onset lone atrial fibrillation. Biomark Med. 2014;8:557–70.
Ma KJ, Li N, Teng SY, et al. Modulation of KCNQ1 current by atrial fibrillation-associated KCNE4 (145E/D) gene polymorphism. Chin Med J (Engl). 2007;120:150–4.
Lemoine MD, Duverger JE, Naud P, et al. Arrhythmogenic left atrial cellular electrophysiology in a murine genetic long QT syndrome model. Cardiovasc Res. 2011;92:67–74.
Christophersen IE, Olesen MS, Liang B, et al. Genetic variation in KCNA5: impact on the atrial-specific potassium current IKur in patients with lone atrial fibrillation. Eur Heart J. 2013;34:1517–25.
Darbar D, Kannankeril PJ, Donahue BS, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35.
Li Q, Huang H, Liu G, Lam K, et al. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem Biophys Res Commun. 2009;380:132–7.
Sun Y, Tong X, Chen H. An atrial-fibrillation-linked connexin40 mutant is retained in the endoplasmic reticulum and impairs the function of atrial gap-junction channels. Dis Model Mech. 2014;7:561–9.
Brauch KM, Chen LY, Olson TM. Comprehensive mutation scanning of LMNA in 268 patients with lone atrial fibrillation. Am J Cardiol. 2009;103:1426–8.
Jiang MH, Su YM, Tang JZ, et al. Angiotensin-converting enzyme gene 2350 G/A polymorphism and susceptibility to atrial fibrillation in Han Chinese patients with essential hypertension. Clin (Sao Paulo). 2013;68:1428–32.
Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17–25.
Husser D, Adams V, Piorkowski C, et al. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–53.
Lubitz SA, Ozcan C, Magnani JW, et al. Genetics of atrial fibrillation: implications for future research directions and personalized medicine. Circ Arrhythm Electrophysiol. 2010;3:291–9.
Liu X, Wang F, Knight AC, Zhao J, et al. Common variants for atrial fibrillation: results from genome-wide association studies. Hum Genet. 2012;131:33–9.
Lubitz SA, Sinner MF, Lunetta KL, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–84.
Liu C, Liu W, Lu MF, et al. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–48.
Wang J, Klysik E, Sood S, et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 2010;107:9753–8.
Mommersteeg MT, Brown NA, Prall OW, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–9.
Kirchhof P, Kahr PC, Kaese S, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–33.
Franco D, Chinchilla A, Daimi H, et al. Modulation of conductive elements by Pitx2 and their impact on atrial arrhythmogenesis. Cardiovasc Res. 2011;91:223–31.
Chinchilla A, Daimi H, Lozano-Velasco E, et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–79.
Lozano-Velasco E, Hernandez-Torres F, Daimi H, et al. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc Res. 2016;109:55–66.
Wang J, Bai Y, Li N, et al. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci U S A. 2014;111:9181–6.
Haissaguerre M, Jais P, Shah DC, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–66.
Tao Y, Zhang M, Li L, et al. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7:23–32.
Ihida-Stansbury K, McKean DM, Gebb SA, et al. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ Res. 2004;94:1507–14.
Skibsbye L, Poulet C, Diness JG, et al. Small-conductance calcium-activated potassium (SK) channels contribute to action potential repolarization in human atria. Cardiovasc Res. 2014;103:156–67.
Macri V, Mahida SN, Zhang ML, et al. A novel trafficking-defective HCN4 mutation is associated with early-onset atrial fibrillation. Heart Rhythm. 2014;11:1055–62.
Martin RI, Babaei MS, Choy MK, et al. Genetic variants associated with risk of atrial fibrillation regulate expression of PITX2, CAV1, MYOZ1, C9orf3 and FANCC. J Mol Cell Cardiol. 2015;85:207–14.
Yi SL, Liu XJ, Zhong JQ, et al. Role of caveolin-1 in atrial fibrillation as an anti-fibrotic signaling molecule in human atrial fibroblasts. PLoS ONE. 2014;9, e85144.
Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212.
Lin H, Dolmatova EV, Morley MP, et al. Gene expression and genetic variation in human atria. Heart Rhythm. 2014;11:266–71.
Gore-Panter SR, Hsu J, Hanna P, et al. Atrial Fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS ONE. 2014;9, e86245.
S. R. Gore-Panter, J. Hsu, J. Barnard, et.al, “PANCR, the PITX2 adjacent noncoding RNA, is expressed in human left atria and regulates PITX2c expression,’ Circ.Arrhythm Electrophysiol., vol. 9, 2016.
Suzuki H, Suzuki A, Maekawa Y, et al. Interplay between two myogenesis-related proteins: TBP-interacting protein 120B and MyoD. Gene. 2012;504:213–9.
Gemel J, Levy AE, Simon AR, et al. Connexin40 abnormalities and atrial fibrillation in the human heart. J Mol Cell Cardiol. 2014;76:159–68.
Pal R, Ramdzan ZM, Kaur S, et al. CUX2 protein functions as an accessory factor in the repair of oxidative DNA damage. J Biol Chem. 2015;290:22520–31.
Rocken C, Peters B, Juenemann G, et al. Atrial amyloidosis: an arrhythmogenic substrate for persistent atrial fibrillation. Circulation. 2002;106:2091–7.
Leone O, Boriani G, Chiappini B, et al. Amyloid deposition as a cause of atrial remodelling in persistent valvular atrial fibrillation. Eur Heart J. 2004;25:1237–41.
Linke RP, Voigt C, Storkel FS, et al. N-terminal amino acid sequence analysis indicates that isolated atrial amyloid is derived from atrial natriuretic peptide. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;55:125–7.
Ma J-F, Yang F, Mahida et al, “TBX5 mutations contribute to early-onset atrial fibrillation in Chinese and Caucasians.” Cardiovascular Research., pp. http://cardiovascres.oxfordjournals.org/content/early/2016/01/12/cvr.cvw003.abstract, 2016.
Souto JC, Pena G, Ziyatdinov A, et al. A genomewide study of body mass index and its genetic correlation with thromboembolic risk. Results from the GAIT project. Thromb Haemost. 2014;112:1036–43.
Adams Jr HP, Bendixen BH, Kappelle LJ, et al. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41.
Lemmens R, Buysschaert I, Geelen V, et al. The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke. 2010;41:1850–7.
Wnuk M, Pera J, Jagiella J, et al. The rs2200733 variant on chromosome 4q25 is a risk factor for cardioembolic stroke related to atrial fibrillation in Polish patients. Neurol Neurochir Pol. 2011;45:148–52.
Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–9.
Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62.
Tsai CT, Chang SN, Chang SH, et al. Renin-angiotensin system gene polymorphisms predict the risk of stroke in patients with atrial fibrillation: a 10-year prospective follow-up study. Heart Rhythm. 2014;11:1384–90.
Stetskaia TA, Bushueva OI, Bulgakova IV, et al. Association of T174M polymorphism of the angiotensinogen gene with the higher risk of cerebral stroke in women. Ter Arkh. 2014;86:66–71.
Auer PL, Nalls M, Meschia JF, et al. Rare and coding region genetic variants associated with risk of ischemic stroke: the NHLBI Exome Sequence Project. JAMA Neurol. 2015;72:781–8.
Tada H, Shiffman D, Smith JG, et al. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45:2856–62. Genetic risk score composed of AF risk SNPs identified subjects at higher risk for future AF and stroke.
Saidi S, Mallat SG, Almawi WY, et al. Association between renin-angiotensin-aldosterone system genotypes and haplotypes and risk of ischemic stroke of atherosclerotic etiology. Acta Neurol Scand. 2009;119:356–63.
Schmidt H, Aulchenko YS, Schweighofer N, et al. Angiotensinogen promoter B-haplotype associated with cerebral small vessel disease enhances basal transcriptional activity. Stroke. 2004;35:2592–7.
Zhao J, Qin X, Li S, et al. Association between the ACE I/D polymorphism and risk of ischemic stroke: an updated meta-analysis of 47,026 subjects from 105 case–control studies. J Neurol Sci. 2014;345:37–47.
Liu Y, Geng PL, Yan FQ, et al. C-reactive protein -717A > G and -286C > T > A gene polymorphism and ischemic stroke. Chin Med J (Engl). 2015;128:1666–70.
Wang Q, Ding H, Tang JR, et al. C-reactive protein polymorphisms and genetic susceptibility to ischemic stroke and hemorrhagic stroke in the Chinese Han population. Acta Pharmacol Sin. 2009;30:291–8.
Y. M. Niu, H. Weng, C. Zhang, et al, “Systematic review by multivariate meta-analyses on the possible role of tumor necrosis factor-alpha gene polymorphisms in association with ischemic stroke,” Neuromolecular Med., Aug 1, 2015.
N. Zhao, X. Liu, Y. Wang, et al, “Association of inflammatory gene polymorphisms with ischemic stroke in a Chinese Han population,” J. Neuroinflammation, vol. 9, pp. 162-2094-9-162, Jul 6, 2012.
Jin J, Li W, Peng L, et al. Relationship between interleukin-10 -1082A/G polymorphism and risk of ischemic stroke: a meta-analysis. PLoS ONE. 2014;9, e94631.
Muino E, Krupinski J, Carrera C, et al. An inflammatory polymorphisms risk scoring system for the differentiation of ischemic stroke subtypes. Mediators Inflamm. 2015;2015:569714.
Li J, Song J, Jiang MH, et al. Interleukin-6 promoter polymorphisms and susceptibility to atrial fibrillation in elderly Han Chinese patients with essential hypertension. J Interferon Cytokine Res. 2012;32:542–7.
Huxley RR, Lopez FL, Folsom AR, et al. Absolute and attributable risks of atrial fibrillation in relation to optimal and borderline risk factors: the Atherosclerosis Risk in Communities (ARIC) study. Circulation. 2011;123:1501–8.
International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9.
Newton-Cheh C, Larson MG, Vasan RS, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–53.
Roberts JD, Davies RW, Lubitz SA, et al. Evaluation of non-synonymous NPPA single nucleotide polymorphisms in atrial fibrillation. Europace. 2010;12:1078–83.
Ren X, Xu C, Zhan C, et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin Chim Acta. 2010;411:481–5.
Y. L. Yang, Y. P. Mo, Y. S. He, et al, “Correlation between renin-angiotensin system gene polymorphisms and essential hypertension in the Chinese Yi ethnic group,” J. Renin Angiotensin Aldosterone Syst., Aug 17, 2015.
L. N. Cosenso-Martin, R. O. Vaz-de-Melo, L. R. Pereira, et.al, “Angiotensin-converting enzyme insertion/deletion polymorphism, 24-h blood pressure profile and left ventricular hypertrophy in hypertensive individuals: a cross-sectional study,” Eur. J. Med. Res., vol. 20, pp. 74-015-0166-9, Sep 4, 2015.
Q. Li, L. Sun, J. Du, et al, “Risk given by AGT polymorphisms in inducing susceptibility to essential hypertension among isolated populations from a remote region of China: a case–control study among the isolated populations,” J. Renin Angiotensin Aldosterone Syst., Sep 20, 2015.
Sun X, Yang J, Hou X, et al. Relationship between -344T/C polymorphism in the aldosterone synthase gene and atrial fibrillation in patients with essential hypertension. J Renin Angiotensin Aldosterone Syst. 2011;12:557–63.
Wanahita N, Messerli FH, Bangalore S, et al. Atrial fibrillation and obesity—results of a meta-analysis. Am Heart J. 2008;155:310–5.
Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60.
Baudrand R, Goodarzi MO, Vaidya A, et al. A prevalent caveolin-1 gene variant is associated with the metabolic syndrome in Caucasians and Hispanics. Metabolism. 2015;64:1674–81.
Pojoga LH, Underwood PC, Goodarzi MO. Variants of the caveolin-1 gene: a translational investigation linking insulin resistance and hypertension. J Clin Endocrinol Metab. 2011;96:E1288–92.
Acknowledgments
This work was supported by the National Institutes of Health grant R01 HL111314 to MKC, the NIH National Center for Research Resources for Case Western Reserve University and Cleveland Clinic Clinical and Translational Science Award UL1-RR024989, the Cleveland Clinic Department of Cardiovascular Medicine philanthropy research funds, and the Tomsich Atrial Fibrillation Research Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Alejandra Gutierrez and Mina K. Chung declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Cardiovascular Genomics
Rights and permissions
About this article
Cite this article
Gutierrez, A., Chung, M.K. Genomics of Atrial Fibrillation. Curr Cardiol Rep 18, 55 (2016). https://doi.org/10.1007/s11886-016-0735-8
Published:
DOI: https://doi.org/10.1007/s11886-016-0735-8